1. Introduction
As an interdisciplinary area, medical image classification is the foundation of automatic disease diagnosis. With the development of deep learning technology, convolutional neural networks (CNNs) [
1,
2,
3,
4,
5,
6,
7] have been widely applied in computer vision tasks, such as image classification [
8,
9], object detection [
10], semantic segmentation [
11], etc. The performance of these tasks was greatly improved with the application of CNNs. However, CNNs have limitations. Firstly, the pooling operation provides some transition invariance and results in the loss of important location information. Secondly, CNNs struggle to learn the part–whole relationship. To address these weaknesses, CapsNet [
12] is proposed to replace the scalar output with vector output for representing different properties, such as the orientation and viewpoints of objects. Different from the translation invariance from the pooling operation, CapsNet can provide translation equivariance. Equivariance is the detection of objects that can transform into each other. Different from CNNs, the knowledge about part–whole relationships is kept in the capsule network, as discussed in [
12,
13]. Capsule networks recognize objects through both local features and part–whole knowledge. For example, a bird, as an object, has several parts, including a head, a trunk, wings, claws, and a tail. When these parts are disturbed, CNNs would still recognize the disturbed object as a bird, while CapsNet can determine that it is not a bird through the part–whole relationship. In the original CapsNet [
12], information is represented in vectorized format, leading to costly calculation of the routing between capsules of different layers. In the matrix CapsNet [
13], vectorized information is replaced by matrix capsules, and routing weights are updated by the expectation-maximization (EM) algorithm.
However, early CapsNets have their drawbacks. First, the low-level features that consist of the capsule are extracted only by shallow convolutional operations. This results in capsules containing very little semantic high-level information. Secondly, low-level convolutional operations lack an attention mechanism, which may import meaningless and redundant information into the capsules. One of the effective ways for performance boosting is to employ a better feature extractor, which can capture richer and more semantic contextual patterns to build capsules. Recent efforts focusing on the improvement of feature extraction for CapsNets were extensively investigated, such as Multi-Scale CapsNet (MS-CapsNet) [
14] and RS-CapsNet [
15].
In this paper, we propose a novel capsule network, named the attentive octave convolutional capsule network (AOC-CapsNet) for medical image classification. In AOC-CapsNet, the traditional convolution operation is replaced by an octave convolution operation. In [
16], the octave convolution operation is proposed to process both higher and lower frequencies in the inputs at the same time. In natural images, higher frequencies correspond to the detailed information that varies greatly in the images, and lower frequencies correspond to the smoothly changing structure in the images. These two types of information are also very important for medical image classification. It is critical to select which kind of information is more important in medical image classification. However, the traditional octave convolutional operation cannot enhance useful information and suppress useless information. In AOC-CapsNet, we adopt a convolutional block attention module (CBAM) to identify and select useful information. The CBAM allows AOC-CapsNet to highlight critical local regions with rich semantic details utilized as distinguishable patterns, leading to a performance gain in the medical image classification task.
Studies on capsule networks [
17,
18] have focused on medical image analysis. A recent benchmark, named MedMNIST [
19], was proposed and used to validate the performance of different models for medical image analysis. MedMNIST is composed of 10 pre-processed open medical image datasets. Similar to the MNIST dataset [
20], classification tasks in MedMNIST are lightweight. The resolution of images in classification tasks is
. Those tasks cover primary medical image modalities and diverse data scales. In this paper, we design comparative experiments on seven datasets in MedMNIST. Through experiments, ResNet [
6], AutoML methods [
21,
22] and methods related to capsules [
12,
13,
14] are compared with the proposed AOC-Caps. In the ablation studies, matrix capsule networks with different convolutional feature extraction layers are compared to determine which type of convolution layer is more suitable for the application of capsule networks in the medical image classification of MedMNIST.
The main contributions of this research are as follows:
An attentive octave convolution operation is proposed. By combining the novel operation with capsule networks, we design an effective classification framework named AOC-CapsNet for medical image classification.
The proposed AOC-CapsNet is validated via extensive experiments on the MedMNIST benchmark and has achieved the state-of-the-art (SOTA) performance in two of the seven tasks.
The rest of this paper is organized as follows.
Section 2 reviews the related research work.
Section 3 explains our proposed method. In
Section 4, comprehensive experiments are conducted to evaluate the effectiveness of the proposed method. Finally, in
Section 5, we conclude the paper.
2. Related Work
Introduced by Hinton [
23], the core idea of “capsule” is to group the neurons into a vector, which is defined as a capsule. In CNNs, the activation of a neuron can be considered the likelihood of detecting a specific feature. Different from feature invariance in CNN, feature equivariance, which is considered the detection of features that can transform into each other, is achieved in capsule networks.
In [
12], the dynamic routing between capsules is applied in the proposed capsule network. The pooling operation is abandoned in [
12] for keeping the location information of features. Although CapsNet with dynamic routing achieved SOTA performance in MNIST and its variant, MultiMNIST, it still has drawbacks, such as huge computational cost and the lack of high-level semantic information. In [
13], the matrix capsule network is constructed by transforming the capsule form from vector to matrix and changing the link mode between capsules of different layers. The coupling coefficients between lower-layer and higher-layer capsules are updated by the EM algorithm. In [
12,
13], all the features used to construct capsules are extracted by a convolution layer. These features are low-level information and cannot effectively recognize complex objects.
To handle the drawbacks explained above, there have been several studies [
14,
15,
17,
24,
25] focusing on applying more powerful feature extraction modules to improve the performance of capsule networks. In [
14], multiple convolutional kernels are used to extract multi-scale features for constructing multi-dimension capsules, and a novel dropout for capsules is proposed. In RS-Caps [
15], the Res2Net block [
26] is used to extract multi-scale features and increase the size of receptive fields of each convolutional layer. What is more, a new linear combination between capsules and routing process is proposed for constructing more effective classification capsules. In HitNet [
24], a new layer called hit-or-miss and a centripetal loss function are designed. HitNet also introduces a data augmentation method that can combine data space and feature space. The most straightforward idea to improve the performance of capsule networks is to increase the number of intermediate capsule layers to obtain deeper capsule networks. However, it was recently proven that directly stacking fully connected capsule layers will result in a decline in performance [
27]. In order to solve this problem, DeepCaps [
25] uses a novel 3D convolution-based dynamic routing algorithm. Furthermore, a class-independent decoder network is also proposed to strengthen the use of reconstruction loss as a regularization term.
Deep learning technology has also been applied in medical image analysis. In [
28], U-Net architecture, which consists of a contracting path to capture context information and an expanding path that enables precise localization, is proposed for biomedical image segmentation. In [
29], an approach based on a volumetric, fully convolutional neural network is proposed for 3D image segmentation. USE-Net [
30], which incorporates squeeze-and-excitation (SE) modules into U-Net, is proposed for magnetic resonance imaging (MRI) segmentation. In [
31], SegNet [
32], which consists of an encoder network, a decoder network followed by a pixel-wise classification layer, U-Net and pix2pix are compared in the experiments on two multi-centric MRI prostate datasets.
3. Attentive Octave Convolutional Capsule Network
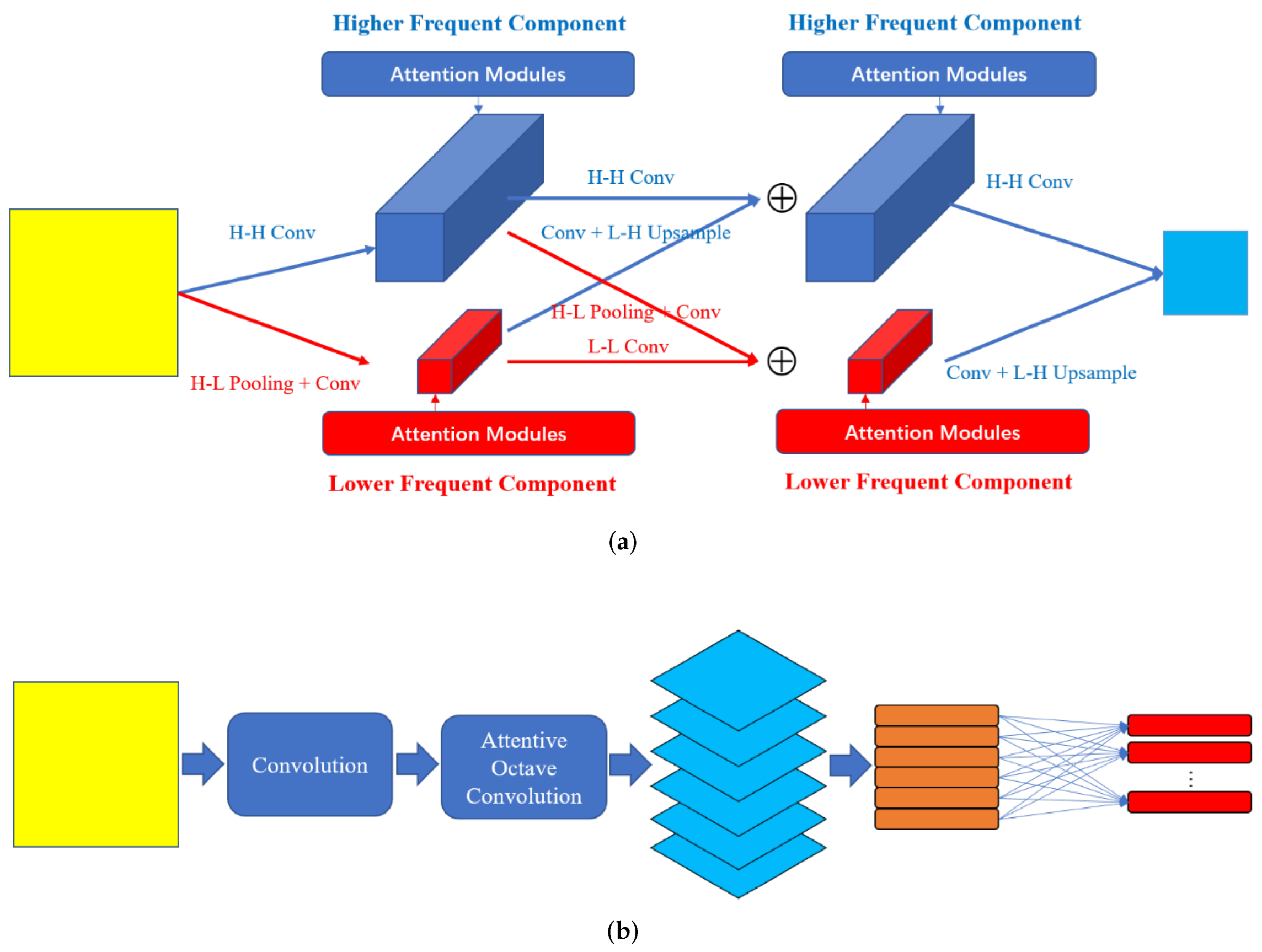
In this section, we introduce our proposed AOC-Caps in detail. As shown in
Figure 1b, input images are fed into a traditional convolution layer followed by batch normalization and RELU operation. The feature maps generated by this convolutional layer are then fed into the attentive octave convolution layer (AOC-Layer). In the AOC-Layer, the higher- and lower-frequencies are processed simultaneously. The useful information is enhanced, and useless information is suppressed in the AOC-Layer. The enhanced feature maps generated by the AOC-Layer are then reshaped into a pose matrix and an activation following the matrix capsule network [
13]. In
Section 3.1, the details of the AOC-Layer are provided. The process of routing and updating in capsule layers is introduced in
Section 3.2. The loss function of the proposed AOC-Caps is described in
Section 3.3.
3.1. Attentive Octave Convolution Layer
In the traditional convolution operation, the input information is processed by convolutional kernels of certain sizes. Convolution operations of different convolutional kernels can obtain information of different frequencies, and there is no effective fusion process between frequencies. The octave convolution operation [
16] is proposed to process different frequencies simultaneously. In octave convolution, the convolution and fusion of two frequencies with a difference of an octave are performed simultaneously without an attention mechanism module. It is very important to select useful information in medical image classification. In order to select information of different frequencies, we add a CBAM [
33] in the AOC-Layer.
Suppose the input is defined as
, where
h and
w are defined as the height and width of the image. The feature maps obtained by the first convolution layer are defined as
, where
c is the channel of the feature maps. In the AOC-Layer, the feature map
F is first divided into two parts
by a convolution operation (H-H Conv in
Figure 1a) and pooling and a convolution operation (H-L Pooling and Conv in
Figure 1a), where
is higher frequency and
is lower frequency. The channels of the feature maps are divided by ratio
. The size of the higher frequency is
and that of the lower frequency is
. In order to obtain lower-frequency information, two pooling operations (average and maximum) are used in the AOC-Layer. Their effects are discussed in detail in
Section 4.5. In order to convert lower-frequency information into higher-frequency information, bilinear interpolation is used to convert lower-resolution feature maps into higher-resolution feature maps. Then, the higher- and lower-frequency communicate with each other by summation, as shown in Formulas (
1) and (
2):
As shown in
Figure 1a, the attention modules are added to the intermediate feature maps with different frequencies to enhance useful information and suppress useless information. Without losing particularity, an intermediate feature map in the AOC-Layer is defined as
, where
is the number of channels of both higher- and lower-frequency parts, and
and
are the height and width of higher- and lower-frequency parts. The attention module sequentially infers a 1D channel attention map
and a 2D spatial attention map
. The selections of channel and spatial information are based on Formulas (
3) and (
4):
where ⊗ denotes the element-wise multiplication. The intermediate feature map
is firstly processed by a channel attention module. The feature map
is pooled along the spatial dimension through average and maximum operations. The average-pooled features and max-pooled features are processed by a multi-layer perception (MLP), with one hidden layer for producing the channel attention vector
. The channel attention enhanced feature map
is then processed by a spatial attention module. Feature map
is pooled along the channel dimension by both average and maximum operations. The average-pooled and max-pooled feature maps are concatenated along the channel dimension to produce
.
is processed by a convolution operation with kernel size 7 followed by a sigmoid function. In the AOC-Layer, the attention modules are plugged into
, and
. The role of attention modules is also discussed in detail in
Section 4.5.
3.2. Capsule Layer
The two commonly used capsule networks are CapsNet with dynamic routing and matrix CapsNet with EM routing. In CapsNet with dynamic routing, the capsule vector is constructed by stacking the neurons with scalar values. In CapsNet with EM routing, the capsule contains a pose matrix and an activation.
The output feature maps of the AOC-Layer are first reshaped into a series of capsules
For one capsule
, its input is the weighted sum of all the prediction vectors
generated by the previous layers. It can be defined as Formulas (
5) and (
6).
where
is the routing coefficient,
is the matrix used for voting, and
is the output vector of the previous capsule layer. The routing coefficients should be computed in dynamic routing [
12] or EM routing [
13]. In dynamic routing,
is a vector.
The process of dynamic routing is shown as follows:
The prior probability between capsule j and capsule i in the previous layer is initialized to be 0;
The routing coefficients can be computed through the softmax function ;
The input to capsule
j is computed by Formula (
5) and then it is squeezed by
;
The is updated by ;
Repeat steps 2 to 4 r times. The value of r is set empirically, usually from 1 to 3.
Different from capsule networks with dynamic routing, capsules in the matrix capsule with EM routing consist of a pose matrix and an activation. A pose matrix defines the translation and the rotation of the objects. The aim of the EM algorithm is to cluster datapoints into different Gaussian distributions. Suppose the pose matrix is a 4 × 4 matrix, i.e., 16 components. Let
be the vote from capsule
i to capsule
j, and
be its
h-th component. The probability density function of a Gaussian is defined as Formula (
7):
It can be applied to compute the probability of
belonging to the capsule
j’s Gaussian model:
Let
be the cost to activate the
h-th component of capsule
j by the
h-th component of capsule
i, where
Whether the capsule
j is activated is determined by the following equation:
where
is the assignment probability of each datapoint to a capsule. , , , and are computed iteratively using the EM algorithm.
3.3. Loss Function
In AOC-Caps, if the dynamic routing is applied, the loss function is defined as
where
if the class
c is present,
is set to be 0.9 and
is set to be 0.1.
If the matrix capsule with EM routing is applied, the loss function has a similar design as in [
13]. The spread loss is used to directly maximize the gap between the activation of the target class and the activation of other classes. The loss function is formed as Formulas (
13) and (
14):
where
is the wrong class and
is the target class.
4. Experiments
4.1. Datasets
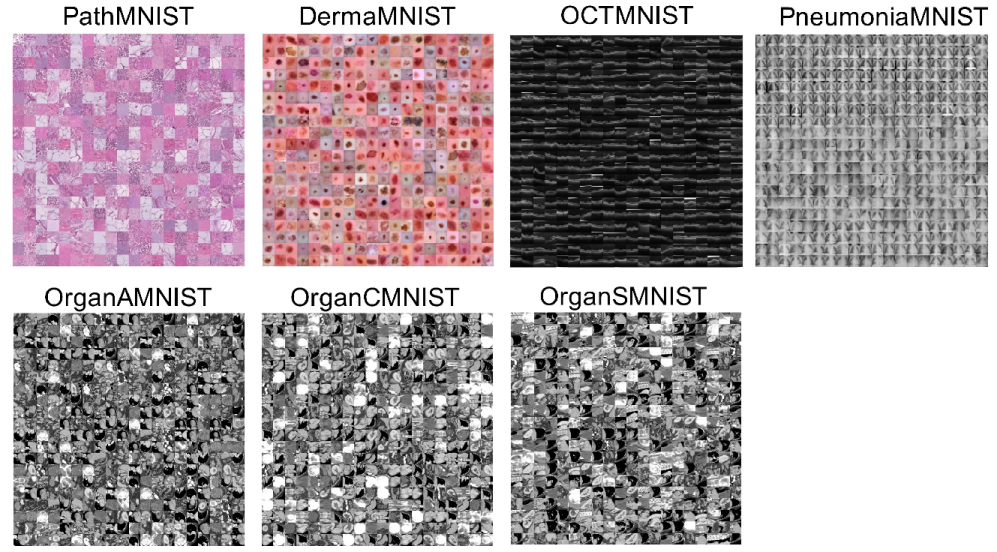
MedMNIST consists of 10 pre-processed datasets. It contains 10 open medical image datasets covering multiple tasks, including multi-class, binary classification, sequential regression, and multi-label. In our experiments, we focus on the multi-class tasks, such as PathMNIST, DermaMNIST, OCTMNIST, PneumoniaMNIST, OrganMNIST_Axial, OrganMNIST_Coronal, and OrganMNIST_Sagittal. In these seven datasets, the height and width of the images are resized to 28.
Figure 2 demonstrates an overview of the seven datasets with samples.
All datasets are divided into a training set, a validation set, and a test set. The number of images in each set is detailed in
Table 1. The models are trained on the training sets, validated on the validation sets after each epoch during training, and finally evaluated on the test sets.
PathMNIST. It is based on a prior study [
34] for predicting survival from colorectal cancer histology slides, which provides a dataset of 100,000 non-overlapping image patches from hematoxylin and eosin-stained histological images, and a test dataset of 7180 images patches from a different clinical center. Nine types of tissues are involved, resulting in a multi-class classification task. The details of these nine categories are introduced in
Table 2.
DermaMNIST. It is based on HAM10000 [
35], a large collection of multi-source dermatoscopic images of common pigmented skin lesions. The dataset consists of 10,015 images labeled as seven different categories, as a multi-class classification task. These seven categories are introduced in
Table 3.
OctMNIST. It is based on a prior dataset [
36] of 109,309 valid optical coherence tomography images for retinal diseases. Four types are involved in this dataset, leading to a multi-class classification task. These four categories are introduced in
Table 4.
PneumoniaMNIST. It is based on a prior dataset [
36] of 5856 pediatric chest X-ray images. This task is a binary class of pneumonia and normal. The source training set is split into training and validation sets with a ratio of 9:1, and its source validation set is used as the test set. These two categories are introduced in
Table 5.
OrganMNIST_(Axial, Coronal, and Sagittal). These three datasets are based on 3D computed tomography (CT) images from the Liver Tumor Segmentation Benchmark [
37]. Bounding-box annotations of 11 body organs from another study are used for obtaining the organ labels. The only differences of OrganMNIST_(Axial, Coronal, and Sagittal) are the views. The 11 categories of each of the three datasets are introduced in
Table 6,
Table 7 and
Table 8.
4.2. Evaluation Metrics
In our experiments, we use accuracy (ACC), area under ROC curve (AUC), precision (PRE), recall (REC) and F1-score (F1) as the evaluation metrics. The formulas of these metrics are shown below:
where
C is the number of classes and
N is the number of total samples.
TP,
TN,
FP, and
FN refer to true positive, true negative, false positive, and false negative.
Accuracy (ACC) is the most commonly used metric among these performance metrics, but it does not indicate the true model performance when the classes are imbalanced. Area under the ROC curve (AUC) is less sensitive to class imbalance than ACC. Precision (PRE) and recall (REC) are related to the positive prediction. In our experiments, we use macro-precision and macro-recall, which are defined by Formulas (
16) and (
18). The F1-score (F1) is a metric that combines both precision and recall.
4.3. Baselines
In our experiments, we use the same baselines as in [
19]. In addition, several methods related to capsule networks are used in the classification tasks of the seven datasets mentioned above.
ResNet18 and ResNet50 [
6]. These two models are trained for 100 epochs, using a cross-entropy loss function and an SGD optimizer with a bath size of 128 and an initial learning rate
.
AutoML Methods. Several AutoML methods [
21,
22] were applied on MedMNIST classification. The experimental settings of three AutoML methods (auto-sklearn [
21], AutoKeras [
22] and Google AutoML Vision) are the same as in [
19]. AutoML methods are designed to search for the optimal hyper-parameter setting or neural architecture to maximize the predictive ability. For example, Auto-sklearn and autoKeras are open-source AutoML tools for both statistical machine learning and deep learning. On the other hand, Google AutoML Vision offers commercial black-box AutoML tools. In this study, the results of AutoML methods in our experiments are directly referenced from [
19].
CapsNet (Dynamic routing) [12]. In [
12], the output feature maps of the convolutional layer with kernel size 9 × 9 and a stride 2 are reshaped into primary capsules. The capsules of the previous layer are routed to classification capsules by agreement. Different from the original setting, we set the iteration number to be 2 instead of 3 for the best performance.
CapsNet (EM routing) [13]. In [
13], the capsule contains a 4 × 4 pose matrix and an activation. The vote for capsules in the next layer is computed by the matrix multiplication between the pose matrix and the trainable transformation matrix. The routing coefficients between capsules and classification capsules are updated through the EM algorithms. Different from the original setting, the kernel size of the first convolutional layer is set to be 3 × 3 for detailed feature representation.
MS-Caps [14]. In [
14], hierarchical features are extracted and reshaped into capsules of different dimensions. The capsules cascade together for the dropout operation. Following the original experimental setting, the Adam optimizing method [
38] is used as the gradient descent algorithm to perform the training. The weight decay factor is set to be 0.00001. The initial learning rate is 0.001 and 0.0001, and the number of iterations is 25 and 50 for converging to the optimal solution quickly.
DeepCaps [25]. In [
25], the Adam optimizer is used with an initial learning rate of 0.001, which is reduced by half after every 20 epochs.
4.4. Implementation Details
In this paper, experiments are implemented by PyTorch on a PC with four GPUs of TITAN X. Different from the experiments in [
19], we conduct experiments only with images of size 28 × 28.
In the experiments, the kernel size of the first convolution layer is set to be 3 × 3 and the stride is set to be 2. In the AOC-Layer, ratios for the split of higher- and lower-frequencies are set to be 0.5. The convolutional kernel size is set to be 3 in the AOC-Layer. In the attention modules, the reduction ratio is set to be 8, and the convolutional kernel size is set to be 3. When the capsule with a dynamic routing framework is applied, the vector dimension of the capsule is set to be 8 and the iteration number is set to be 2. When the matrix capsule with the EM routing framework is applied, the pose matrix is set to be a 4 × 4 matrix. The batch size is set to be 96 and the initial learning rate is 3 ×. The training epoch is set to be 120.
In our experiments, all metrics are implemented by a python module called Torchmetrics.
4.5. Ablation Study
To verify the effectiveness of each design in AOC-Caps, we conduct experiments for the ablation study. The ablation study focuses on (1) different forms of capsules and different routing methods; (2) types of pooling operations in the AOC-Layer; (3) the effect of attention modules.
To test different forms of capsules and different routing methods, we choose the pooling operation type to be maximal and plug attention modules as introduced in
Section 3.1. It should be noted that in dynamic routing, we do not add the decode term. As shown in
Table 9, the AOC-Caps with matrix and EM routing outperforms the one with dynamic routing. It can be seen from
Table 9 that different routing methods have a great impact on the results. For example, in the OrganMNIST_Axial dataset, the accuracy with EM routing is 93.1%, while the accuracy with dynamic routing is 80.2%. The AOC-Caps with EM routing also achieves better precision and recall, both of which are more than 10% higher.
To test the effectiveness of different pooling operations, such as average and maximum, we keep the capsule form to matrix and EM routing. The attention modules are plugged into the AOC-Layer as introduced in
Section 3.1. As shown in
Table 10, the AOC-Caps with max-pooling outperforms the one with avg-pooling. What is more, the type of pooling operations has much less impact on the results than the routing methods.
To test the effectiveness of the attention modules, we keep the capsule form to matrix and EM routing. The pooling type is set to be maximal. As shown in
Table 11, the AOC-Caps with the attention module outperforms the one without the attention module. Attention modules have a greater impact on the results than the pooling operation type and less than the routing method. In
Table 11, the AOC-Caps with attention modules achieves higher accuracy, precision and recall.
The results in
Table 9,
Table 10 and
Table 11 have demonstrated the effectiveness of EM routing, max-pooling in the AOC-Layer, and attention modules. By putting them together, we find the optimal combination to implement AOC-Caps based on the following comparative experiments.
4.6. Comparative Experiments
The results of the comparative experiments are reported in
Table 12, in which the results of ResNet18, ResNet50, Auto-sklearn, AutoKeras, and Google AutoML Vision are from [
19]. For the three AutoML models, the original paper does not provide metrics in Pre, Rec, and F1, which are marked as dashes in
Table 12.
Table 12 shows the performance of the comparative models on seven datasets in MedMNIST. In terms of accuracy (ACC), AOC-Caps achieves the best accuracy on DermaMNIST and OrganMNIST_Axial datasets, and ranks second on other datasets. CapsNet with dynamic routing and EM routing demonstrate worse performance than other methods, due to the shallow feature extraction network. However, because the data of OrganMNIST_(Axial, Coronal, and Sagittal) are collected from 3D images, the viewpoints are different. Therefore, the CapsNet with the EM routing with transformation invariance can obtain good accuracy, even with the shallow feature extraction network.
Due to the class imbalance in certain datasets of MedMNIST, it is necessary to consider different metrics. In terms of precision (PRE) and recall (REC), the performance of all methods is lower than accuracy on datasets DermaMNIST and OrganMNIST_Sagittal because of the data imbalance between different categories in DermaMNIST and OrganMNIST_Sagittal. In this case, AOC-Caps still achieves better results. However, it should be pointed out that there is no corresponding solution to this data imbalance problem in our experiments.
In our comparative experiments, a model with deeper layers does not necessarily achieve higher accuracy. Consider ResNet18 as an example. It outperforms ResNet50 on datasets such as DermaMNIST, OCTMNIST, OrganMNIST_Axial, and OrganMNIST_Sagittal. As part of the reason, the low resolution of the image data does not require a deep network model to extract high-level features.
5. Conclusions
In this paper, we proposed a novel attentive octave convolutional capsule network (AOC-Caps) for medical image classification. In the AOC-Layer, the octave convolution and attention modules (CBAM) are used for communicating and selecting the higher- and lower-frequency information. The output feature maps of the AOC-Layer are used as the foundation for constructing capsules. The experiments have verified the effectiveness of the AOC-Layer. Because the images of certain datasets come from 3D images and the viewpoint is likely to change, the capsule routing method with transformation invariance, such as matrix CapsNet with the EM routing, can obtain higher accuracy. By combining the AOC-Layer and matrix CapsNet with the EM routing, AOC-Caps could achieve better performance than most baselines in the experiments.
However, the AOC-Layer in AOC-Caps is still a shallow convolutional network. Although AOC-Caps has achieved better results than DeepCaps on the classification tasks in MedMNIST, the deep network structure is still necessary when handling high-resolution medical images. This may be due to the smaller image size of the dataset and less detailed information. In future studies, we will consider high-resolution medical images of different diseases and investigate the effect of AOC-Caps in the case of deep structures. Furthermore, it is necessary to consider how to solve the problem of class imbalance in medical image analysis.