Isolation of Jannaschia sedimins sp. nov. from East Coast of China: Bacterial Taxonomy and Antimicrobial Resistance Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and Sampling
2.2. Bacterial Strains, Isolation, and Cultivation
2.3. Phenotypic and Physiological Characterization
2.4. Phylogenetic Analysis and Genomic Characterization
2.5. Chemotaxonomic Characterization
3. Results and Discussion
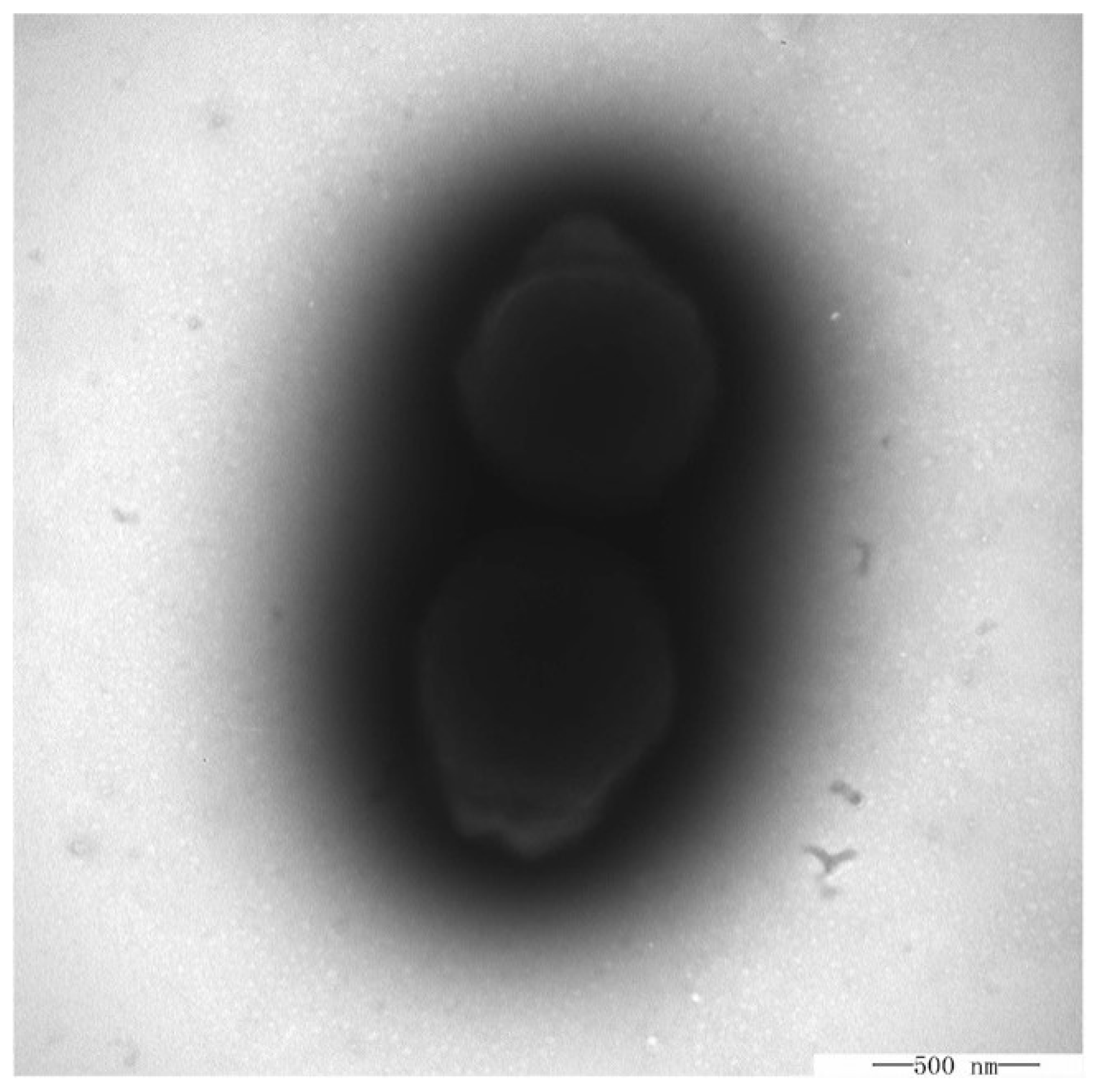
3.1. Morphological, Physiologial, and Biochemical Characteristics
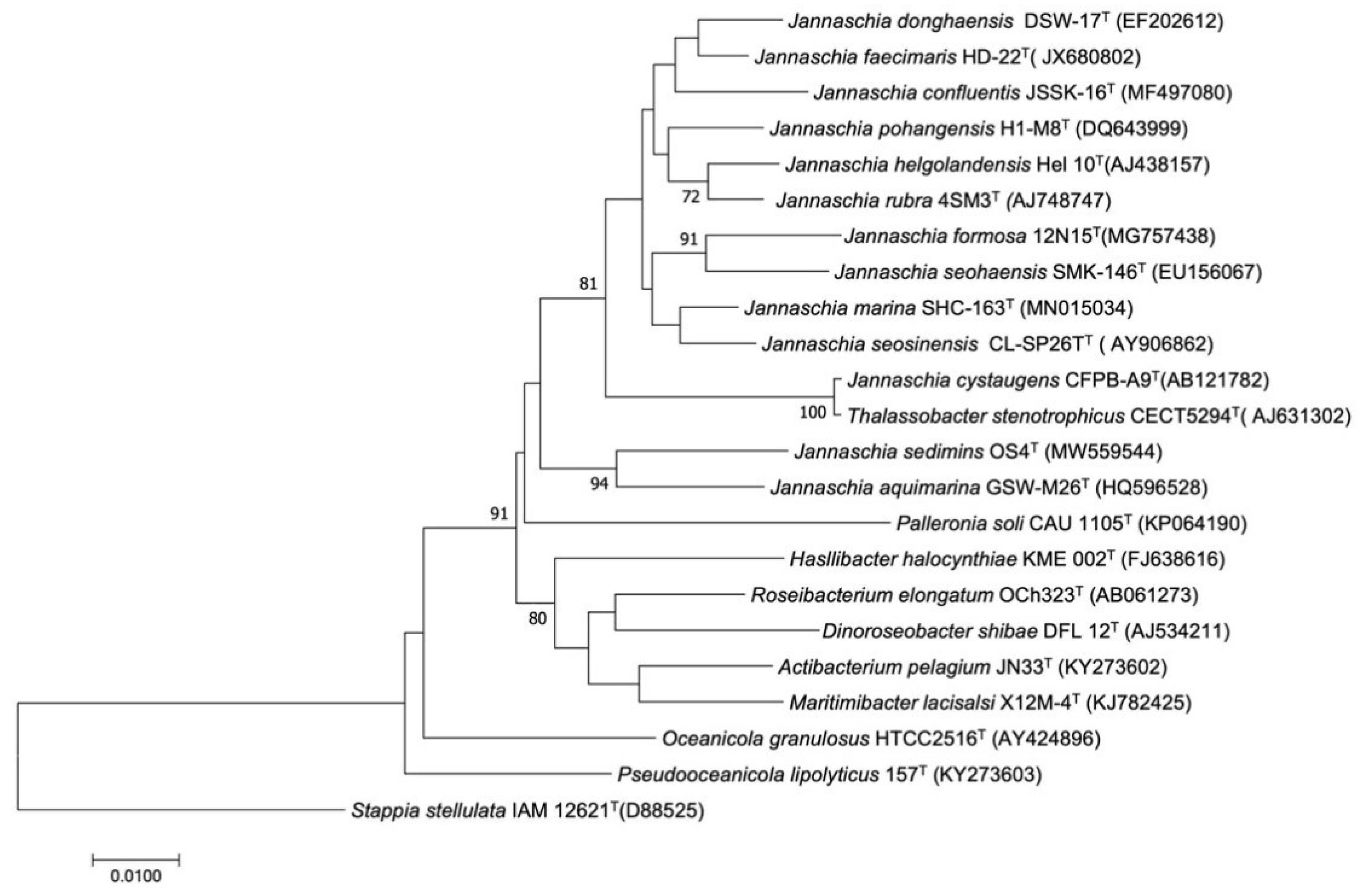
3.2. Phylogenetic Analysis
3.3. Chemotaxonomic Characteristics
3.4. Description of Jannaschia Sediminis sp. nov.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fair, R.J.; Tor, Y. Antibiotics and bacterial resistance in the 21st century. Perspect. Med. Chem. 2014, 6, S14459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miethke, M.; Pieroni, M.; Weber, T.; Brönstrup, M.; Hammann, P.; Halby, L.; Arimondo, P.B.; Glaser, P.; Aigle, B.; Bode, H.B. Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 2021, 5, 726–749. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, S.; Pruden, A.; Virta, M.; Zhang, T. Antibiotic resistance in aquatic systems. Front. Microbiol. 2017, 8, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michael, I.; Rizzo, L.; McArdell, C.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for the release of antibiotics in the environment: A review. Water Res. 2013, 47, 957–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner-Döbler, I.; Rheims, H.; Felske, A.; Pukall, R.; Tindall, B.J. Jannaschia helgolandensis gen. nov., sp. nov., a novel abundant member of the marine Roseobacter clade from the North Sea. Int. J. Syst. Evol. Microbiol. 2003, 53, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Adachi, M.; Kanno, T.; Okamoto, R.; Shinozaki, A.; Fujikawa-Adachi, K.; Nishijima, T. Jannaschia cystaugens sp. nov., an Alexandrium (Dinophyceae) cyst formation-promoting bacterium from Hiroshima Bay, Japan. Int. J. Syst. Evol. Microbiol. 2004, 54, 1687–1692. [Google Scholar] [CrossRef] [PubMed]
- Macián, M.; Arahal, D.; Garay, E.; Ludwig, W.; Schleifer, K.; Pujalte, M. Jannaschia rubra sp. nov., a red-pigmented bacterium isolated from sea water. Int. J. Syst. Evol. Microbiol. 2005, 55, 649–653. [Google Scholar] [CrossRef] [Green Version]
- Choi, D.H.; Yi, H.; Chun, J.; Cho, B.C. Jannaschia seosinensis sp. nov., isolated from hypersaline water of a solar saltern in Korea. Int. J. Syst. Evol. Microbiol. 2006, 56, 45–49. [Google Scholar] [CrossRef] [Green Version]
- Jung, Y.-T.; Yoon, J.-H. Jannaschia faecimaris sp. nov., isolated from a tidal flat sediment. Int. J. Syst. Evol. Microbiol. 2014, 64, 945–951. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; He, M.; Lai, Q.; Xu, Y.; Shang, C. Jannaschia marina sp. nov., isolated from the gut of a gastropod, Onchidium reevesii. Int. J. Syst. Evol. Microbiol. 2021, 71, 004756. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Wang, C.; Wang, X.-T.; Mu, D.-S.; Du, Z.-J. Jannaschia formosa sp. nov., isolated from marine saltern sediment. Int. J. Syst. Evol. Microbiol. 2019, 69, 2037–2042. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Choi, S.J.; Won, S.-M.; Yoon, J.-H. Jannaschia confluentis sp. nov., isolated from the junction between the ocean and a freshwater spring. Int. J. Syst. Evol. Microbiol. 2018, 68, 669–674. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.-H.; Kang, S.-J.; Park, S.; Oh, T.-K. Jannaschia donghaensis sp. nov., isolated from seawater of the East Sea, Korea. Int. J. Syst. Evol. Microbiol. 2007, 57, 2132–2136. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.-Y.; Yoo, S.-H.; Weon, H.-Y.; Jeon, Y.-A.; Hong, S.-B.; Go, S.-J.; Stackebrandt, E.; Kwon, S.-W. Jannaschia pohangensis sp. nov., isolated from seashore sand in Korea. Int. J. Syst. Evol. Microbiol. 2008, 58, 496–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, J.-H.; Kang, S.-J.; Park, S.; Oh, K.-H.; Oh, T.-K. Jannaschia seohaensis sp. nov., isolated from a tidal flat sediment. Int. J. Syst. Evol. Microbiol. 2010, 60, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Yoon, J.-H. Jannaschia aquimarina sp. nov., isolated from seawater. Int. J. Syst. Evol. Microbiol. 2012, 62, 2631–2636. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.-X.; Liu, Q.-Q.; Zhou, Y.-X.; Chen, G.-J.; Du, Z.-J. Alkalimarinus sediminis gen. nov., sp. nov., isolated from marine sediment. Int. J. Syst. Evol. Microbiol. 2015, 65, 3511–3516. [Google Scholar] [CrossRef] [PubMed]
- Smibert, R. Phenotypic characterization. Methods Gen. Mol. Bacteriol. 1994, 607–654. [Google Scholar]
- Bowman, J.P. Description of Cellulophaga algicola sp. nov., isolated from the surfaces of Antarctic algae, and reclassification of Cytophaga uliginosa (ZoBell and Upham 1944) Reichenbach 1989 as Cellulophaga uliginosa comb. nov. Int. J. Syst. Evol. Microbiol. 2000, 50, 1861–1868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2020. [Google Scholar]
- Du, Z.-J.; Wang, Y.; Dunlap, C.; Rooney, A.P.; Chen, G.-J. Draconibacterium orientale gen. nov., sp. nov., isolated from two distinct marine environments, and proposal of Draconibacteriaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2014, 64, 1690–1696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stackebrandt, E.; Goodfellow, M. Nucleic Acid Techniques in Bacterial Systematics; Wiley: Hoboken, NJ, USA, 1991. [Google Scholar]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes de novo assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [Green Version]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids; MIDI, Inc.: Newark, NY, USA, 1990; Available online: http://natasha.eng.usf.edu/gilbert/courses/Biotransport%20Phenomena/pdf/bacteria_gc_1.pdf (accessed on 11 February 2022).
- Tindall, B.J.; Sikorski, J.; Smibert, R.A.; Krieg, N.R. Phenotypic characterization and the principles of comparative systematics. Methods Gen. Mol. Microbiol. 2007, 330–393. [Google Scholar] [CrossRef]
- Kroppenstedt, R.M. Separation of bacterial menaquinones by HPLC using reverse phase (RP18) and a silver loaded ion exchanger as stationary phases. J. Liq. Chromatogr. 1982, 5, 2359–2367. [Google Scholar] [CrossRef]
- Minnikin, D.; O’donnell, A.; Goodfellow, M.; Alderson, G.; Athalye, M.; Schaal, A.; Parlett, J. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 1984, 2, 233–241. [Google Scholar] [CrossRef]

| Characteristics | OS4T | 14858T | GSW-M26T |
|---|---|---|---|
| Cell size (µm) | 0.5–0.8 × 0.8–2.0 | 0.7–1.1 × 1.9–3.2 | 0.5–0.7 × 0.5–4.0 |
| pH range for growth | 7.0–8.5 (8.0) | 7.0–8.0 (7.5) | 5.5–8.0 (7.0–8.0) |
| NaCl range (%) | 0–5.0 (2.0) | 1–7.0 (2.0) | 0–7.0 (2) |
| Growth temperature (°C) | 20–37 (25) | 15–30 (25) | 15–37 (30) |
| Catalase | + | + | + |
| Oxidase | + | + | + |
| Hydrolysis of | |||
| Starch | - | + | + |
| Alginate | + | - | - |
| Tween (20, 40, 60, 80) | - | - | + |
| Casein | + | - | - |
| Utilization of: | |||
| Sucrose | + | - | + |
| yesD-xylose | - | + | + |
| D-glucose | - | - | + |
| D-fructose | + | + | |
| Aesculin | w | - | - |
| D-mannose | - | - | + |
| 5-ketogluconate | - | + | + |
| Enzyme activity | |||
| alkaline phosphatase | + | + | + |
| esterase(C4) | + | + | + |
| esterase lipase (C8) | + | W | + |
| lipase (C14) | + | - | + |
| leucine arylamidase | + | - | + |
| valine arylamidase | + | - | + |
| cystine arylamidase | W | - | W |
| Trypsin | - | - | - |
| α-chymotrypsin | - | - | - |
| acid phosphatase | + | - | + |
| naphtol-AS-BI-phosphohydrolase | - | - | W |
| α-galactosidase | - | - | + |
| β-galactosidase | - | - | - |
| β-glucuronidase | - | - | - |
| α-glucosidase | + | - | + |
| β-glucosidase | - | - | + |
| Fatty Acid | OS4T | 14858T | GSW-M26T |
|---|---|---|---|
| Saturated | |||
| C16:0 | 1.6 | 1.5 | - |
| C18:0 | 9.9 | - | - |
| Branched chain | |||
| iso-C10:0 | - | - | - |
| iso-C14:0 | - | 1.1 | - |
| iso-C15:0 | - | 47.9 | 4.9 |
| anteiso-C15:0 | - | 35.3 | 50.7 |
| iso-C16:0 | - | 1.9 | 20.5 |
| iso-C17:0 | - | 2 | 1.9 |
| anteiso-C17:0 | - | 3.6 | 19.4 |
| Hydroxy | |||
| C10:0 3OH | 5.4 | - | - |
| Monounsaturated | |||
| C20:1 ω7c | 2.9 | - | - |
| Summed features | |||
| C18:1 ω6c/ω7c | 73.3 | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sha, S.; Li, S.; Zhao, L.; Zhao, J.; Chen, G. Isolation of Jannaschia sedimins sp. nov. from East Coast of China: Bacterial Taxonomy and Antimicrobial Resistance Analysis. Appl. Sci. 2022, 12, 2883. https://doi.org/10.3390/app12062883
Sha S, Li S, Zhao L, Zhao J, Chen G. Isolation of Jannaschia sedimins sp. nov. from East Coast of China: Bacterial Taxonomy and Antimicrobial Resistance Analysis. Applied Sciences. 2022; 12(6):2883. https://doi.org/10.3390/app12062883
Chicago/Turabian StyleSha, Sha, Shuqian Li, Lihua Zhao, Jinxin Zhao, and Guanjun Chen. 2022. "Isolation of Jannaschia sedimins sp. nov. from East Coast of China: Bacterial Taxonomy and Antimicrobial Resistance Analysis" Applied Sciences 12, no. 6: 2883. https://doi.org/10.3390/app12062883
APA StyleSha, S., Li, S., Zhao, L., Zhao, J., & Chen, G. (2022). Isolation of Jannaschia sedimins sp. nov. from East Coast of China: Bacterial Taxonomy and Antimicrobial Resistance Analysis. Applied Sciences, 12(6), 2883. https://doi.org/10.3390/app12062883







