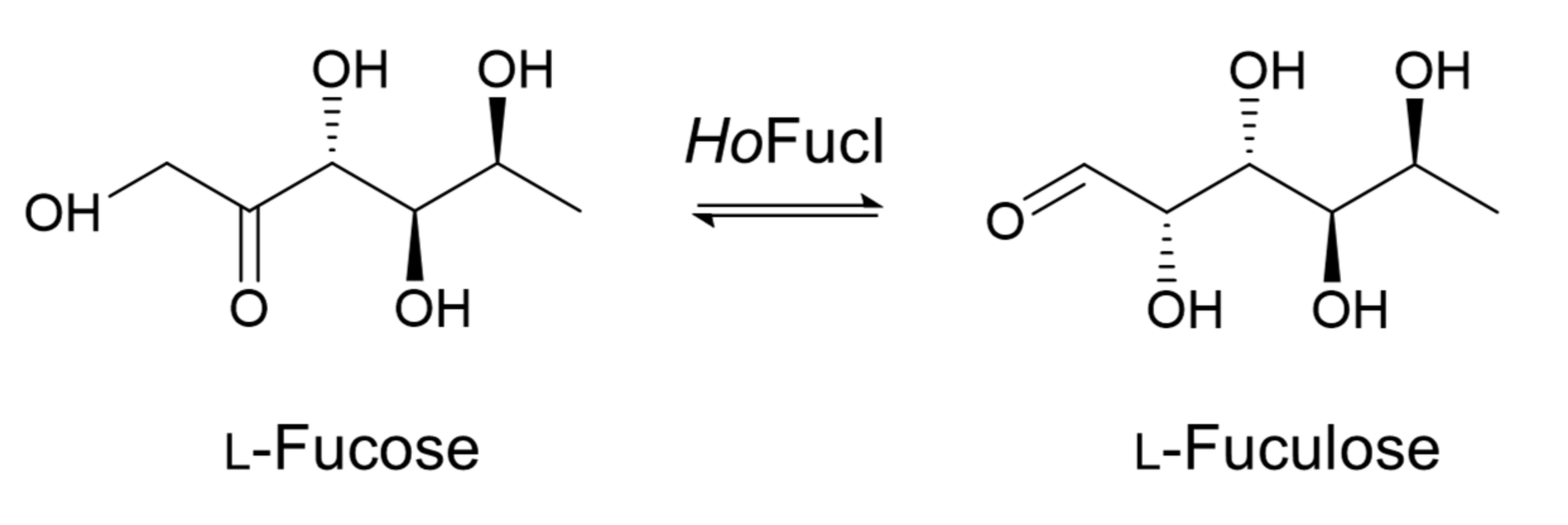
l-Fucose Synthesis Using a Halo- and Thermophilic l-Fucose Isomerase from Polyextremophilic Halothermothrix orenii
Abstract
:1. Introduction
2. Materials and Methods
2.1. Expression and Purification of HoFucI
2.2. Enzyme Assay
2.3. Effects of Various Biochemical Conditions on HoFucI Activity
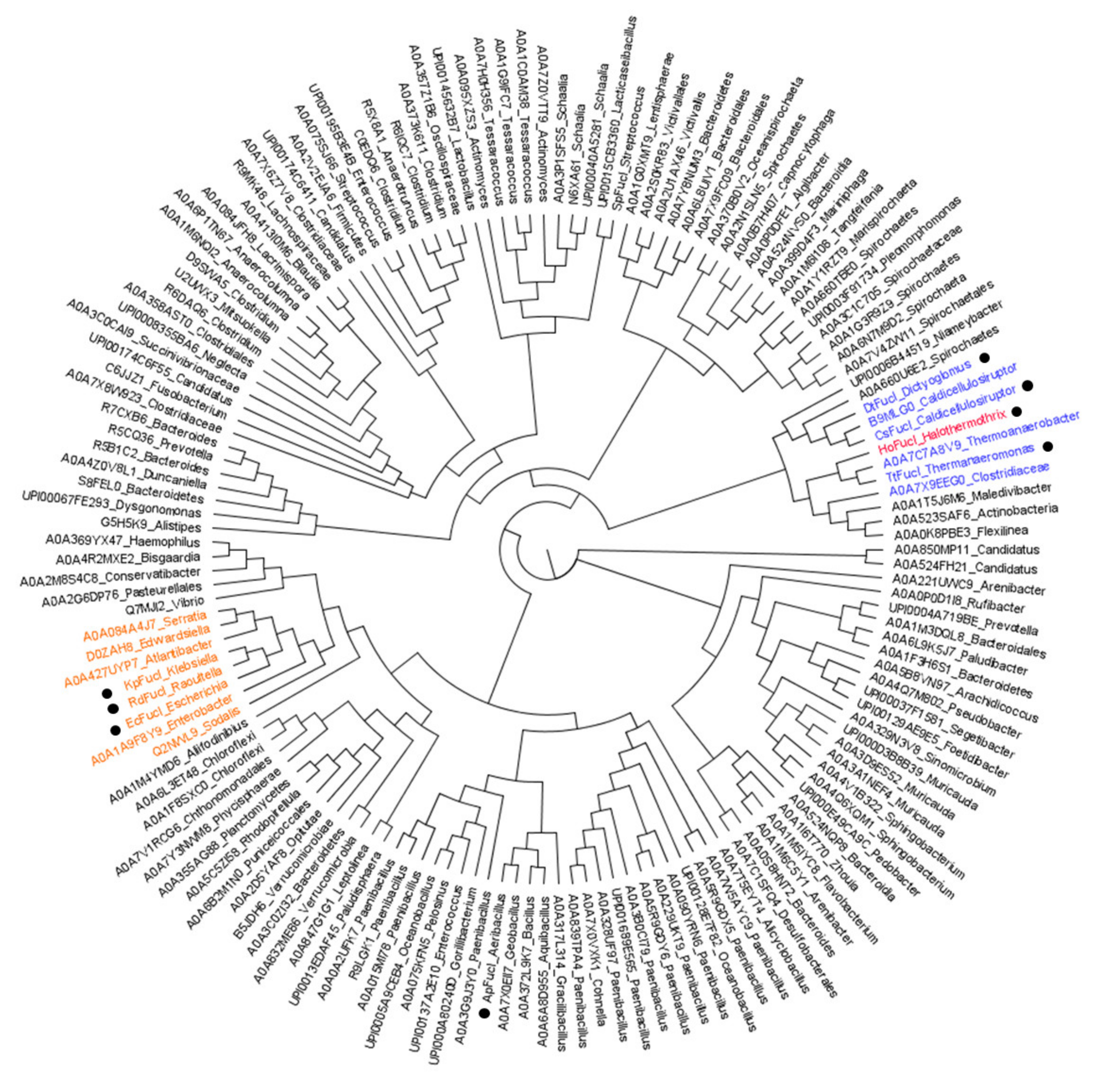
2.4. Phylogenetic Tree
2.5. Sequence Alignment and Homology Modeling
3. Results
3.1. Oligomeric State of HoFucI
3.2. Effect of Salinity on HoFucI Activity
3.3. Effect of Temperature and pH on HoFucI Activity
3.4. Effect of Metal Ions on HoFucI Activity
3.5. Amino Acid Composition of HoFucI
3.6. Phylogenetic Tree of l-FucIs
3.7. Comparison of the Specific Activities of l-FucIs against l-Fuculose
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Green, M.; Cohen, S.S. Enzymatic conversion of l-fucose to l-fuculose. J. Biol. Chem. 1956, 219, 557–568. [Google Scholar] [CrossRef]
- Kim, I.J.; Kim, D.H.; Nam, K.H.; Kim, K.H. Enzymatic synthesis of l-fucose from l-fuculose using a fucose isomerase from Raoultella sp. and the biochemical and structural analyses of the enzyme. Biotechnol. Biofuels 2019, 12, 282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, I.J.; Kim, K.H. Thermophilic l-fucose isomerase from Thermanaeromonas toyohensis for l-fucose synthesis from l-fuculose. Process Biochem. 2020, 96, 131–137. [Google Scholar] [CrossRef]
- Baldomà, L.; Aguilar, J. Metabolism of l-fucose and l-rhamnose in Escherichia coli: Aerobic–anaerobic regulation of l-lactaldehyde dissimilation. J. Bacteriol. 1988, 170, 416–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, S.H.; Lim, Y.R.; Kim, Y.S.; Oh, D.K. Molecular characterization of a thermostable l-fucose isomerase from Dictyoglomus turgidum that isomerizes l-fucose and d-arabinose. Biochimie 2012, 94, 1926–1934. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.H.; Oh, D.K. Characterization of a recombinant l-fucose isomerase from Caldicellulosiruptor saccharolyticus that isomerizes l-fucose, d-arabinose, d-altrose, and l-galactose. Biotechnol. Lett. 2010, 32, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Péterszegi, G.; Fodil-Bourahla, I.; Robert, A.M.; Robert, L. Pharmacological properties of fucose. Applications in age-related modifications of connective tissues. Biomed. Pharmacother. 2003, 57, 240–245. [Google Scholar] [CrossRef]
- Péterszegi, G.; Isnard, N.; Robert, A.M.; Robert, L. Studies on skin aging. Preparation and properties of fucose-rich oligo- and polysaccharides. Effect on fibroblast proliferation and survival. Biomed. Pharmacother. 2003, 57, 187–194. [Google Scholar] [CrossRef]
- Robert, C.; Robert, A.M.; Robert, L. Effect of a preparation containing a fucose-rich polysaccharide on periorbital wrinkles of human voluntaries. Skin Res. Technol. 2005, 11, 47–52. [Google Scholar] [CrossRef]
- Ale, M.T.; Maruyama, H.; Tamauchi, H.; Mikkelsen, J.D.; Meyer, A.S. Fucose-containing sulfated polysaccharides from brown seaweeds inhibit proliferation of melanoma cells and induce apoptosis by activation of caspase-3 in vitro. Mar. Drugs 2011, 9, 2605–2621. [Google Scholar] [CrossRef] [Green Version]
- Sutherland, I.W. Structural studies on colanic acid, the common exopolysaccharide found in the Enterobacteriaceae, by partial acid hydrolysis. Oligosaccharides from colanic acid. Biochem. J. 1969, 115, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Thurl, S.; Munzert, M.; Henker, J.; Boehm, G.; Müller-Werner, B.; Jelinek, J.; Stahl, B. Variation of human milk oligosaccharides in relation to milk groups and lactational periods. Br. J. Nutr. 2010, 104, 1261–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gesson, J.-P.; Jacquesy, J.-C.; Mondon, M.; Petit, P. A short synthesis of l-fucose and analogs from d-mannose. Tetrahedron Lett. 1992, 33, 3637–3640. [Google Scholar] [CrossRef]
- Sarbajna, S.; Das, S.K.; Roy, N. A novel synthesis of l-fucose from d-galactose. Carbohydr. Res. 1995, 270, 93–96. [Google Scholar] [CrossRef]
- Suzuki, S.; Kunihiko, W. Method for Producing l-fuculose and Method for Producing l-fucose. U.S. Patent US/0026504 A1, 1 February 2007. Available online: https://patentimages.storage.googleapis.com/b7/43/40/840ebb8a3ce304/US20070026504A1.pdf (accessed on 30 March 2022).
- Wong, C.-H.; Alajarin, R.; Moris-Varas, F.; Blanco, O.; Garcia-Junceda, E. Enzymic synthesis of l-fucose and analogs. J. Org. Chem. 1995, 60, 7360–7363. [Google Scholar] [CrossRef]
- Iqbal, M.W.; Riaz, T.; Hassanin, H.A.M.; Ni, D.; Mahmood Khan, I.; Rehman, A.; Mahmood, S.; Adnan, M.; Mu, W. Characterization of a novel d-arabinose isomerase from Thermanaeromonas toyohensis and its application for the production of d-ribulose and l-fuculose. Enzyme Microb. Technol. 2019, 131, 109427. [Google Scholar] [CrossRef]
- Mavromatis, K.; Ivanova, N.; Anderson, I.; Lykidis, A.; Hooper, S.D.; Sun, H.; Kunin, V.; Lapidus, A.; Hugenholtz, P.; Patel, B.; et al. Genome analysis of the anaerobic thermohalophilic bacterium Halothermothrix orenii. PLoS ONE 2009, 4, e4192. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, A.; Pletschke, B.I. Review of the enzymatic machinery of Halothermothrix orenii with special reference to industrial applications. Enzyme Microb. Technol. 2014, 55, 159–169. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal omega. Curr. Protoc. Bioinform. 2014, 48, 3131–3136. [Google Scholar] [CrossRef]
- Gouet, P.; Courcelle, E.; Stuart, D.I.; Métoz, F. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics 1999, 15, 305–308. [Google Scholar] [CrossRef] [Green Version]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Prot. Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Seemann, J.E.; Schulz, G.E. Structure and mechanism of l-fucose isomerase from Escherichia coli. J. Mol. Biol. 1997, 273, 256–268. [Google Scholar] [CrossRef] [PubMed]
- Siglioccolo, A.; Paiardini, A.; Piscitelli, M.; Pascarella, S. Structural adaptation of extreme halophilic proteins through decrease of conserved hydrophobic contact surface. BMC Struct. Biol. 2011, 11, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukuchi, S.; Yoshimune, K.; Wakayama, M.; Moriguchi, M.; Nishikawa, K. Unique amino acid composition of proteins in halophilic bacteria. J. Mol. Biol. 2003, 327, 347–357. [Google Scholar] [CrossRef]
- Ortega, G.; Laín, A.; Tadeo, X.; López-Méndez, B.; Castaño, D.; Millet, O. Halophilic enzyme activation induced by salts. Sci. Rep. 2011, 1, 6. [Google Scholar] [CrossRef] [PubMed]
- Lanyi, J.K. Salt-dependent properties of proteins from extremely halophilic bacteria. Bacteriol. Rev. 1974, 38, 272–290. [Google Scholar] [CrossRef] [PubMed]
- Zaccai, G.; Eisenberg, H. Halophilic proteins and the influence of solvent on protein stabilization. Trends Biochem. Sci. 1990, 15, 333–337. [Google Scholar] [CrossRef]
- Paul, S.; Bag, S.K.; Das, S.; Harvill, E.T.; Dutta, C. Molecular signature of hypersaline adaptation: Insights from genome and proteome composition of halophilic prokaryotes. Genome Biol. 2008, 9, R70. [Google Scholar] [CrossRef] [Green Version]
- Esclapez, J.; Pire, C.; Bautista, V.; Martínez-Espinosa, R.M.; Ferrer, J.; Bonete, M.J. Analysis of acidic surface of Haloferax mediterranei glucose dehydrogenase by site-directed mutagenesis. FEBS Lett. 2007, 581, 837–842. [Google Scholar] [CrossRef] [Green Version]
- Roberts, M.F. Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Syst. 2005, 1, 5. [Google Scholar] [CrossRef] [Green Version]
- Weinisch, L.; Kühner, S.; Roth, R.; Grimm, M.; Roth, T.; Netz, D.J.A.; Pierik, A.J.; Filker, S. Identification of osmoadaptive strategies in the halophile, heterotrophic ciliate Schmidingerothrix salinarum. PLoS Biol. 2018, 16, e2003892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sivakumar, N.; Li, N.; Tang, J.W.; Patel, B.K.; Swaminathan, K. Crystal structure of AmyA lacks acidic surface and provide insights into protein stability at poly-extreme condition. FEBS Lett. 2006, 580, 2646–2652. [Google Scholar] [CrossRef] [PubMed]
- Pancsa, R.; Kovacs, D.; Tompa, P. Misprediction of structural disorder in Halophiles. Molecules 2019, 24, 479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elevi Bardavid, R.; Oren, A. The amino acid composition of proteins from anaerobic halophilic bacteria of the order Halanaerobiales. Extremophiles 2012, 16, 567–572. [Google Scholar] [CrossRef]



| Metal Ions | Concentration (mM) | ||
|---|---|---|---|
| 0 | 1 | 10 | |
| EDTA | 100.0 ± 5.1 1,2 | 69.7 ± 24.6 | 80.8 ± 61.9 |
| Mn2+ | 555.7 ± 78.7 | 346.5 ± 7.4 | |
| Mg2+ | 252.1 ± 32.1 | 488.1 ± 19.8 | |
| Co2+ | 523.1 ± 74.2 | 112.8 ± 19.3 | |
| Amino Acid | HoFucI (%) | RdFucI (%) | TtFucI (%) | Halophiles (%) | Thermophiles (%) | Significance |
|---|---|---|---|---|---|---|
| Alanine | 8.5 2 | 11.0 | 9.7 | 10.0 | 8.0 | High frequency in halophiles |
| Aspartic acid | 5.3 | 5.4 | 5.0 | 6.0 | 4.5 | Important for halostability |
| Histidine | 1.8 | 2.7 | 1.7 | 2.0 | 1.8 | Low frequency in thermophiles |
| Lysine | 5.3 | 4.2 | 4.3 | 3.0 | 6.5 | Low frequency in halophiles |
| Glutamine | 3.0 | 4.1 | 2.8 | 5.0 | 3.0 | Important for halostability, but low frequency in thermophiles |
| Glutamate | 7.1 | 6.9 | 7.0 | 6.8 | 7.2 | Important for halostability |
| Threonine | 5.5 | 5.4 | 5.7 | 6.2 | 5.0 | Important for halostability, but low frequency in thermophiles |
| Specific Activity (U/mg) | Condition | |
|---|---|---|
| HoFucI [this study] | 106.2 ± 8.7 | 60 °C, pH 7, w/o NaCl |
| 151.1 ± 11.9 | 60 °C, pH 7, w/0.5 M NaCl | |
| RdFucI [2] | 64.0 ± 3.4 | 40 °C, pH 7, w/o NaCl |
| TtFucI [3] | 199.8 ± 0.3 | 70 °C, pH 7, w/o NaCl |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, I.J.; Kim, K.H. l-Fucose Synthesis Using a Halo- and Thermophilic l-Fucose Isomerase from Polyextremophilic Halothermothrix orenii. Appl. Sci. 2022, 12, 4029. https://doi.org/10.3390/app12084029
Kim IJ, Kim KH. l-Fucose Synthesis Using a Halo- and Thermophilic l-Fucose Isomerase from Polyextremophilic Halothermothrix orenii. Applied Sciences. 2022; 12(8):4029. https://doi.org/10.3390/app12084029
Chicago/Turabian StyleKim, In Jung, and Kyoung Heon Kim. 2022. "l-Fucose Synthesis Using a Halo- and Thermophilic l-Fucose Isomerase from Polyextremophilic Halothermothrix orenii" Applied Sciences 12, no. 8: 4029. https://doi.org/10.3390/app12084029
APA StyleKim, I. J., & Kim, K. H. (2022). l-Fucose Synthesis Using a Halo- and Thermophilic l-Fucose Isomerase from Polyextremophilic Halothermothrix orenii. Applied Sciences, 12(8), 4029. https://doi.org/10.3390/app12084029







