Abstract
Since the rise of deep learning (DL) in the mid-2010s, cardiac magnetic resonance (CMR) image segmentation has achieved state-of-the-art performance. Despite achieving inter-observer variability in terms of different accuracy performance measures, visual inspections reveal errors in most segmentation results, indicating a lack of reliability and robustness of DL segmentation models, which can be critical if a model was to be deployed into clinical practice. In this work, we aim to bring attention to reliability and robustness, two unmet needs of cardiac image segmentation methods, which are hampering their translation into practice. To this end, we first study the performance accuracy evolution of CMR segmentation, illustrate the improvements brought by DL algorithms and highlight the symptoms of performance stagnation. Afterwards, we provide formal definitions of reliability and robustness. Based on the two definitions, we identify the factors that limit the reliability and robustness of state-of-the-art deep learning CMR segmentation techniques. Finally, we give an overview of the current set of works that focus on improving the reliability and robustness of CMR segmentation, and we categorize them into two families of methods: quality control methods and model improvement techniques. The first category corresponds to simpler strategies that only aim to flag situations where a model may be incurring poor reliability or robustness. The second one, instead, directly tackles the problem by bringing improvements into different aspects of the CMR segmentation model development process. We aim to bring the attention of more researchers towards these emerging trends regarding the development of reliable and robust CMR segmentation frameworks, which can guarantee the safe use of DL in clinical routines and studies.
1. Introduction
Cardiovascular diseases (CVDs) are the leading cause of death globally and a major contributor to disability [1]. In 2019, an estimate of 17.9 million people died from CVDs, representing 32% of all global deaths and 38% of premature deaths (under the age of 70) due to non-communicable diseases [2]. It is projected that, by 2035, the number of people with CVD will increase by 30%, reaching over 130 million people and a prevalence rate of 45.1% [3]. As a consequence, there are important efforts in place to improve prevention, early diagnosis and management of CVDs [4].
In this context, cardiovascular magnetic resonance (CMR) imaging has been positioned as a reference for quantitative cardiac analysis, due to its non-invasive nature and its superior spatiotemporal resolution that allows imaging the cardiac chambers and great vessels with a great level of detail [5]. Quantitative cardiac analysis from CMR requires an accurate segmentation of the heart. Manual delineation of the cardiac anatomical structures can take a trained expert around 20 min per subject, which is lengthy, monotonous, and prone to subjective errors [6]. Therefore, alongside the advances in CMR imaging, there has been a substantial part of research devoted to the development of techniques for automatic CMR segmentation [7,8,9].
Before the emergence of deep learning (DL), traditional techniques, such as thresholding, edge-based and region-based approaches, model-based (e.g., active shape and appearance models) and atlas-based segmentation methods, represented the state-of-the-art performance in CMR segmentation [7]. The main drawback of traditional techniques is that they require significant user expertise, in the form of feature engineering, encoded prior knowledge or posterior user intervention, to reach good accuracy.
Over the last ten years, benefiting from advanced computer hardware and greater availability of public datasets, DL-based techniques emerged as the reference method for CMR segmentation [9], outperforming previous approaches and demonstrating the capacity to reproduce the analysis of experts [10]. In fact, DL currently represents a real chance of developing CMR segmentation frameworks to assist, automate and accelerate routine clinical procedures and large-scale population studies. Nevertheless, despite their success and high reported accuracy, they still lack the necessary reliability and robustness to be safely translated into practice. As highlighted by recent studies [11], unlike experts, even the top-performing DL methods sometimes generate anatomically impossible segmentation results. If a model were to be deployed in clinical practice, such segmentation errors would represent a risk. With DL algorithms unable to provide guarantees on the quality of their results, the task of inspecting, detecting errors, correcting them and validating the segmentation results is left to the responsibility of an expert. The development of additional mechanisms to enable their use in subsequent quantitative cardiac analyses is highly desirable.
The goal of this paper is threefold. Firstly, we motivate the need to shift research from targeting high accuracy to new performance goals by showing that the accuracy objective has currently been met. Second, we provide formal definitions of robustness and reliability and summarize the major challenges that DL-based CMR segmentation methods face when trying to meet these two criteria. Finally, we present a review of the current and ongoing research for reliable and robust CMR segmentation.
The remainder of the paper is organized as follows: Section 2 motivates this work by illustrating the improvements brought by DL-based algorithms in CMR segmentation over the last decade. Section 3 formalizes the concepts of reliability and robustness and presents the challenges faced by DL-based methods that hinder the reliability and robustness of the CMR segmentations. Section 4 reviews current methods addressing reliability and robustness and categorizes the proposed solutions into two families, Quality Control (QC) and Model Improvement (MI) techniques. Although sharing the same objective, QC techniques are typically external tools that do not require any modification in model architecture or training procedure, allowing an effortless integration into state-of-the-art segmentation pipelines. MI techniques, instead, are harder to integrate into existing pipelines, as their functioning is related to an inner modification of the models. Finally, discussion and conclusions are presented in Section 6.
2. Evolution of CMR Segmentation Performance (2009–2021)
We motivate the need to shift from a focus on accuracy, as the main performance criterion, towards other criteria, i.e., reliability and robustness, by studying the evolution of CMR segmentation methods’ accuracy over approximately a decade. To this end, we focus on fully-automated cardiac segmentation methods from short-axis (SA) CMR acquisitions. SA CMR segmentation has been widely studied, thanks to the large number of labelled SA CMR datasets available through multiple segmentation challenges and within the UK Biobank [12], a large-scale biomedical database containing in-depth genetic and health information from half a million participants.
We analyze the performance of 50 CMR segmentation methods, published since 2009, the year where the Sunnybrook Cardiac MR Left Ventricle Segmentation Challenge (https://www.cardiacatlas.org/studies/sunnybrook-cardiac-data/, accessed on 7 April 2022). took place. This challenge is the first ever reported CMR segmentation challenge. A large number of the here-reported works were developed in the context of this and four other CMR segmentation challenges. In chronological order, these are: the LV Segmentation Challenge (http://www.cardiacatlas.org/challenges/lv-segmentation-challenge, accessed on 7 April 2022) in 2011 [13], the Right Ventricle (RV) Segmentation Challenge (https://rvsc.projets.litislab.fr, accessed on 7 April 2022) in 2012 [14], the Automated Cardiac Diagnosis Challenge (https://www.creatis.insa-lyon.fr/Challenge/acdc, accessed on 7 April 2022) in 2017 [11] (ACDC), and the Multi-Centre, Multi-Vendor & Multi-Disease Cardiac Image Segmentation Challenge (https://www.ub.edu/mnms, accessed on 7 April 2022) in 2020 [15] (M&Ms).
Table 1 presents the SA CMR segmentation methods considered in our study and specifies the cardiac structures each method extracts, i.e., the left ventricle (LV), the right ventricle (RV) and left ventricular myocardium (MYO). Figure 1 presents SA CMR segmentation methods’ progress in performance measured with the Dice Score Coefficient (DSC). The methods are discriminated per segmented cardiac structure (LV, RV and MYO). Furthermore, we differentiate between DL-based (blue) and non-DL methods (orange).

Table 1.
Fully automated SA CMR segmentation methods published between 2009 and 2021 with the segmented structure of interest (LV, RV or MYO). ALL denotes that a method segments the three cardiac sub-structures.
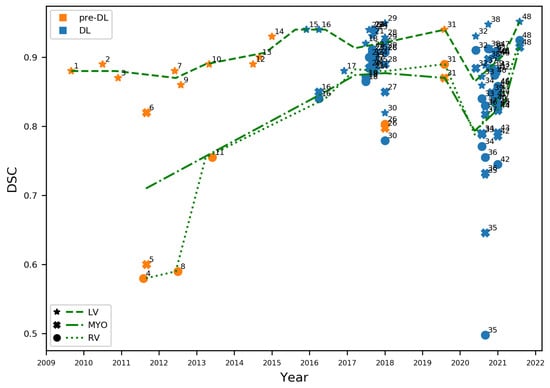
Figure 1.
Dice Score Coefficients (DSCs) obtained between 2009 and 2021 for LV, RV, and MYO. Methods that do not use deep learning appear in orange, DL-based methods in blue. Green lines indicate the performance trend over the years, estimated as an average of DSCs within a window of 290 days. Interpretation of numbered labels in Table 1.
We observe that, up to 2015, methods were exclusively not DL-based, mostly focused on LV segmentation, and with an important performance gap between the LV and the RV and MYO. The latter may be explained by the LV’s relatively lower variability in shape than the other cardiac structures. In 2015, in the context of the Kaggle Second Annual Data Science Bowl (https://www.kaggle.com/c/second-annual-data-science-bowl, accessed on 7 April 2022), the top-performing methods relied on deep learning technologies (https://github.com/woshialex/diagnose-heart, accessed on 7 April 2022). After this milestone, the scientific community shifted quickly towards DL. After 2016, only one non-DL CMR segmentation method [19] has been reported.
An immediate consequence of this change of techniques is the jump in performance for all cardiac structures. This is more evident for MYO and RV, which had the lowest DSCs, improving from average DSCs of 0.71 and 0.64, respectively before 2015, to both achieving 0.85 after 2015. LV segmentation reports an improvement from 0.88 average DSC to 0.91. Since then, the number of methods has exploded. However, performance improvements have stalled and, in some cases, deteriorated. This is the case of the general performance in the M&Ms Challenge [15], which assessed how well methods could cope with changes in the properties of the input images (e.g., different origins, scanner vendors and protocols). The result was a drop in the performance, as observed from the RV trend line or the very low performing methods (e.g., point 34) in Figure 1.
Finally, while most DL-based methods in Figure 1 report a very high accuracy, close to the inter-observer variability, Bernard et al. [11] demonstrated that DL-based methods, even the best performing ones [25], produced CMR segmentations with implausible anatomical configurations. The authors go then to suggest the adoption of new performance evaluation metrics that are more resilient to abnormalities. In the following, we show that the problems here identified, i.e., performance drops or implausible segmentations, can be addressed by accounting for reliability and robustness.
3. Robustness and Reliability: New Challenges in CMR Segmentation
In this section, we first provide formal definitions of reliability and robustness. Based on these definitions, we then identify the main factors that can hinder the reliability and robustness of DL-based CMR segmentation methods.
3.1. Definitions
The literature offers several definitions for reliability and robustness, as they can have slightly different interpretations associated with the domain where they are used, or they are often interchangeably used with related terms, such as stability [63] or safety [64]. In this work, we consider a CMR segmentation method as a computer system, thus we adhere to the following definitions from the IEEE Standard Glossary of Software Engineering Terminology [65].
3.1.1. Reliability
The ability of a system to perform its required functions under some stated conditions for a specified period of time.
3.1.2. Robustness
The degree to which a system can function correctly in the presence of invalid inputs.
3.2. Challenges to Reliable Segmentation
Following the definitions in Section 3.1, we identify two factors that can hinder the reliability of a DL-based segmentation method: overfitting and loss formulation.
3.2.1. Overfitting
The first and most basic condition that a reliable segmentation model should meet is that its performance is consistent from training to testing. Failing to do so is commonly referred to as overfitting or poor generalization. Two main factors are linked to overfitting: model complexity and data collection. Model complexity is related to the number of parameters in a model (e.g., the number of weights in a network), whereas data collection refers to the task of collecting and pre-processing data to train a model. In this study, we assume that the best architectures for fulfilling segmentation in the presence of an adequate number of training samples have already been identified. Therefore, we consider that overfitting can only be caused by poor data collection. In other words, the CMR segmentation methods presented in Section 2 should have a consistent training vs. testing performance as long as good data collection is guaranteed.
The data collection process that can guarantee the reliability of the model during testing needs to meet two conditions. First, it requires collecting a large number of samples. Being CMR segmentation typically fulfilled in a supervised manner, this also implies that the collected samples require annotations. Second, the collected data should be representative of the phenomenon under study. Failing to do so is commonly known as data bias.
3.2.2. Loss Formulation
State-of-the-art CMR segmentation is performed through supervised learning techniques. During supervised training, the loss functions measure the dissimilarity between the ground truth and the predicted segmentation. There is a vast offer of loss functions for medical image segmentation (e.g., the cross-entropy loss, the soft-Dice loss) [66], which can be used independently or combining multiple losses together. An inherent disadvantage of most of these loss functions is that they are typically pixel-wise objective functions, which measure dissimilarity in terms of correctly classified pixels over the total. This formulation does not optimize the model towards the final problem task since it does not reward segmentation results that better reflect the anatomy, i.e., the shape of the heart. Instead, it favors similarity among pixel intensities and, eventually, it leads to incomplete and unrealistic segmentation results both at training and at inference. In particular, predictions may contain holes inside the structures, abnormal concavities, or duplicated regions, typically located in the most basal and apical slices [67]. Being caused by intrinsic limitations of DL-based algorithms, anatomical failures can occur at inference without any possibility of inferring the quality of the model outcome. Therefore, the model becomes unpredictable, intractable for model verification, and ultimately unreliable.
3.3. Challenges to Robust Segmentation
Robustness is associated with performance in face of invalid inputs. We identify two sources that can lead to invalid inputs, thus affecting the robustness of a DL-based segmentation method: domain shift and data acquisition.
3.3.1. Domain Shift
Domain shift, or distribution shift, refers to a change in the data distribution between the one observed at training dataset, and the one the model encounters at inference, i.e., when deployed. Domain shift represents a critical risk for supervised deployed models because it has been shown that the inference error increases proportionally to the difference between samples from the two distributions [68]. In a strict sense, domain shifted data do not constitute an invalid input because it is still representative of the phenomenon under study. In this work, we follow a computer system approach where we consider domain shifted data as deviated from the “specifications” in which the model is developed or trained. As such, it does not affect reliability. However, the model is expected to perform well even in the presence of the domain shifted data, i.e., they should be robust. In CMR segmentation, this drift can be caused by numerous factors, such as changes in demographics, modalities, acquisition protocols and scanner vendors or simply anatomical variability or, even, an adversarial attack that may alter the statistical properties of the input [69]. The M&Ms challenge [15] was designed to assess the capacity of existing methods to cope with CMR domain shift. The result was an overall drop in performance showing a lack of robustness in existing methods.
3.3.2. Data Acquisition
Data acquisition may deteriorate the quality of an image and its visual appearance, but differently from domain shift, it does not alter the image’s statistical properties. Several factors affect the quality of a CMR image during its acquisition. Some of them are under the control of the clinician (e.g., the number of acquired slices), some depend on the subject being scanned (e.g., bulk or respiratory motion), and some are out of control (e.g., arrhythmias, blood flow or magnetic field inhomogeneities) [70]. When the quality is compromised, CMR images may contain artifacts like ghosting, blurring and smearing. During manual labelling, these images can be discarded for training. At inference, low-quality input images may not be possible to discard. Potentially, they could be the only information available for a patient. However, these low-quality inputs images may lead to poor segmentation results, if the segmentation model is not capable of handling invalid inputs.
4. Methods for Improved Reliability and Robustness
Two different approaches have arisen aiming to improve the reliability and the robustness of state-of-the-art DL-based segmentation methods. We distinguish between techniques limited to identify failures of the segmentation model, which hinder its reliability or robustness, and techniques that adopt countermeasures to improve the segmentation performance. In the former case, which we denote quality control (QC), the developed tools raise a flag when the system (i.e., the segmentation model) under analysis incurs into a lack of reliability or robustness, without necessarily explaining the cause or source of failure. In the latter case, models are improved in their architecture, acting on the sources of failures to eradicate them, and as a result to increase reliability and robustness. We denote this category as model improvement (MI) techniques.
4.1. Quality Control Techniques
QC techniques grade the quality of either input CMR images or segmentation outputs, allowing for recognizing anomalous scenarios, but without performing any action to correct the identified problem. Therefore, they improve reliability and/or robustness by signalling the identified anomalies to the users for them to act upon the problem. Most of these frameworks are not conceived to depend on a specific segmentation architecture, but they can adapt to the different segmentation pipelines available in the literature.
We identify two types of QC techniques, depending on when they are used. We denote as pre-analysis QC [71,72,73,74,75,76,77] those methods that act exclusively on the inputs of a DL-based model, i.e., before the model is executed, thus aiming specifically to improve robustness. Post-analysis QC [76,77,78,79,80,81,82,83,84,85,86,87,88] refers to those methods that act on the outputs of the model to detect a malfunction, thus addressing reliability. Pre- and post-analysis mechanisms are not mutually exclusive. They can be combined in an end-to-end framework. Moreover, pre-analysis QC tools can be combined with further processing steps that mitigate the erroneous detected inputs.
4.1.1. Pre-Analysis QC Tools
Pre-analysis QC tools aim to identify erroneous inputs, addressing robustness by discarding them from the segmentation pipeline. The first barrier to overcome by this type of methods is to define quality itself. Some methods aim to detect predefined types of artifacts using learning-based approaches [73], heuristic techniques [71] or a combination of both [72,75]. Other works, instead, follow a more qualitative definition that is based on a cardiologist’s input [74,76,77]. In this category, machine learning classifiers provided with a set of qualitative labels (e.g., good/bad, discard/keep) are trained to emulate experts criteria, aiming to flag low quality. At inference, these models automatically retrieve the binary feedback, which replaces experts’ decisions in high-throughput pipelines.
In one of the first QC works, Miao et al. [71] assess a perceptual difference model that quantitatively evaluates image quality of large volumes of magnetic resonance images to rate different image reconstruction algorithms. Lorch et al. [72] use box-, line-, histogram-, and texture-based features to train a random decision forest algorithm to distinguish between motion-corrupted and artifact-free images. Zhang et al. [73] aim to identify missing apical and/or basal LV slices in CMR images by using generative adversarial networks (GANs). This is achieved in two stages. First, adversarial examples are generated and exploited to extract high-level features from the CMR images. The features are then used to detect missing basal and apical slices. Such process improves not only robustness to adversarial examples, but also generalization performance for original examples. Oksuz et al. [74] exploit different levels of k-space synthetic corruption to detect CMR images with low perceptual quality, defined as the mean of the individual ratings assigned by human observers. The authors use a data augmentation technique to handle the severe class imbalance between good-quality and motion-corrupted images, training two deep learning architectures to increase their robustness in the classification task. In [70,75], Tarroni et al. present a quality control pipeline for CMR images in the UK Biobank dataset, capable of detecting three problematic scenarios to warn a human operator. The scenarios are low heart coverage, high inter-slice motion and low cardiac image contrast.
Finally, some recent works have succeeded at integrating QC tools within a more complex cardiac analysis pipeline. Machado et al. [76] use a ResNet [89] to classify CMR images as analyzable or non-analyzable. The network is trained with a dataset of 225 images labelled by an expert cardiologist. Those considered as analyzable move in forward in a cardiac analysis pipeline (see Section 4.1.2). Ruijsink et al. [77] present a DL-based pipeline for automated analysis of cardiac function. Inside the pipeline, two convolutional neural networks (CNNs) are trained to perform pre-analysis QC: a two-dimensional CNN with a recurrent long short-term memory layer for motion artifacts detection, and a two-dimensional CNN for detecting erroneous planning of the 4-chamber view. Flagged images are discarded from the subsequent segmentation step that serves as input to the cardiac function analysis.
4.1.2. Post-Analysis QC Tools
Post-analysis QC tools focus on the assessment of the segmentation outputs of a model. In this sense, we consider these tools as targeting reliability, as the quality of the segmentation output is the final indicator of the model’s performance.
Methods under this category follow two main approaches to performance assessment. They act either as binary classifiers, assigning correct/incorrect labels to a segmentation, or as regressors, which attempt to infer well-known validation metrics, such as the Dice Score or the Hausdorff Distance (HD), or uncertainty estimates.
Among regressors, Kohlberger et al. [82] train an SVM regressor from DSCs measured against ground truth to build confidence measures and rank candidate segmentation models against each other. Valindria et al. [83] propose the Reverse Classification Accuracy (RCA), a registration-based method relying on the spatial overlap between predicted segmentations and reference atlases as a pseudo-measure of the performance of a segmentation model on new data. The technique has been extensively validated in the UK Biobank [84], despite being computationally expensive at inference time or prone to failure at the registration stage [90].
Robinson et al. [85] rely on a CNN to predict the DSC of unseen segmented data. The authors are the first to observe that it is difficult to obtain a balanced set of labelled data reflecting the complete feasible distribution of DSCs. Hann et al. [86] use an ensemble of neural networks to segment the LV from T1 magnetic resonance, while providing an estimate of the DSC of the predicted segmentation using multiple linear regression. Fournel et al. [87] question the usefulness of 3D DSCs as the sole measure of segmentation quality, as it excludes specific information related to the single slices, which is actually fundamental when analysing the base and the apex. The authors overcome this limitation by performing simultaneously quality control at 2D-level and 3D-level using a CNN capable of predicting both 3D and 2D DSCs. Galati and Zuluaga [88] use a convolutional autoencoder that reconstructs input segmentation masks into pseudo ground truth masks. Pseudo DSC and HD are then measured between the segmentations and their reconstructions that act as surrogate measures of the quality of the segmentation results.
Among the classifiers, Albà et al. [78] use statistical, pattern and fractal descriptors in a random forest classifier, which detect segmentation failures to be corrected or removed from subsequent analyses. Puyol-Antón et al. [79] use the uncertainty information captured in the evidence lower bound (ELBO) produced by a Bayesian CNN to identify incorrect segmentations, which can be rejected or flagged for revision by an expert. In [80], segmentation uncertainty is first assessed at the voxel level by using the multi-class entropy and Monte Carlo dropout. After deriving uncertainty maps, a CNN is trained to detect image regions containing local segmentation failures that potentially need correction by an expert. The authors differentiate tolerated errors, which lay within the range of inter-observer variability, and the segmentation failures, which are flagged to be corrected by an expert. Gonzalez et al. [81] propose combining self-supervision loss terms and post hoc uncertainty estimations into a reliable and lightweight novelty score that allows anomalous samples’ identification.
The RCA [83], a regressor approach, has been embedded into the method proposed in [76], where the authors build a cardiac analysis pipeline that integrates both pre- (see Section 4.1.1) and post-analysis QC. For the latter, they estimate several quality metrics between pairs of segmentations, before and after being processed by RCA. Based on these values, an SVM binary classifier is trained to discriminate between poor and good quality segmentations. As [76], Ruijsink et al. [77] integrate pre- and post-analysis QC in a unified end-to-end pipeline. When dealing with post-analysis, they attempt to determine inconsistencies by making comparisons between long and short-axis views, LV and RV volumes, end-diastole and end-systole phases. They implement two support vector machine (SVM) classification algorithms to detect abnormalities in the obtained volume and strain curves.
Table 2 summarizes the main characteristics of the reported post-analysis QC tools. In addition to the distinction among classifiers and regressors (Regression), we highlight whether a proposed method formulates the problem in a traditional supervised manner, thus requiring QC labels (no QC labels). Given the cost of data labelling, it can be disadvantageous to require QC labels on top of the labels required to train the segmentation algorithm. Classification methods typically exploit qualitative (e.g., correct/incorrect) labels, whereas regressors require quantitative labels (e.g., DSC), which can be difficult to obtain [85]. To avoid these, a final set of methods avoid the use of QC labels by considering alternative self-supervised techniques or registration-based approaches as the RCA. Finally, Table 2 also highlights whether a given method allows the identification of the specific areas of segmentation failure, or it just gives an estimation of the general quality (detection).

Table 2.
Post-analysis QC methods and their three main characteristics: performing regression or classification(regression), the need of quality control labels (no QC labels) and if they detect the element causing the error within the image (detection).
4.2. Model Improvement Techniques
We denote model improvement (MI) techniques as those methods that directly address the limitations of DL-based approaches leading to poor reliability or robustness. Differently from QC techniques, where an external algorithmic tool flags problematic situations, MI techniques solve the lack of reliability or robustness by explicitly correcting the model. Another key difference w.r.t. QC tools, which can be plugged in most of the segmentation models as an external module, is that MI techniques imply modifications to the models or the overall analysis pipelines. In the following, we first present MI techniques for improved reliability and robustness classifying them based on the specific problem they tackle (Section 3). The section concludes with an ablation analysis of the presented MI techniques to illustrate their contributions to the performance of CMR segmentation methods.
4.2.1. Overfitting
As discussed in Section 3.2.1, the necessary complexity of DL-based models to guarantee a high-performance accuracy has been established. Therefore, MI techniques to reduce overfitting firstly consist of strategies to enlarge the available datasets, when further data collection is not possible. Chen et al. [91] apply geometrical operations to the source training data in order to simulate various possible data distributions across different domains. This data augmentation strategy was also adopted by Full et al. [45] in the context of the M&Ms Challenge.
Other MI techniques assume it is not possible to sufficiently increase (artificially or through further data collection) the size of the training set that it avoids overfitting and propose to control the complexity of the highly complex models through regularization. Among them, Khened et al. [21] present a DenseNet-based FCN architecture with long skip and short-cut connections to increase parameter efficiency. Guo et al. [92] integrate continuous kernel cut and bound optimization into a CNN, building a unified max-flow framework with improved generalization capabilities.
4.2.2. Loss Formulation
MI techniques mitigating the lack of reliability induced by typical loss functions aim at re-formulating the training procedure through the definition of additional objective losses that take into account anatomical constraints. Many of these works rely on shape priors, embedding prior expertise knowledge into the segmentation model. A second set of works takes inspiration from control theory, proposing automatic correction schemes that make use of high-level feedback systems.
Shape Priors
Zotti et al. [93] extend the well-established U-net architecture [94] through the formulation of a probabilistic framework, which allows the embedding of a cardiac shape prior, in the form of a 3D volume encoding the probability of a voxel to belong to a certain “cardiac class” (LV, RV, or MYO), and the definition of a loss function tailored to the cardiac anatomy. Clough et al. [95] propose a loss function that measures the topological correspondence between predicted segmentations and prior shape knowledge. This is done by using the differentiable properties of persistent homology, which compares topologies in terms of their Betti numbers. Wyburg et al. [96] enforce topology preservation by combining a segmentation network with spatial transformers and diffeomorphic displacement fields. In this way, the network learns to warp a binary prior, completing the segmentation task with the desired topological characteristics.
Automatic Correction
Girum et al. [67] formulate the segmentation problem as a two systems task: the first is a U-Net inspired encoder–decoder CNN predicting segmentations from the input images, the second is a fully convolutional network (FCN) working as a context feedback system. Once fed with segmentations, the FCN outputs encoded features which are integrated back into the decoder of the CNN. This context feedback loop helps the model extract high-level image features and fix uncertainties over time.
Ruijsink et al. [97] build from their previously proposed QC technique [77] to embed anatomical awareness into CMR segmentation models. The authors assume that the QC information provided by the QC tool encapsulates expertise biophysical knowledge that can be used to provide feedback to the network. As such, predictions flagged as high quality by the QC tool are fed back into the network model to reinforce its anatomical awareness. Painchaud et al. [98] present a segmentation framework that guarantees anatomical criteria by warping the predictions of a given model towards the closest anatomically valid cardiac shape with the use of a constrained Variational Autoencoder (cVAE). This warping step acts as the correction procedure, effectively leading to a reduced number of anatomical errors in the segmentation results. Finally, Galati and Zuluaga [99] use the information from an autoencoder-based post-analysis QC tool as a proxy of a model’s performance in unseen cardiac images [88]. The QC tool allows the automatic identification of Out-of-Distribution (OoD) data, which cause failures of the segmentation model. The information is then used as feedback to refine the training of the segmentation model, thus adapting to the OoD data.
4.2.3. Data Acquisition
Methods trying to mitigate data acquisition problems to improve the robustness of CMR segmentation models have mostly focused on improving the image quality at the image reconstruction phase. Among these, Schlemper et al. [100] propose two different methods to segment the heart directly from the k-space of dynamic MRI data, bypassing middle reconstruction stages. The first method relies on an end-to-end synthesis network that exploits the spatiotemporal redundancy of the input to generate the segmentations directly from the input k-space. The second method is conceived for heavily undersampled and aliased images, where there may be a loss of geometrical information and the first approach fails. It uses an autoencoder and a predictor network. The autoencoder is trained to encode and decode segmentations. The predictor learns to map undersampled images to latent encodings. The predicted encodings are used by the autoencoder to decode the corresponding segmentation maps. Huang et al. [101] propose a method that takes as input the undersampled k-space data from CMR scans to solve the reconstruction and segmentation problems simultaneously. The reconstruction is derived from the fast iterative shrinkage-thresholding algorithm (FISTA), while the segmentation is based on a U-Net architecture. Combining the two modules into a joint single-step, the reconstructed image becomes a set of differentiable parameters for the segmentation module itself, allowing the two to mutually benefit from each other through backpropagation. Finally, Oksuz et al. [102] propose to detect, correct and segment CMR images with motion artifacts, integrating reconstruction and segmentation in a unique framework, which combines a spatiotemporal 2D+time CNN for artifact detection, a convolutional recurrent neural network for reconstruction and a classical U-net for segmentation. The full framework is trained by incorporating terms from all three subnetworks into an overall loss function.
4.2.4. Domain Shift
Domain adaptation is the umbrella term used to refer to the techniques addressing the domain shift problem [103,104]. Within our work, we consider domain adaptation as an MI technique that aims at improving robustness to domain-shifted inputs. It consists of combining labelled source domain data, i.e., data from the original training distribution, with target domain one, i.e., the domain shifted data, typically in an unsupervised manner that avoids labelling the target domain, where in principle no annotated data are available.
Different alternatives have been explored to improve the generalization capacity of CMR segmentation models to an unseen domain, where the unseen domain can be a different image modality, such as computed tomography [105,106,107], a different magnetic resonance sequence, such as late gadolinium enhancement [108], or the same modality with varying statistical properties (e.g., different vendors and/or centers) [99]. Chen et al. [105,106] present an unsupervised domain adaptation framework, named SIFA. This framework adapts a segmentation network to an unlabeled domain by aligning source and target domains from both image and feature perspectives. Adversarial learning is enforced at multiple levels in the pipeline, guiding the two adaptive perspectives through a shared feature encoder to exploit their mutual benefits. Ouyang et al. [107] introduce an unsupervised domain adaptation method specifically designed to compensate for the drawback of domain adversarial training when only a small number of target samples is available. This result is achieved by introducing prior regularization on a shared domain-invariant latent space of the source and target domain images, which is exploited during segmentation. Chen et al. [108] tackle the problem of domain adaptation by using a common feature generator to fuse the feature spaces of source and target data into a combined feature domain. This new space is kept domain-invariant via indirect double-sided adversarial learning.
4.2.5. Ablation Analysis of MI Techniques
We analyzed the reported performance accuracy of the different MI techniques and their ablated versions. By ablated version, we refer to the backbone architecture of each method without MI. Figure 2 summarizes the reported DSC and HD of the different methods. We observe a clear trend of improvement when using MI: there is an DSC increase, whereas the HD is reduced. Although the reported methods use different backbone architectures, configurations and datasets, which limit a direct comparison, there is a clear trend that suggests that MI techniques addressing robustness and reliability do have a positive impact in the performance of CMR segmentation methods.
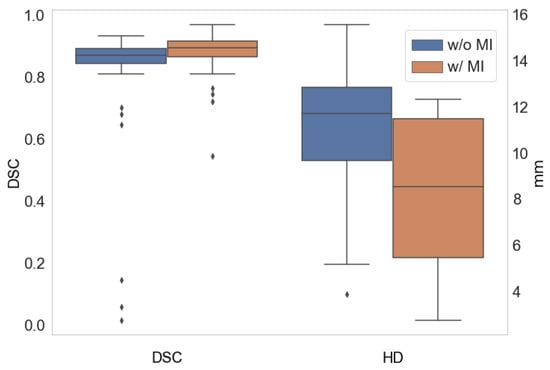
Figure 2.
Average DSC (left) and HD (right) with (w/) the use of MI techniques and without (w/o) them.
5. Discussion
After tracing DL history for CMR segmentation (Section 2), we have highlighted the shortcomings that currently prevent this technology from meeting some of the requirements to be safely deployed and used in clinical routine and cardiac analysis pipelines [109]. In this work, we focus on two main factors: a lack of reliability and robustness of many state-of-the-art methods. After providing formal definitions for the two terms, we have identified and discuss the elements that lead to poor reliability and/or robustness and we presented a wide range of works that have recently been published tackling both problems in CMR segmentation.
In this survey, we proposed to categorize the existing literature into two families: quality control and model improvement techniques. Quality control techniques can be seen as simpler strategies that only aim at flagging situations where a model may be incurring poor reliability or robustness, without aiming to fix the problem. Their main advantage is that these methods are typically external modules that can be promptly attached to an existing segmentation pipeline. However, they leave the problem to the expert, who needs to decide how to address the identified situation. Therefore, QC tools contribute to reducing the analysis time for the expert and providing some safety guarantees, through the generation of alerts, but do not contribute to improving CMR segmentation performance.
Model improvement techniques, instead, bring specific improvements in several aspects of the segmentation model development process, with the final goal of addressing the limitations of DL models that lead to poor reliability or robustness. As such, these type of methods are not only capable of identifying a potential problem, as QC tools do, but they can also act on it and aim to fix it. This being a more complex problem to tackle, it may explain why the number of existing QC methods is larger than MI techniques. A second possible explanation to this may be that the development of QC techniques has been strongly driven by the need to fully automate the processing pipelines of large databases, such as the UK Biobank.
A current limiting factor to further research on new QC and MI techniques addressing robustness and reliability is the lack of a common and well-established framework for their evaluation. QC techniques use different types of outputs, such as quantitative scores or a wide range of qualitative labels, with no clear mapping among them. MI techniques, as discussed in Section 4.2.5, rely on different backbone architectures and configurations that cannot be directly compared. The heterogeneity of existing solutions for both categories of methods challenges an objective and consistent evaluation. Moreover, as demonstrated by Bernard et al. [11], current performance measures, such as the DSC or HD, are not well-suited to identify errors which are associated with poor reliability and robustness. Progress in the field should therefore be accompanied with the investigation of better evaluation strategies.
6. Conclusions
In this paper, we present an overview of the state-of-the-art methods in CMR segmentation deep learning techniques, focusing on the changes of performances preceding and succeeding their rise. As we show, DL models have reached their maturity, achieving performance comparable to experts. Therefore, efforts to develop new models that optimize performance accuracy seem unnecessary. Instead, we observe that works specifically tackling reliability and robustness are rather limited and the field is quite young. We hope that our review can increase the awareness of these two important challenges of CMR segmentation and more research work will focus on developing methods that can efficiently solve them, thus enabling the translation of accurate, reliable, and robust CMR segmentation pipelines into the clinic.
Author Contributions
Conceptualization and methodology, F.G. and M.A.Z.; investigation, F.G.; resources, M.A.Z.; data curation, F.G.; writing—original draft preparation, review and editing, F.G., S.O. and M.A.Z.; visualization, F.G.; supervision, project administration, and funding acquisition, M.A.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by the French government, through the 3IA Côte d’Azur Investments in the future project managed by the National Research Agency (ANR) (ANR-19-P3IA-0002).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
We have uploaded at https://zenodo.org/record/6451469 (accessed on 7 April 2022) the data and code needed to build Figure 1 and Figure 2.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P.; et al. Global burden of cardiovascular diseases and risk factors, 1990–2019: Update from the GBD 2019 study. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Cardiovascular Diseases (CVDs) Fact Sheet. 2021. Available online: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 1 February 2022).
- Nelson, S.; Whitsel, L.; Khavjou, O.; Phelps, D.; Leib, A. Projections of Cardiovascular Disease Prevalence and Costs. Available online: https://www.heart.org/-/media/Files/Get-Involved/Advocacy/CVD-Predictions-Through-2035.pdf (accessed on 7 April 2022).
- World Health Organization. Global Action Plan for the Prevention and Control of NCDs 2013–2020; WHO: Geneva, Switzerland, 2013. [Google Scholar]
- Van der Geest, R.J.; Reiber, J.H. Quantification in cardiac MRI. J. Magn. Reson. Imaging 1999, 10, 602–608. [Google Scholar] [CrossRef]
- Bai, W.; Sinclair, M.; Tarroni, G.; Oktay, O.; Rajchl, M.; Vaillant, G.; Lee, A.M.; Aung, N.; Lukaschuk, E.; Sanghvi, M.M.; et al. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. J. Cardiovasc. Magn. Reson. 2018, 20, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petitjean, C.; Dacher, J.N. A review of segmentation methods in short axis cardiac MR images. Med. Image Anal. 2011, 15, 169–184. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, X. Challenges and methodologies of fully automatic whole heart segmentation: A review. J. Healthc. Eng. 2013, 4, 371–407. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Qin, C.; Qiu, H.; Tarroni, G.; Duan, J.; Bai, W.; Rueckert, D. Deep learning for cardiac image segmentation: A review. Front. Cardiovasc. Med. 2020, 7, 25. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in Medical Image Analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [Green Version]
- Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.A.; Cetin, I.; Lekadir, K.; Camara, O.; Ballester, M.A.G.; et al. Deep Learning Techniques for Automatic MRI Cardiac Multi-Structures Segmentation and Diagnosis: Is the Problem Solved? IEEE Trans. Med. Imaging 2018, 37, 2514–2525. [Google Scholar] [CrossRef]
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.; et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLoS Med. 2015, 12, e1001779. [Google Scholar] [CrossRef] [Green Version]
- Suinesiaputra, A.; Cowan, B.R.; Al-Agamy, A.O.; Elattar, M.A.; Ayache, N.; Fahmy, A.S.; Khalifa, A.M.; Medrano-Gracia, P.; Jolly, M.; Kadish, A.H.; et al. A collaborative resource to build consensus for automated left ventricular segmentation of cardiac MR images. Med. Image Anal. 2014, 18, 50–62. [Google Scholar] [CrossRef] [Green Version]
- Petitjean, C.; Zuluaga, M.A.; Bai, W.; Dacher, J.; Grosgeorge, D.; Caudron, J.; Ruan, S.; Ayed, I.B.; Cardoso, M.J.; Chen, H.; et al. Right ventricle segmentation from cardiac MRI: A collation study. Med. Image Anal. 2015, 19, 187–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campello, V.M.; Gkontra, P.; Izquierdo, C.; Martín-Isla, C.; Sojoudi, A.; Full, P.M.; Maier-Hein, K.; Zhang, Y.; He, Z.; Ma, J.; et al. Multi-Centre, Multi-Vendor and Multi-Disease Cardiac Segmentation: The M&Ms Challenge. IEEE Trans. Med. Imaging 2021, 40, 3543–3554. [Google Scholar] [PubMed]
- Jolly, M.; Xue, H.; Grady, L.J.; Guehring, J. Combining Registration and Minimum Surfaces for the Segmentation of the Left Ventricle in Cardiac Cine MR Images. In Medical Image Computing and Computer-Assisted Intervention-MICCAI 2009, Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, 12th International Conference, London, UK, 20–24 September 2009; Lecture Notes in Computer Science Series; Springer: Berlin/Heidelberg, Germany, 2009; Volume 5762, pp. 910–918. [Google Scholar]
- Baumgartner, C.F.; Koch, L.M.; Pollefeys, M.; Konukoglu, E. An Exploration of 2D and 3D Deep Learning Techniques for Cardiac MR Image Segmentation. In Statistical Atlases and Computational Models of the Heart, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, ACDC and MMWHS Challenges-8th International Workshop, STACOM 2017, Held in Conjunction with MICCAI 2017, Quebec City, QC, Canada, 10–14 September 2017; Lecture Notes in Computer Science Series; Springer: Cham, Switzerland, 2017; Volume 10663, pp. 111–119. [Google Scholar]
- Huang, S.; Liu, J.; Lee, L.C.; Venkatesh, S.K.; Teo, L.L.S.; Au, C.; Nowinski, W.L. An Image-Based Comprehensive Approach for Automatic Segmentation of Left Ventricle from Cardiac Short Axis Cine MR Images. J. Digit. Imaging 2011, 24, 598–608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grinias, E.; Tziritas, G. Fast Fully-Automatic Cardiac Segmentation in MRI Using MRF Model Optimization, Substructures Tracking and B-Spline Smoothing. In Statistical Atlases and Computational Models of the Heart, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, ACDC and MMWHS Challenges-8th International Workshop, STACOM 2017, Held in Conjunction with MICCAI 2017, Quebec City, QC, Canada, 10–14 September 2017; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2017; Volume 10663, pp. 91–100. [Google Scholar]
- Schaerer, J.; Casta, C.; Pousin, J.; Clarysse, P. A dynamic elastic model for segmentation and tracking of the heart in MR image sequences. Med. Image Anal. 2010, 14, 738–749. [Google Scholar] [CrossRef]
- Khened, M.; Kollerathu, V.A.; Krishnamurthi, G. Fully convolutional multi-scale residual DenseNets for cardiac segmentation and automated cardiac diagnosis using ensemble of classifiers. Med. Image Anal. 2019, 51, 21–45. [Google Scholar] [CrossRef] [Green Version]
- Ou, Y.; Sotiras, A.; Paragios, N.; Davatzikos, C. DRAMMS: Deformable registration via attribute matching and mutual-saliency weighting. Med. Image Anal. 2011, 15, 622–639. [Google Scholar] [CrossRef]
- Jang, Y.; Hong, Y.; Ha, S.; Kim, S.; Chang, H. Automatic Segmentation of LV and RV in Cardiac MRI. In Statistical Atlases and Computational Models of the Heart, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, ACDC and MMWHS Challenges-8th International Workshop, STACOM 2017, Held in Conjunction with MICCAI 2017, Quebec City, QC, Canada, 10–14 September 2017; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2017; Volume 10663, pp. 161–169. [Google Scholar]
- Margeta, J.; Geremia, E.; Criminisi, A.; Ayache, N. Layered Spatio-temporal Forests for Left Ventricle Segmentation from 4D Cardiac MRI Data. In Statistical Atlases and Computational Models of the Heart, Imaging and Modelling Challenges, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, Toronto, ON, Canada, 22 September 2011; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2011; Volume 7085, pp. 109–119. [Google Scholar]
- Isensee, F.; Jaeger, P.F.; Full, P.M.; Wolf, I.; Engelhardt, S.; Maier-Hein, K.H. Automatic Cardiac Disease Assessment on cine-MRI via Time-Series Segmentation and Domain Specific Features. In Statistical Atlases and Computational Models of the Heart, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, ACDC and MMWHS Challenges-8th International Workshop, STACOM 2017, Held in Conjunction with MICCAI 2017, Quebec City, QC, Canada, 10–14 September 2017; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2017; Volume 10663, pp. 120–129. [Google Scholar]
- Jolly, M.; Guetter, C.; Lu, X.; Xue, H.; Guehring, J. Automatic Segmentation of the Myocardium in Cine MR Images Using Deformable Registration. In Statistical Atlases and Computational Models of the Heart, Imaging and Modelling Challenges, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, Toronto, ON, Canada, 22 September 2011; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2011; Volume 7085, pp. 98–108. [Google Scholar]
- Yang, X.; Bian, C.; Yu, L.; Ni, D.; Heng, P. Class-Balanced Deep Neural Network for Automatic Ventricular Structure Segmentation. In Statistical Atlases and Computational Models of the Heart, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, ACDC and MMWHS Challenges-8th International Workshop, STACOM 2017, Held in Conjunction with MICCAI 2017, Quebec City, QC, Canada, 10–14 September 2017; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2017; Volume 10663, pp. 152–160. [Google Scholar]
- Liu, H.; Hu, H.; Xu, X.; Song, E. Automatic Left Ventricle Segmentation in Cardiac MRI Using Topological Stable-State Thresholding and Region Restricted Dynamic Programming. Acad. Radiol. 2012, 19, 723–731. [Google Scholar] [CrossRef]
- Attar, R.; Pereañez, M.; Gooya, A.; Zhang, X.A.L.; de Vila, M.H.; Lee, A.M.; Aung, N.; Lukaschuk, E.; Sanghvi, M.M.; Fung, K.; et al. Quantitative CMR population imaging on 20,000 subjects of the UK Biobank imaging study: LV/RV quantification pipeline and its evaluation. Med. Image Anal. 2019, 56, 26–42. [Google Scholar] [CrossRef]
- Wang, C.W.; Peng, C.W.; Chen, H.C. A simple and fully automatic right ventricle segmentation method for 4-dimensional cardiac MR images. In Proceedings of the MICCAI RV Segmentation Challenge, Nice, France, 1–5 October 2012. [Google Scholar]
- Calisto, M.G.B.; Lai-Yuen, S.K. AdaEn-Net: An ensemble of adaptive 2D-3D Fully Convolutional Networks for medical image segmentation. Neural Netw. 2020, 126, 76–94. [Google Scholar] [CrossRef]
- Constantinides, C.; Roullot, E.; Lefort, M.; Frouin, F. Fully automated segmentation of the left ventricle applied to cine MR images: Description and results on a database of 45 Subjects. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBC 2012, San Diego, CA, USA, 28 August–1 September 2012; IEEE: Piscataway, NJ, USA, 2012; pp. 3207–3210. [Google Scholar]
- Scannell, C.M.; Chiribiri, A.; Veta, M. Domain-Adversarial Learning for Multi-Centre, Multi-Vendor, and Multi-Disease Cardiac MR Image Segmentation. In Statistical Atlases and Computational Models of the Heart, M&Ms and EMIDEC Challenges, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, 11th International Workshop, STACOM 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, 4 October 2020; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2020; Volume 12592, pp. 228–237. [Google Scholar]
- Hu, H.; Liu, H.; Gao, Z.; Huang, L. Hybrid segmentation of left ventricle in cardiac MRI using gaussian-mixture model and region restricted dynamic programming. Magn. Reson. Imaging 2013, 31, 575–584. [Google Scholar] [CrossRef]
- Liu, X.; Thermos, S.; Chartsias, A.; O’Neil, A.; Tsaftaris, S.A. Disentangled Representations for Domain-Generalized Cardiac Segmentation. In Statistical Atlases and Computational Models of the Heart, M&Ms and EMIDEC Challenges, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, 11th International Workshop, STACOM 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, 4 October 2020; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2020; Volume 12592, pp. 187–195. [Google Scholar]
- Zuluaga, M.A.; Cardoso, M.J.; Modat, M.; Ourselin, S. Multi-atlas Propagation Whole Heart Segmentation from MRI and CTA Using a Local Normalised Correlation Coefficient Criterion. In Statistical Atlases and Computational Models of the Heart, M&Ms and EMIDEC Challenges, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, 11th International Workshop, STACOM 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, 4 October 2020; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2020; Volume 7945, pp. 174–181. [Google Scholar]
- Li, L.; Zimmer, V.A.; Ding, W.; Wu, F.; Huang, L.; Schnabel, J.A.; Zhuang, X. Random Style Transfer Based Domain Generalization Networks Integrating Shape and Spatial Information. In Statistical Atlases and Computational Models of the Heart, M&Ms and EMIDEC Challenges, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, 11th International Workshop, STACOM 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, 4 October 2020; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2020; Volume 12592, pp. 208–218. [Google Scholar]
- Ngo, T.A.; Carneiro, G. Fully Automated Non-rigid Segmentation with Distance Regularized Level Set Evolution Initialized and Constrained by Deep-Structured Inference. In Proceedings of the 2014 IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 3118–3125. [Google Scholar]
- Huang, X.; Chen, Z.; Yang, X.; Liu, Z.; Zou, Y.; Luo, M.; Xue, W.; Ni, D. Style-Invariant Cardiac Image Segmentation with Test-Time Augmentation. In Statistical Atlases and Computational Models of the Heart, M&Ms and EMIDEC Challenges, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, 11th International Workshop, STACOM 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, 4 October 2020; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2020; Volume 12592, pp. 305–315. [Google Scholar]
- Queiros, S.F.; Barbosa, D.; Heyde, B.; Morais, P.; Vilaça, J.L.; Friboulet, D.; Bernard, O.; D’hooge, J. Fast automatic myocardial segmentation in 4D cine CMR datasets. Med. Image Anal. 2014, 18, 1115–1131. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, J.; Menze, B.H. Generalisable Cardiac Structure Segmentation via Attentional and Stacked Image Adaptation. In Statistical Atlases and Computational Models of the Heart, M&Ms and EMIDEC Challenges, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, 11th International Workshop, STACOM 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, 4 October 2020; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2020; Volume 12592, pp. 297–304. [Google Scholar]
- Tufvesson, J.; Hedström, E.; Steding-Ehrenborg, K.; Carlsson, M.; Arheden, H.; Heiberg, E. Validation and development of a new automatic algorithm for time resolved segmentation of the left ventricle in magnetic resonance imaging. J. Cardiovasc. Magn. Reson. 2015, 17, 1–3. [Google Scholar] [CrossRef] [Green Version]
- Simantiris, G.; Tziritas, G. Cardiac MRI Segmentation With a Dilated CNN Incorporating Domain-Specific Constraints. IEEE J. Sel. Top. Signal Process. 2020, 14, 1235–1243. [Google Scholar] [CrossRef]
- Avendi, M.R.; Kheradvar, A.; Jafarkhani, H. A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Med. Image Anal. 2016, 30, 108–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Full, P.M.; Isensee, F.; Jäger, P.F.; Maier-Hein, K. Studying Robustness of Semantic Segmentation Under Domain Shift in Cardiac MRI. In Statistical Atlases and Computational Models of the Heart, M&Ms and EMIDEC Challenges, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, 11th International Workshop, STACOM 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, 4 October 2020; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2020; Volume 12592, pp. 238–249. [Google Scholar]
- Tran, P.V. A Fully Convolutional Neural Network for Cardiac Segmentation in Short-Axis MRI. arXiv 2016, arXiv:1604.00494. [Google Scholar]
- Ma, J. Histogram Matching Augmentation for Domain Adaptation with Application to Multi-centre, Multi-vendor and Multi-disease Cardiac Image Segmentation. In Statistical Atlases and Computational Models of the Heart, M&Ms and EMIDEC Challenges, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, 11th International Workshop, STACOM 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, 4 October 2020; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2020; Volume 12592, pp. 177–186. [Google Scholar]
- Tan, L.K.; Liew, Y.M.; Lim, E.; McLaughlin, R.A. Cardiac left ventricle segmentation using convolutional neural network regression. In Proceedings of the 2016 IEEE EMBS Conference on Biomedical Engineering and Sciences (IECBES), Kuala Lumpur, Malaysia, 4–8 December 2016; pp. 490–493. [Google Scholar]
- Zhang, Y.; Yang, J.; Hou, F.; Liu, Y.; Wang, Y.; Tian, J.; Zhong, C.; Zhang, Y.; He, Z. Semi-supervised Cardiac Image Segmentation via Label Propagation and Style Transfer. In Statistical Atlases and Computational Models of the Heart, M&Ms and EMIDEC Challenges, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, 11th International Workshop, STACOM 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, 4 October 2020; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2020; Volume 12592, pp. 219–227. [Google Scholar]
- Patravali, J.; Jain, S.; Chilamkurthy, S. 2D-3D Fully Convolutional Neural Networks for Cardiac MR Segmentation. In Statistical Atlases and Computational Models of the Heart, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, ACDC and MMWHS Challenges-8th International Workshop, STACOM 2017, Held in Conjunction with MICCAI 2017, Quebec City, QC, Canada, 10–14 September 2017; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2017; Volume 10663, pp. 130–139. [Google Scholar]
- Carscadden, A.; Noga, M.; Punithakumar, K. A Deep Convolutional Neural Network Approach for the Segmentation of Cardiac Structures from MRI Sequences. In Statistical Atlases and Computational Models of the Heart, M&Ms and EMIDEC Challenges, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, 11th International Workshop, STACOM 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, 4 October 2020; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2020; Volume 12592, pp. 250–258. [Google Scholar]
- Tan, L.K.; Liew, Y.M.; Lim, E.; McLaughlin, R.A. Convolutional neural network regression for short-axis left ventricle segmentation in cardiac cine MR sequences. Med. Image Anal. 2017, 39, 78–86. [Google Scholar] [CrossRef]
- Khader, F.; Schock, J.; Truhn, D.; Morsbach, F.; Haarburger, C. Adaptive Preprocessing for Generalization in Cardiac MR Image Segmentation. In Statistical Atlases and Computational Models of the Heart, M&Ms and EMIDEC Challenges, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, 11th International Workshop, STACOM 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, 4 October 2020; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2020; Volume 12592, pp. 269–276. [Google Scholar]
- Wolterink, J.M.; Leiner, T.; Viergever, M.A.; Isgum, I. Automatic Segmentation and Disease Classification Using Cardiac Cine MR Images. In Statistical Atlases and Computational Models of the Heart, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, ACDC and MMWHS Challenges-8th International Workshop, STACOM 2017, Held in Conjunction with MICCAI 2017, Quebec City, QC, Canada, 10–14 September 2017; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2017; Volume 10663, pp. 101–110. [Google Scholar]
- Saber, M.; Abdelrauof, D.; Elattar, M. Multi-center, Multi-vendor, and Multi-disease Cardiac Image Segmentation Using Scale-Independent Multi-gate UNET. In Statistical Atlases and Computational Models of the Heart, M&Ms and EMIDEC Challenges, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, 11th International Workshop, STACOM 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, 4 October 2020; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2020; Volume 12592, pp. 259–268. [Google Scholar]
- Rohé, M.; Sermesant, M.; Pennec, X. Automatic Multi-Atlas Segmentation of Myocardium with SVF-Net. In Statistical Atlases and Computational Models of the Heart, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, ACDC and MMWHS Challenges-8th International Workshop, STACOM 2017, Held in Conjunction with MICCAI 2017, Quebec City, QC, Canada, 10–14 September 2017; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2017; Volume 10663, pp. 170–177. [Google Scholar]
- Kong, F.; Shadden, S.C. A Generalizable Deep-Learning Approach for Cardiac Magnetic Resonance Image Segmentation Using Image Augmentation and Attention U-Net. In Statistical Atlases and Computational Models of the Heart, M&Ms and EMIDEC Challenges, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, 11th International Workshop, STACOM 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, 4 October 2020; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2020; Volume 12592, pp. 287–296. [Google Scholar]
- Zotti, C.; Luo, Z.; Humbert, O.; Lalande, A.; Jodoin, P.M. GridNet with Automatic Shape Prior Registration for Automatic MRI Cardiac Segmentation. In Statistical Atlases and Computational Models of the Heart, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, ACDC and MMWHS Challenges-8th International Workshop, STACOM 2017, Held in Conjunction with MICCAI 2017, Quebec City, QC, Canada, 10–14 September 2017; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2017; Volume 10663, pp. 73–81. [Google Scholar]
- Acero, J.C.; Sundaresan, V.; Dinsdale, N.K.; Grau, V.; Jenkinson, M. A 2-Step Deep Learning Method with Domain Adaptation for Multi-Centre, Multi-Vendor and Multi-Disease Cardiac Magnetic Resonance Segmentation. In Statistical Atlases and Computational Models of the Heart, M&Ms and EMIDEC Challenges, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, 11th International Workshop, STACOM 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, 4 October 2020; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2020; Volume 12592, pp. 196–207. [Google Scholar]
- Khened, M.; Varghese, A.; Krishnamurthi, G. Densely Connected Fully Convolutional Network for Short-Axis Cardiac Cine MR Image Segmentation and Heart Diagnosis Using Random Forest. In Statistical Atlases and Computational Models of the Heart, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, ACDC and MMWHS Challenges-8th International Workshop, STACOM 2017, Held in Conjunction with MICCAI 2017, Quebec City, QC, Canada, 10–14 September 2017; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2017; Volume 10663, pp. 140–151. [Google Scholar]
- Parreño, M.; Paredes, R.; Albiol, A. Deidentifying MRI Data Domain by Iterative Backpropagation. In Statistical Atlases and Computational Models of the Heart, M&Ms and EMIDEC Challenges, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, 11th International Workshop, STACOM 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, 4 October 2020; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2020; Volume 12592, pp. 277–286. [Google Scholar]
- Zhou, R.; Guo, F.; Azarpazhooh, M.R.; Hashemi, S.; Cheng, X.; Spence, J.D.; Ding, M.; Fenster, A. Deep Learning-Based Measurement of Total Plaque Area in B-Mode Ultrasound Images. IEEE J. Biomed. Health Inform. 2021, 25, 2967–2977. [Google Scholar] [CrossRef] [PubMed]
- Bousquet, O.; Elisseeff, A. Stability and generalization. J. Mach. Learn. Res. 2002, 2, 499–526. [Google Scholar]
- Bahr, N.J. System Safety Engineering and Risk Assessment: A Practical Approach; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- IEEE Standards Coordinating Committee. IEEE Standard Glossary of Software Engineering Terminology. CA IEEE Comput. Soc. 1990, 169, 132. [Google Scholar]
- Ma, J.; Chen, J.; Ng, M.; Huang, R.; Li, Y.; Li, C.; Yang, X.; Martel, A.L. Loss odyssey in medical image segmentation. Med. Image Anal. 2021, 71, 102035. [Google Scholar] [CrossRef]
- Girum, K.B.; Créhange, G.; Lalande, A. Learning With Context Feedback Loop for Robust Medical Image Segmentation. IEEE Trans. Med. Imaging 2021, 40, 1542–1554. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Feng, J.; Saenko, K. Return of frustratingly easy domain adaptation. In Proceedings of the AAAI Conference on Artificial Intelligence, Phoenix, AZ, USA, 12–17 February 2016; Volume 30. [Google Scholar]
- Gao, R.; Liu, F.; Zhang, J.; Han, B.; Liu, T.; Niu, G.; Sugiyama, M. Maximum Mean Discrepancy Test is Aware of Adversarial Attacks. In Proceedings of the 38th International Conference on Machine Learning, ICML 2021, Virtual Event, 18–24 July 2021; Volume 139, pp. 3564–3575. [Google Scholar]
- Tarroni, G.; Oktay, O.; Bai, W.; Schuh, A.; Suzuki, H.; Passerat-Palmbach, J.; de Marvao, A.; O’Regan, D.P.; Cook, S.; Glocker, B.; et al. Large-scale Quality Control of Cardiac Imaging in Population Studies: Application to UK Biobank. Sci. Rep. 2020, 10, 2408. [Google Scholar] [CrossRef] [Green Version]
- Miao, J.; Huo, D.; Wilson, D.L. Quantitative image quality evaluation of MR images using perceptual difference models. Med. Phys. 2008, 35, 2541–2553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorch, B.; Vaillant, G.; Baumgartner, C.; Bai, W.; Rueckert, D.; Maier, A. Automated detection of motion artefacts in MR imaging using decision forests. J. Med. Eng. 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Gooya, A.; Frangi, A.F. Semi-supervised Assessment of Incomplete LV Coverage in Cardiac MRI Using Generative Adversarial Nets. In Simulation and Synthesis in Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2017; Volume 10557, pp. 61–68. [Google Scholar]
- Öksüz, I.; Ruijsink, B.; Puyol-Antón, E.; Clough, J.R.; Cruz, G.; Bustin, A.; Prieto, C.; Botnar, R.M.; Rueckert, D.; Schnabel, J.A.; et al. Automatic CNN-based detection of cardiac MR motion artefacts using k-space data augmentation and curriculum learning. Med. Image Anal. 2019, 55, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Tarroni, G.; Oktay, O.; Bai, W.; Schuh, A.; Suzuki, H.; Passerat-Palmbach, J.; de Marvao, A.; O’Regan, D.P.; Cook, S.; Glocker, B.; et al. Learning-Based Quality Control for Cardiac MR Images. IEEE Trans. Med. Imaging 2019, 38, 1127–1138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machado, I.; Puyol-Antón, E.; Hammernik, K.; Cruz, G.; Ugurlu, D.; Ruijsink, B.; Castelo-Branco, M.; Young, A.; Prieto, C.; Schnabel, J.A.; et al. Quality-Aware Cine Cardiac MRI Reconstruction and Analysis from Undersampled K-Space Data. In Statistical Atlases and Computational Models of the Heart. Multi-Disease, Multi-View, and Multi-Center Right Ventricular Segmentation in Cardiac MRI Challenge, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, 12th International Workshop, STACOM 2021, Held in Conjunction with MICC, 12th International Workshop, Strasbourg, France, 27 September 2021; Springer: Berlin/Heidelberg, Germany, 2022; pp. 12–20. [Google Scholar]
- Ruijsink, B.; Puyol-Antón, E.; Oksuz, I.; Sinclair, M.; Bai, W.; Schnabel, J.A.; Razavi, R.; King, A.P. Fully Automated, Quality-Controlled Cardiac Analysis From CMR: Validation and Large-Scale Application to Characterize Cardiac Function. JACC Cardiovasc. Imaging 2020, 13, 684–695. [Google Scholar] [CrossRef] [PubMed]
- Albà, X.; Lekadir, K.; Pereañez, M.; Medrano-Gracia, P.; Young, A.A.; Frangi, A.F. Automatic initialization and quality control of large-scale cardiac MRI segmentations. Med. Image Anal. 2018, 43, 129–141. [Google Scholar] [CrossRef] [Green Version]
- Puyol-Antón, E.; Ruijsink, B.; Baumgartner, C.F.; Masci, P.G.; Sinclair, M.; Konukoglu, E.; Razavi, R.; King, A.P. Automated quantification of myocardial tissue characteristics from native T1 mapping using neural networks with uncertainty-based quality-control. J. Cardiovasc. Magn. Reson. 2020, 22, 60. [Google Scholar] [CrossRef]
- Sander, J.; de Vos, B.D.; Isgum, I. Automatic segmentation with detection of local segmentation failures in cardiac MRI. Sci. Rep. 2020, 10, 21769. [Google Scholar] [CrossRef]
- González, C.; Mukhopadhyay, A. Self-supervised Out-of-distribution Detection for Cardiac CMR Segmentation. In Proceedings of the Medical Imaging with Deep Learning, Lübeck, Germany, 7–9 July 2021; Volume 143, pp. 205–218. [Google Scholar]
- Kohlberger, T.; Singh, V.K.; Alvino, C.V.; Bahlmann, C.; Grady, L.J. Evaluating Segmentation Error without Ground Truth. In Medical Image Computing and Computer-Assisted Intervention-MICCAI 2012, Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, 15th International Conference, Nice, France, 1–5 October 2012; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2012; Volume 7510, pp. 528–536. [Google Scholar]
- Valindria, V.V.; Lavdas, I.; Bai, W.; Kamnitsas, K.; Aboagye, E.O.; Rockall, A.G.; Rueckert, D.; Glocker, B. Reverse Classification Accuracy: Predicting Segmentation Performance in the Absence of Ground Truth. IEEE Trans. Med. Imaging 2017, 36, 1597–1606. [Google Scholar] [CrossRef] [Green Version]
- Robinson, R.; Valindria, V.V.; Bai, W.; Oktay, O.; Kainz, B.; Suzuki, H.; Sanghvi, M.M.; Aung, N.; Paiva, J.M.; Zemrak, F.; et al. Automated quality control in image segmentation: Application to the UK Biobank cardiovascular magnetic resonance imaging study. J. Cardiovasc. Magn. Reson. 2019, 21, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, R.; Oktay, O.; Bai, W.; Valindria, V.V.; Sanghvi, M.M.; Aung, N.; Paiva, J.M.; Zemrak, F.; Fung, K.; Lukaschuk, E.; et al. Real-Time Prediction of Segmentation Quality. In Medical Image Computing and Computer Assisted Intervention-MICCAI 2018, Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, 21st International Conference, Granada, Spain, 16–20 September 2018; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2018; Volume 11070, pp. 578–585. [Google Scholar]
- Hann, E.; Popescu, I.A.; Zhang, Q.; Gonzales, R.A.; Barutçu, A.; Neubauer, S.; Ferreira, V.M.; Piechnik, S.K. Deep neural network ensemble for on-the-fly quality control-driven segmentation of cardiac MRI T1 mapping. Med. Image Anal. 2021, 71, 102029. [Google Scholar] [CrossRef] [PubMed]
- Fournel, J.; Bartoli, A.; Bendahan, D.; Guye, M.; Bernard, M.; Rauseo, E.; Khanji, M.Y.; Petersen, S.E.; Jacquier, A.; Ghattas, B. Medical image segmentation automatic quality control: A multi-dimensional approach. Med. Image Anal. 2021, 74, 102213. [Google Scholar] [CrossRef] [PubMed]
- Galati, F.; Zuluaga, M.A. Efficient Model Monitoring for Quality Control in Cardiac Image Segmentation. In Functional Imaging and Modeling of the Heart, Proceedings of the International Conference on Functional Imaging and Modeling of the Heart, 11th International Conference, FIMH 2021, Stanford, CA, USA, 21–25 June 2021; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2021; pp. 101–111. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Zuluaga, M.A.; Burgos, N.; Mendelson, A.F.; Taylor, A.M.; Ourselin, S. Voxelwise atlas rating for computer assisted diagnosis: Application to congenital heart diseases of the great arteries. Med. Image Anal. 2015, 26, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Bai, W.; Davies, R.H.; Bhuva, A.N.; Manisty, C.H.; Augusto, J.B.; Moon, J.C.; Aung, N.; Lee, A.M.; Sanghvi, M.M.; et al. Improving the generalizability of convolutional neural network-based segmentation on CMR images. Front. Cardiovasc. Med. 2020, 7, 105. [Google Scholar] [CrossRef]
- Guo, F.; Ng, M.; Goubran, M.; Petersen, S.E.; Piechnik, S.K.; Neubauer, S.; Wright, G. Improving cardiac MRI convolutional neural network segmentation on small training datasets and dataset shift: A continuous kernel cut approach. Med. Image Anal. 2020, 61, 101636. [Google Scholar] [CrossRef]
- Zotti, C.; Luo, Z.; Lalande, A.; Jodoin, P.M. Convolutional Neural Network With Shape Prior Applied to Cardiac MRI Segmentation. IEEE J. Biomed. Health Inform. 2019, 23, 1119–1128. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention-MICCAI 2015, Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention 18th International Conference, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Clough, J.; Byrne, N.; Oksuz, I.; Zimmer, V.A.; Schnabel, J.A.; King, A. A Topological Loss Function for Deep-Learning based Image Segmentation using Persistent Homology. arXiv 2020, arXiv:1910.01877. [Google Scholar] [CrossRef]
- Wyburd, M.K.; Dinsdale, N.K.; Namburete, A.I.L.; Jenkinson, M. TEDS-Net: Enforcing Diffeomorphisms in Spatial Transformers to Guarantee Topology Preservation in Segmentations. In Medical Image Computing and Computer Assisted Intervention-MICCAI 2021, Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 250–260. [Google Scholar]
- Ruijsink, B.; Puyol-Antón, E.; Li, Y.; Bai, W.; Kerfoot, E.; Razavi, R.; King, A.P. Quality-Aware Semi-supervised Learning for CMR Segmentation. In Statistical Atlases and Computational Models of the Heart, M&Ms and EMIDEC Challenges, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, 11th International Workshop, STACOM 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, 4 October 2020; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2020; Volume 12592, pp. 97–107. [Google Scholar]
- Painchaud, N.; Skandarani, Y.; Judge, T.; Bernard, O.; Lalande, A.; Jodoin, P.M. Cardiac Segmentation With Strong Anatomical Guarantees. IEEE Trans. Med. Imaging 2020, 39, 3703–3713. [Google Scholar] [CrossRef]
- Galati, F.; Zuluaga, M.A. Using Out-of-Distribution Detection for Model Refinement in Cardiac Image Segmentation. In Statistical Atlases and Computational Models of the Heart. Multi-Disease, Multi-View, and Multi-Center Right Ventricular Segmentation in Cardiac MRI Challenge, Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, 12th International Workshop, STACOM 2021, Held in Conjunction with MICC, 12th International Workshop, Strasbourg, France, 27 September 2021; Springer: Berlin/Heidelberg, Germany, 2022; pp. 374–382. [Google Scholar]
- Schlemper, J.; Oktay, O.; Bai, W.; de Castro, D.C.; Duan, J.; Qin, C.; Hajnal, J.V.; Rueckert, D. Cardiac MR Segmentation from Undersampled k-space Using Deep Latent Representation Learning. In Medical Image Computing and Computer Assisted Intervention-MICCAI 2018, Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, 21st International Conference, Granada, Spain, 16–20 September 2018; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2018; Volume 11070, pp. 259–267. [Google Scholar]
- Huang, Q.; Yang, D.; Yi, J.; Axel, L.; Metaxas, D.N. FR-Net: Joint Reconstruction and Segmentation in Compressed Sensing Cardiac MRI. In Functional Imaging and Modeling of the Heart, Proceedings of the International Conference on Functional Imaging and Modeling of the Heart, 10th International Conference, FIMH 2019, Bordeaux, France, 6–8 June 2019; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2018; Volume 11504, pp. 352–360. [Google Scholar]
- Oksuz, I.; Clough, J.R.; Ruijsink, B.; Anton, E.P.; Bustin, A.; Cruz, G.; Prieto, C.; King, A.P.; Schnabel, J.A. Deep Learning-Based Detection and Correction of Cardiac MR Motion Artefacts During Reconstruction for High-Quality Segmentation. IEEE Trans. Med. Imaging 2020, 39, 4001–4010. [Google Scholar] [CrossRef] [PubMed]
- Torralba, A.; Efros, A.A. Unbiased look at dataset bias. In Proceedings of the 2011 IEEE Conference on Computer Vision and Pattern Recognition, Washington, DC, USA, 20–25 June 2011; pp. 1521–1528. [Google Scholar]
- Saito, K.; Kim, D.; Sclaroff, S.; Darrell, T.; Saenko, K. Semi-supervised domain adaptation via minimax entropy. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Korea, 27 October–2 November 2019; pp. 8050–8058. [Google Scholar]
- Chen, C.; Dou, Q.; Chen, H.; Qin, J.; Heng, P.A. Synergistic Image and Feature Adaptation: Towards Cross-Modality Domain Adaptation for Medical Image Segmentation. Proc. AAAI Conf. Artif. Intell. 2019, 33, 865–872. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Dou, Q.; Chen, H.; Qin, J.; Heng, P.A. Unsupervised Bidirectional Cross-Modality Adaptation via Deeply Synergistic Image and Feature Alignment for Medical Image Segmentation. IEEE Trans. Med. Imaging 2020, 39, 2494–2505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouyang, C.; Kamnitsas, K.; Biffi, C.; Duan, J.; Rueckert, D. Data efficient unsupervised domain adaptation for cross-modality image segmentation. In Medical Image Computing and Computer Assisted Intervention-MICCAI 2019, Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, 22nd International Conference, Shenzhen, China, 13–17 October 2019; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2019; pp. 669–677. [Google Scholar]
- Chen, J.; Zhang, H.; Zhang, Y.; Zhao, S.; Mohiaddin, R.; Wong, T.; Firmin, D.; Yang, G.; Keegan, J. Discriminative consistent domain generation for semi-supervised learning. In Medical Image Computing and Computer Assisted Intervention-MICCAI 2019, Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, 22nd International Conference, Shenzhen, China, 13–17 October 2019; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2019; pp. 595–604. [Google Scholar]
- Floridi, L. Establishing the rules for building trustworthy AI. Nat. Mach. Intell. 2019, 1, 261–262. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).