Abstract
This article proposes a two-phase hybrid method to train RBF neural networks for classification and regression problems. During the first phase, a range for the critical parameters of the RBF network is estimated and in the second phase a genetic algorithm is incorporated to locate the best RBF neural network for the underlying problem. The method is compared against other training methods of RBF neural networks on a wide series of classification and regression problems from the relevant literature and the results are reported.
1. Introduction
In machine learning, many practical problems appear such as classification and regression problems. A good programming tool that can be used to tackle this problem is radial basis function (RBF) networks [1]. These networks typically are expressed as a function:
where is the input pattern, the vector is called the weight vector and is the predicted value of the network. RBF networks are feedforward neural networks [2] with three computational layers:
- The input layer, where the problem is presented in the form of patterns to the neural network.
- The processing layer, where a computation is performed using the Gaussian processing units . These units can have many forms in the relevant literature but the most used form is the Gaussian function expressed as:The value depends only on the distance of vector from some other vector , which typically is called centroid.
- The output layer where the output of every function is multiplied by a corresponding weight value .
RBF networks have been used in many classification and regression problems from the areas of physics [3,4,5,6], medicine [7,8,9], solution of differential equations [10,11], chemistry [12,13,14], economics [15,16,17], digital communications [18,19], etc. Furthermore, recently, RBF networks have been used in more difficult problems such as the authentication assurance of meat products [20], trajectory tracking for electrohydraulic servo systems, identification of geographical origin for foods [21], prediction of solution gas–oil ratio of crude oil systems [22], prediction of occurrences of haloketones in tap water [23], health monitoring [24], etc. Because of the extensive use of RBF networks, many methods have been proposed in the recent literature to enhance them. There are methods that parallelize the RBF networks [25,26], methods that improve the initialization of the RBF parameters [27,28,29], methods that alter the architecture of the network [30,31,32], methods aimed to locate the best set of RBF parameters with global optimization techniques [33,34,35], etc. This article transforms the problem of RBF training into an optimization problem and applies a modified genetic algorithm technique to solve it. The global optimization problem is defined as:
where m is the total number of input patterns and is the output for pattern . The suggested approach has two phases: firstly, reasonable bounds for the RBF parameters are estimated using the k-means [36] algorithm and in the second phase, the modified algorithm is used to solve the problem of Equation (3) inside the bounds located in the first phase.
2. Method Description
The proposed method can be divided into two main phases: during the first phase, an approximation for the bound of RBF parameters is made using the k-means algorithm; in the second phase, the optimization problem is solved using a modified genetic algorithm. These phases are outlined in detail in the following subsections.
2.1. Bound Location Phase
The proposed genetic algorithm has chromosomes with dimension , where d is the dimension of the input problem, i.e., the dimension of the vector in Equation (3), and k is the total number of processing units of the RBF network. The layout of each chromosome is presented in Table 1. Every center in Equation (1) is a vector of dimension d and an additional parameter is also reserved for the parameter of every function. The centroids and the corresponding variances are estimated using the k-means algorithm that is described in Algorithm 1. The value for every is calculated as:

Table 1.
The layout of the chromosomes in the proposed genetic algorithm.
After the estimation of and , the vectors with dimension are constructed. These vectors will serve as the bounds for the chromosomes of the genetic population. These vectors are constructed using the following procedure:
- Setm = 0
- Set
- Fordo
- (a)
- Fordo
- i.
- Set = , =
- ii.
- Set
- (b)
- EndFor
- (c)
- Set,
- (d)
- Set
- EndFor
| Algorithm 1: The k-means algorithm. |
|
2.2. Main Algorithm
The genetic algorithm used here is based on the algorithm denoted as GA in the Kaelo and Ali’s paper [37], with a modified stopping rule as proposed in [38]. The basic steps of the main algorithm are given below:
- Initialization Step
- (a)
- Read the train set with m patterns of d dimension.
- (b)
- Setk the number of nodes for the RBF network.
- (c)
- Estimate the vectors using the procedure of Section 2.1.
- (d)
- Initialize a genetic population of random chromosomes inside .
- (e)
- Set the selection rate , the mutation rate , and the maximum number of generations.
- Evaluation StepFor every chromosome g calculate the fitness using the procedure defined in Section 2.3.
- Genetic operations stepDuring this step three genetic operations are performed: selection, crossover and mutation.
- (a)
- Selection procedure. Firstly, the chromosomes are sorted with relevance to their corresponding fitness value. The best are transferred without change to the next generation and the remaining ones are substituted by offspring created through the crossover procedure. In the crossover procedure, the mating parent are selected using a tournament selection for every parent. The tournament selection is as follows:
- Select a set of chromosomes from the population.
- Return the chromosome with the best fitness value in that subset.
- (b)
- Crossover procedure. For every pair of selected parents create two new offspring and :with a random number and [37]. This crossover scheme will be able to better explore the search space of the train error.
- (c)
- Mutation procedure. For every element of each chromosome, create a random number . If , then change that element randomly. The mutation is performed in a way similar to other approaches of genetic algorithms [38] and it is described in Section 2.5.
- (d)
- Replace the worst chromosomes in the population with the generated offsprings.
- Termination Check Step
- (a)
- Set
- (b)
- Terminate if the termination criteria of Section 2.4 are satisfied, else Goto Evaluation Step.
2.3. Fitness Evaluation
In this step, a valid RBF network , is created using the chromosome g and is subsequently trained using the typical training procedure for RBF networks. The main steps to calculate the fitness of a chromosome g are the following:
- Decode the chromosome g to the parts (centers and variances) of the RBF network as defined by the layout of Table 1.
- Calculate the output vectors by solving the induced system of equations:
- (a)
- Set the matrix of k weights, and .
- (b)
- Solve:The matrix is the pseudo-inverse of , with
- Set
2.4. Stopping Rule
Define as the best chromosome in the population and define as the variance of best fitness at generation iter. If fitness has not improved for a number of generations, then probably the algorithm should terminate. Hence, the termination rule is defined as:
where is the last generation where a new minimum was found.
2.5. Mutation Procedure
Let be the chromosome to be mutated. The proposed mutation procedure modifies with probability and the resulting element is given by
where t is a random number with . The function is given by:
where r is a random number in and b controls the change of element . In the proposed algorithm the value was used.
3. Experiments
In order to evaluate the performance of the proposed method, comparative experiments were performed on a series of well-known classification and regression datasets from the relevant literature.
3.1. Experimental Setup
The RBF network was coded in ANSI C++, using the Armadillo library [39] and the optimization was performed using the genetic optimization method of the optimization package OPTIMUS, that is freely available from https://github.com/itsoulos/OPTIMUS/ (accessed on 18 January 2021). Furthermore, to have more reliability in the results the commonly used method of 10-fold cross-validation was used, which means that the original data were randomly partitioned into 10 equally sized subsamples. Subsequently, 10 independent experiments were conducted: in each experiment one subsample was used as the testing data and all the others as the training data. The average error on the test data was the total test error. All the experiments were executed 30 times with different initialization for the random generator each time. The random generator used was the function drand48() of the C programming language. The execution environment was an Intel Xeon E5-2630 multicore machine using the OpenMP library [40] for parallelization and the Ubuntu Linux operating system. The parameters for the genetic algorithm are displayed in Table 2. The parameters of the method were chosen so that there is a balance between speed and efficiency of the method.

Table 2.
Experimental parameters.
3.2. Experimental Datasets
The classification problems used for the experiments were found in most cases on two Internet databases:
- UCI dataset repository, https://archive.ics.uci.edu/ml/index.php (accessed on 18 January 2021)
- Keel repository, https://sci2s.ugr.es/keel/datasets.php (accessed on 18 January 2021) [41].
The following classification datasets were used:
- The Alcohol dataset, which is related to alcohol consumption [42].
- The Appendicitis dataset, proposed in [43].
- The Australian dataset [44], which refers to credit card applications.
- The Balance dataset [45], which is used to predict psychological states.
- The Cleveland dataset, used in various papers to detect heart disease [46,47].
- The Dermatology dataset [48], which is used for differential diagnosis of erythematosquamous diseases.
- The Glass dataset, which contains glass component analysis and has been used in a variety of papers [49,50].
- The Hayes roth dataset [51].
- The Heart dataset [52], used to detect heart disease.
- The HouseVotes dataset [53], which is about votes in the U.S. House of Representatives.
- The Ionosphere dataset, which contains data from the Johns Hopkins Ionosphere database and has been studied in a number of papers [54,55].
- The Liverdisorder dataset [56], used to detect liver disorders in people using blood analysis.
- The Mammographic dataset [57], used to identify the severity of a mammographic mass lesions.
- The Parkinson dataset, which is composed of a range of biomedical voice measurements from 31 people, 23 with Parkinson’s disease (PD) [58].
- The Pima dataset [59], used to detect the presence of diabetes.
- The Popfailures dataset [60], related to climate model simulation crashes.
- The Regions2 dataset, which was created from liver biopsy images of patients with hepatitis C [61].
- The Ring dataset [62], a 20-dimension problem with two classes, where each class is drawn from a multivariate normal distribution.
- The Saheart dataset [63], used to detect heart disease.
- The Segment dataset [64], which contains patterns from a database of seven outdoor images (classes).
- The Sonar dataset [65]. The task is to discriminate between sonar signals bounced off a metal cylinder.
- The Spiral dataset, which is an artificial dataset containing 1000 two-dimensional examples that belong to two classes.
- The Tae dataset [66], which concerns evaluations of teaching performance.
- The Thyroid dataset, which concerns thyroid disease records [67].
- The Wdbc dataset [68], which contains data from breast tumors.
- The Wine dataset, used to detect through chemical analysis the origin of wines in various research papers [69,70].
- The EEG dataset from [71], which consists of five sets (denoted as Z, O, N, F and S), each containing 100 single-channel EEG segments each lasting 23.6 s. With different combinations of these sets the produced datasets are Z_F_S, ZO_NF_S, ZONF_S.
- The Zoo dataset [72], where the task is to classify animals in seven predefined classes.
The regression datasets are in most cases available from the StatLib URL ftp://lib.stat.cmu.edu/datasets/index.html (accessed on 18 January 2021):
- The Abalone dataset [73], which can be used to obtain a model to predict the age of abalone from physical measurements.
- The Airfoil dataset, which is used by NASA for a series of aerodynamic and acoustic tests [74].
- The Anacalt dataset [75], which contains information about the decisions taken by a supreme court.
- The BK dataset [76], used to estimate the points in a basketball game.
- The BL dataset, which can be downloaded from StatLib and contains data from an experiment on the effects of machine adjustments on the time it takes to count bolts.
- The Concrete dataset, used to measure the concrete compressive strength [77].
- The Housing dataset, taken from the StatLib library, which is maintained at Carnegie Mellon University, and described in [78].
- The Laser dataset, used in laser experiments. It has been obtained from the Santa Fe Time Series Competition Data repository.
- The MB dataset, available from Smoothing Methods in Statistics [77].
- The NT dataset, which contains data from [79] that examined whether the true mean body temperature is 98.6 F.
- The Quake dataset, whose objective is to approximate the strength of a earthquake. It has been obtained from the Bilkent University Function Approximation Repository.
3.3. Experimental Results
The results for the classification datasets are listed in Table 3 and for the regression datasets the results are reported in Table 4. For the first case, the average classification error is reported and for the case of regression datasets, the total test error is reported. The column KRBF denotes the classic RBF training method, GRBF denotes the method proposed in [80] and the column “proposed” denotes the proposed method. The KBF simply consists of two phases: in the first phase, the centers and variances are estimated through the k-means algorithm and in the second phase, a system of equations is solved to obtain the weights of the RBF network.

Table 3.
Classification error for different datasets.

Table 4.
Regression error for different datasets.
From the experimental results, it is clear that the proposed method is significantly superior to other methods in almost all datasets. In the proposed method, the appropriate initialization interval was found for the parameters of RBF using k-means. A parallel genetic algorithm was then applied to this previous value range, creating a variety of neural networks. This combination of techniques obviously has very good results as it combines a very efficient clustering method and an excellent optimization method that is ideally parallelized. Of course, the new method requires much more execution time, due to the presence of the genetic algorithm, but the parallel execution of the software drastically reduces this time. Furthermore, in order to study the effectiveness of the selection of parameter F an additional experiment was conducted, where the best fitness of the genetic algorithm is plotted for the Wine problem. The outcome of this experiment is graphically outlined in Figure 1. The graph shows that the behavior of the proposed method does not change significantly for different values of the parameter F. Furthermore, the plot for the Wine dataset of best, worst and average fitness for is shown in Figure 2. An additional experiment was performed to evaluate the effect of the parameter change k on the results. In Figure 3, the plot for different values of k for the Housing dataset is outlined and in Figure 4 the same experiment is shown for the Z_F_S classification dataset. Of course, from the value onwards, the error falls but not at the same rate. The proposed value was used in all datasets in order to have a balance between the speed and the efficiency of the method. Finally, the classification performance was evaluated based on two evaluation metrics: precision and recall for some datasets as shown in Table 5. In these results, the precision of the proposed method showed the best classification performance under different datasets. We found the same trends in recall metric in two of three datasets (spiral and EEG datasets).
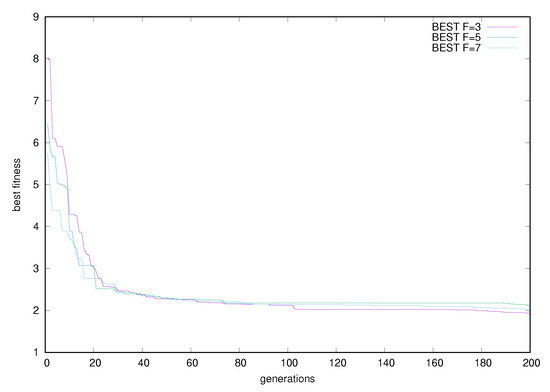
Figure 1.
Plot of best fitness for the Wine problem for different values of parameter F.
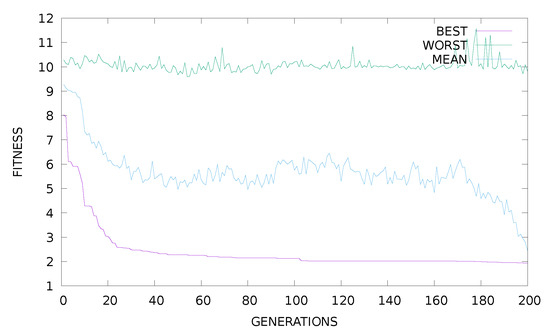
Figure 2.
Plot of best, worst and average fitness for the Wine dataset and .
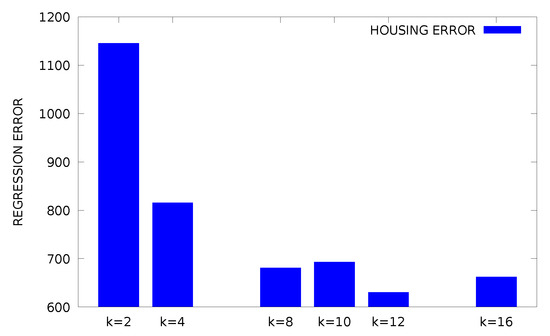
Figure 3.
Experiments with different values of k for the Housing dataset.
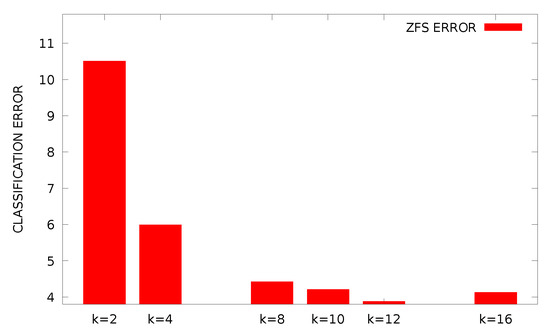
Figure 4.
Classification error of Z_F_S dataset for different values of k.

Table 5.
Comparison of precision and recall between the traditional RBF and the proposed method for some datasets.
4. Conclusions
A two-phase method was proposed in this article to train RBF neural networks for classification and regression problems. Firstly, a commonly used clustering method was used to estimate an interval for the critical parameters of the neural network. Subsequently, a parallel genetic algorithm was incorporated to locate the best RBF network with good generalization capabilities. The used algorithm was coded using ANSI C++ and open source libraries such as the Armadillo library and the OpenMP library for parallelization. Future research may include:
- Using parallel methods for the k-means clustering phase of the method.
- Dynamic selection of k in k-means algorithm.
- More advanced stopping rules for the genetic algorithm.
- Replacing the genetic algorithm with other optimization methods such as particle swarm optimization, ant colony optimization, etc.
Author Contributions
I.G.T., A.T. and E.K. conceived of the idea and methodology and supervised the technical part regarding the software for the RBF trainig method. I.G.T. conducted the experiments, employing several datasets, and provided the comparative experiments. A.T. performed the statistical analysis. E.K. and all other authors prepared the manuscript. E.K. and I.G.T. organized the research team and A.T. supervised the project. All authors have read and agreed to the published version of the manuscript.
Funding
We acknowledge support of this work from the project “Immersive Virtual, Augmented and Mixed Reality Center of Epirus” (MIS 5047221) which is implemented under the Action “Reinforcement of the Research and Innovation Infrastructure”, funded by the Operational Programme “Competitiveness, Entrepreneurship and Innovation” (NSRF 2014-2020) and co-financed by Greece and the European Union (European Regional Development Fund).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The experiments of this research work were performed at the high performance computing system established at Knowledge and Intelligent Computing Laboratory, Department of Informatics and Telecommunications, University of Ioannina, acquired with the project “Educational Laboratory equipment of TEI of Epirus” with MIS 5007094 funded by the Operational Programme “Epirus” 2014–2020, by ERDF and national funds.
Conflicts of Interest
All authors declare that they have no conflict of interest.
References
- Park, J.; Sandberg, I.W. Universal Approximation Using Radial-Basis-Function Networks. Neural Comput. 1991, 3, 246–257. [Google Scholar] [CrossRef]
- Bishop, C.M. Neural networks and their applications. Rev. Sci. Instrum. 1994, 65, 1803–1832. [Google Scholar] [CrossRef] [Green Version]
- Teng, P. Machine-learning quantum mechanics: Solving quantum mechanics problems using radial basis function networks. Phys. Rev. E 2018, 98, 033305. [Google Scholar] [CrossRef] [Green Version]
- Jovanović, R.; Sretenovic, A. Ensemble of radial basis neural networks with K-means clustering for heating energy consumption prediction. FME Trans. 2017, 45, 51–57. [Google Scholar] [CrossRef] [Green Version]
- Alexandridis, A.; Chondrodima, E.; Efthimiou, E.; Papadakis, G.; Vallianatos, F.; Triantis, D. Large Earthquake Occurrence Estimation Based on Radial Basis Function Neural Networks. IEEE Trans. Geosci. Remote Sens. 2014, 52, 5443–5453. [Google Scholar] [CrossRef]
- Gorbachenko, V.I.; Zhukov, M.V. Solving boundary value problems of mathematical physics using radial basis function networks. Comput. Math. Math. Phys. 2017, 57, 145–155. [Google Scholar] [CrossRef]
- Wang, Y.P.; Dang, J.W.; Li, Q.; Li, S. Multimodal medical image fusion using fuzzy radial basis function neural networks. In Proceedings of the 2007 International Conference on Wavelet Analysis and Pattern Recognition, Ningbo, China, 9–12 July 2017; pp. 778–782. [Google Scholar]
- Mehrabi, S.; Maghsoudloo, M.; Arabalibeik, H.; Noormand, R.; Nozari, Y. Congestive heart failure, Chronic obstructive pulmonary disease, Clinical decision support system, Multilayer perceptron neural network and radial basis function neural network. Expert Syst. Appl. 2009, 36, 6956–6959. [Google Scholar] [CrossRef]
- Veezhinathan, M.; Ramakrishnan, S. Detection of Obstructive Respiratory Abnormality Using Flow–Volume Spirometry and Radial Basis Function Neural Networks. J. Med. Syst. 2007, 31, 461. [Google Scholar] [CrossRef]
- Mai-Duy, N.; Tran-Cong, T. Numerical solution of differential equations using multiquadric radial basis function networks. Neural Netw. 2001, 14, 185–199. [Google Scholar] [CrossRef] [Green Version]
- Mai-Duy, N. Solving high order ordinary differential equations with radial basis function networks. Int. J. Numer. Meth. Eng. 2005, 62, 824–852. [Google Scholar] [CrossRef] [Green Version]
- Wan, C.; Harrington, P.d.B. Self-Configuring Radial Basis Function Neural Networks for Chemical Pattern Recognition. J. Chem. Inf. Comput. Sci. 1999, 39, 1049–1056. [Google Scholar] [CrossRef]
- Yao, X.; Zhang, X.; Zhang, R.; Liu, M.; Hu, Z.; Fan, B. Prediction of enthalpy of alkanes by the use of radial basis function neural networks. Comput. Chem. 2001, 25, 475–482. [Google Scholar] [CrossRef]
- Shahsavand, A.; Ahmadpour, A. Application of optimal RBF neural networks for optimization and characterization of porous materials. Comput. Chem. Eng. 2005, 29, 2134–2143. [Google Scholar] [CrossRef]
- Momoh, J.A.; Reddy, S.S. Combined Economic and Emission Dispatch using Radial Basis Function. In Proceedings of the 2014 IEEE PES General Meeting|Conference & Exposition, National Harbor, MD, USA, 27–31 July 2014; pp. 1–5. [Google Scholar] [CrossRef]
- Guo, J.-J.; Luh, P.B. Selecting input factors for clusters of Gaussian radial basis function networks to improve market clearing price prediction. IEEE Trans. Power Syst. 2003, 18, 665–672. [Google Scholar]
- Falat, L.; Stanikova, Z.; Durisova, M.; Holkova, B.; Potkanova, T. Application of Neural Network Models in Modelling Economic Time Series with Non-constant Volatility. Procedia Econ. Financ. 2015, 34, 600–607. [Google Scholar] [CrossRef] [Green Version]
- Laoudias, C.; Kemppi, P.; Panayiotou, C.G. Localization Using Radial Basis Function Networks and Signal Strength Fingerprints in WLAN. In Proceedings of the GLOBECOM 2009—2009 IEEE Global Telecommunications Conference, Honolulu, HI, USA, 30 November–4 December 2009; pp. 1–6. [Google Scholar]
- Chen, S.; Mulgrew, B.; Grant, P.M. A clustering technique for digital communications channel equalization using radial basis function networks. IEEE Trans. Neural Netw. 1993, 4, 570–590. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Yang, Y.; Shi, M. Chemometrics in Tandem with Hyperspectral Imaging for Detecting Authentication of Raw and Cooked Mutton Rolls. Foods 2021, 10, 2127. [Google Scholar] [CrossRef]
- Li, X.; Jiang, H.; Jiang, X.; Shi, M. Identification of Geographical Origin of Chinese Chestnuts Using Hyperspectral Imaging with 1D-CNN Algorithm. Agriculture 2021, 11, 1274. [Google Scholar] [CrossRef]
- Fath, A.H.; Madanifar, F.; Abbasi, M. Implementation of multilayer perceptron (MLP) and radial basis function (RBF) neural networks to predict solution gas-oil ratio of crude oil systems. Petroleum 2020, 6, 80–91. [Google Scholar] [CrossRef]
- Deng, Y.; Zhou, X.; Shen, J.; Xiao, G.; Hong, H.; Lin, H.; Wu, F.; Liao, B.Q. New methods based on back propagation (BP) and radial basis function (RBF) artificial neural networks (ANNs) for predicting the occurrence of haloketones in tap water. Sci. Total Environ. 2021, 772, 145534. [Google Scholar] [CrossRef]
- Aqdam, H.R.; Ettefagh, M.M.; Hassannejad, R. Health monitoring of mooring lines in floating structures using artificial neural networks. Ocean Eng. 2018, 164, 284–297. [Google Scholar] [CrossRef]
- Arenas, M.G.; Parras-Gutierrez, E.; Rivas, V.M.; Castillo, P.A.; del Jesus, M.J.; Merelo, J.J. Parallelizing the Design of Radial Basis Function Neural Networks by Means of Evolutionary Meta-algorithms. In Proceedings of the Bio-Inspired Systems: Computational and Ambient Intelligence, IWANN 2009, Salamanca, Spain, 10–12 June 2009; Cabestany, J., Sandoval, F., Prieto, A., Corchado, J.M., Eds.; Lecture Notes in Computer Science. Springer: Berlin/Heidelberg, Germany, 2009; Volume 5517. [Google Scholar]
- Brandstetter, A.; Artusi, A. Radial Basis Function Networks GPU-Based Implementation. IEEE Trans. Neural Netw. 2008, 19, 2150–2154. [Google Scholar] [CrossRef] [Green Version]
- Kuncheva, L.I. Initializing of an RBF network by a genetic algorithm. Neurocomputing 1997, 14, 273–288. [Google Scholar] [CrossRef]
- Kubat, M. Decision trees can initialize radial-basis function networks. IEEE Trans. Neural Netw. 1998, 9, 813–821. [Google Scholar] [CrossRef]
- Franco, D.G.B.; Steiner, M.T.A. New Strategies for Initialization and Training of Radial Basis Function Neural Networks. IEEE Lat. Am. Trans. 2017, 15, 1182–1188. [Google Scholar] [CrossRef]
- Ricci, E.; Perfetti, R. Improved pruning strategy for radial basis function networks with dynamic decay adjustment. Neurocomputing 2006, 69, 1728–1732. [Google Scholar] [CrossRef]
- Bortman, M.; Aladjem, M. A Growing and Pruning Method for Radial Basis Function Networks. IEEE Trans. Neural Netw. 2009, 20, 1039–1045. [Google Scholar] [CrossRef]
- Huang, G.-B.; Saratchandran, P.; Sundararajan, N. A generalized growing and pruning RBF (GGAP-RBF) neural network for function approximation. IEEE Trans. Neural Netw. 2005, 16, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Y.; Qin, Z.; Jia, J. A PSO-Based Subtractive Clustering Technique for Designing RBF Neural Networks. In Proceedings of the 2008 IEEE Congress on Evolutionary Computation (IEEE World Congress on Computational Intelligence), Hong Kong, 1–6 June 2008; pp. 2047–2052. [Google Scholar]
- Esmaeili, A.; Mozayani, N. Adjusting the parameters of radial basis function networks using Particle Swarm Optimization. In Proceedings of the 2009 IEEE International Conference on Computational Intelligence for Measurement Systems and Applications, Hong Kong, 11–13 May 2009; pp. 179–181. [Google Scholar]
- O’Hora, B.; Perera, J.; Brabazon, A. Designing Radial Basis Function Networks for Classification Using Differential Evolution. In Proceedings of the 2006 IEEE International Joint Conference on Neural Network Proceedings, Vancouver, BC, USA, 16–21 July 2006; pp. 2932–2937. [Google Scholar]
- MacQueen, J. Some methods for classification and analysis of multivariate observations. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Davis Davis, CA, USA, 27 December 1965–7 January 1966; Volume 1, pp. 281–297. [Google Scholar]
- Kaelo, P.; Ali, M.M. Integrated crossover rules in real coded genetic algorithms. Eur. J. Oper. 2007, 176, 60–76. [Google Scholar] [CrossRef]
- Tsoulos, I.G. Modifications of real code genetic algorithm for global optimization. Appl. Math. Comput. 2008, 203, 598–607. [Google Scholar] [CrossRef]
- Sanderson, C.; Curtin, R. Armadillo: A template-based C++ library for linear algebra. J. Open Source Softw. 2016, 1, 26. [Google Scholar] [CrossRef]
- Chandra, R.; Dagum, L.; Kohr, D.; Maydan, D.; McDonald, J.; Menon, R. Parallel Programming in OpenMP; Morgan Kaufmann Publishers Inc.: San Francisco, CA, USA, 2001. [Google Scholar]
- Alcalá-Fdez, J.; Fernandez, A.; Luengo, J.; Derrac, J.; García, S.; Sánchez, L.; Herrera, F. KEEL Data-Mining Software Tool: Data Set Repository, Integration of Algorithms and Experimental Analysis Framework. J. Mult.-Valued Log. Soft Comput. 2011, 17, 255–287. [Google Scholar]
- Tzimourta, K.D.; Tsoulos, I.; Bilero, T.; Tzallas, A.T.; Tsipouras, M.G.; Giannakeas, N. Direct Assessment of Alcohol Consumption in Mental State Using Brain Computer Interfaces and Grammatical Evolution. Inventions 2018, 3, 51. [Google Scholar] [CrossRef] [Green Version]
- Weiss, S.M.; Kulikowski, C.A. Computer Systems That Learn: Classification and Prediction Methods from Statistics, Neural Nets, Machine Learning, and Expert Systems; Morgan Kaufmann Publishers Inc.: San Francisco, CA, USA, 1991. [Google Scholar]
- Quinlan, J.R. Simplifying Decision Trees. Int. Man–Mach. Stud. 1987, 27, 221–234. [Google Scholar] [CrossRef] [Green Version]
- Shultz, T.; Mareschal, D.; Schmidt, W. Modeling Cognitive Development on Balance Scale Phenomena. Mach. Learn. 1994, 16, 59–88. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.H.; Jiang, Y. NeC4.5: Neural ensemble based C4.5. IEEE Trans. Knowl. Data Eng. 2004, 16, 770–773. [Google Scholar] [CrossRef]
- Setiono, R.; Leow, W.K. FERNN: An Algorithm for Fast Extraction of Rules from Neural Networks. Appl. Intell. 2000, 12, 15–25. [Google Scholar] [CrossRef]
- Demiroz, G.; Govenir, H.A.; Ilter, N. Learning Differential Diagnosis of Eryhemato-Squamous Diseases using Voting Feature Intervals. Artif. Intell. Med. 1998, 13, 147–165. [Google Scholar]
- Valentini, G.; Masulli, F. NEURObjects: An object-oriented library for neural network development. Neurocomputing 2002, 48, 623–646. [Google Scholar] [CrossRef] [Green Version]
- Denoeux, T. A neural network classifier based on Dempster-Shafer theory. IEEE Trans. Syst. Man Cybern. Part A Syst. Hum. 2000, 30, 131–150. [Google Scholar] [CrossRef] [Green Version]
- Hayes-Roth, B.; Hayes-Roth, F. Concept learning and the recognition and classification of exemplars. J. Verbal Learn. Verbal Behav. 1977, 16, 321–338. [Google Scholar] [CrossRef]
- Kononenko, I.; Šimec, E.; Robnik-Šikonja, M. Overcoming the Myopia of Inductive Learning Algorithms with RELIEFF. Appl. Intell. 1997, 7, 39–55. [Google Scholar] [CrossRef]
- French, R.M.; Chater, N. Using noise to compute error surfaces in connectionist networks: A novel means of reducing catastrophic forgetting. Neural Comput. 2002, 14, 1755–1769. [Google Scholar] [CrossRef] [PubMed]
- Dy, J.G.; Brodley, C.E. Feature Selection for Unsupervised Learning. J. Mach. Learn. Res. 2004, 5, 845–889. [Google Scholar]
- Perantonis, S.J.; Virvilis, V. Input Feature Extraction for Multilayered Perceptrons Using Supervised Principal Component Analysis. Neural Process. Lett. 1999, 10, 243–252. [Google Scholar] [CrossRef]
- Garcke, J.; Griebel, M. Classification with sparse grids using simplicial basis functions. Intell. Data Anal. 2002, 6, 483–502. [Google Scholar] [CrossRef] [Green Version]
- Elter, M.; Schulz-Wendtland, R.; Wittenberg, T. The prediction of breast cancer biopsy outcomes using two CAD approaches that both emphasize an intelligible decision process. Med. Phys. 2007, 34, 4164–4172. [Google Scholar] [CrossRef]
- Little, M.A.; McSharry, P.E.; Hunter, E.J.; Spielman, J.; Ramig, L.O. Suitability of dysphonia measurements for telemonitoring of Parkinson’s disease. IEEE Trans. Biomed. Eng. 2009, 56, 1015. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.W.; Everhart, J.E.; Dickson, W.C.; Knowler, W.C.; Johannes, R.S. Using the ADAP learning algorithm to forecast the onset of diabetes mellitus. In Proceedings of the Symposium on Computer Applications and Medical Care, Washington, DC, USA, 6–9 November 1988; pp. 261–265. [Google Scholar]
- Lucas, D.D.; Klein, R.; Tannahill, J.; Ivanova, D.; Brandon, S.; Domyancic, D.; Zhang, Y. Failure analysis of parameter-induced simulation crashes in climate models. Geosci. Model Dev. 2013, 6, 1157–1171. [Google Scholar] [CrossRef] [Green Version]
- Giannakeas, N.; Tsipouras, M.G.; Tzallas, A.T.; Kyriakidi, K.; Tsianou, Z.E.; Manousou, P.; Hall, A.; Karvounis, E.C.; Tsianos, V.; Tsianos, E. A clustering based method for collagen proportional area extraction in liver biopsy images. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Milan, Italy, 25–29 August 2015; pp. 3097–3100. [Google Scholar]
- Breiman, L. Bias, Variance and Arcing Classifiers; Tec. Report 460; Statistics Department, University of California: Berkeley, CA, USA, 1996. [Google Scholar]
- Hastie, T.; Tibshirani, R. Non-parametric logistic and proportional odds regression. J. R. Stat. Soc. Ser. C (Appl. Stat.) 1987, 36, 260–276. [Google Scholar] [CrossRef]
- Dash, M.; Liu, H.; Scheuermann, P.; Tan, K.L. Fast hierarchical clustering and its validation. Data Knowl. Eng. 2003, 44, 109–138. [Google Scholar] [CrossRef]
- Gorman, R.P.; Sejnowski, T.J. Analysis of Hidden Units in a Layered Network Trained to Classify Sonar Targets. Neural Netw. 1988, 1, 75–89. [Google Scholar] [CrossRef]
- Lim, T.S.; Loh, W.Y. A Comparison of Prediction Accuracy, Complexity, and Training Time of Thirty-Three Old and New Classification Algorithms. Mach. Learn. 2000, 40, 203–228. [Google Scholar] [CrossRef]
- Quinlan, J.R.; Compton, P.J.; Horn, K.A.; Lazurus, L. Inductive knowledge acquisition: A case study. In Proceedings of the Second Australian Conference on Applications of Expert Systems, Sydney, Australia, 14–16 May 1986. [Google Scholar]
- Wolberg, W.H.; Mangasarian, O.L. Multisurface method of pattern separation for medical diagnosis applied to breast cytology. Proc. Natl. Acad. Sci. USA 1990, 87, 9193–9196. [Google Scholar] [CrossRef] [Green Version]
- Raymer, M.; Doom, T.E.; Kuhn, L.A.; Punch, W.F. Knowledge discovery in medical and biological datasets using a hybrid Bayes classifier/evolutionary algorithm. IEEE Trans. Syst. Man Cybern. Part B Cybern. 2003, 33, 802–813. [Google Scholar] [CrossRef]
- Zhong, P.; Fukushima, M. Regularized nonsmooth Newton method for multi-class support vector machines. Optim. Methods Softw. 2007, 22, 225–236. [Google Scholar] [CrossRef]
- Andrzejak, R.G.; Lehnertz, K.; Mormann, F.; Rieke, C.; David, P.; Elger, C.E. Indications of nonlinear deterministic and finite-dimensional structures in time series of brain electrical activity: Dependence on recording region and brain state. Phys. Rev. E 2001, 64, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Koivisto, M.; Sood, K. Exact Bayesian Structure Discovery in Bayesian Networks. J. Mach. Learn. Res. 2004, 5, 549–573. [Google Scholar]
- Nash, W.J.; Sellers, T.L.; Talbot, S.R.; Cawthor, A.J.; Ford, W.B. The Population Biology of Abalone (_Haliotis_ species) in Tasmania. I. Blacklip Abalone (_H. rubra_) from the North Coast and Islands of Bass Strait; Technical Report No. 48; Sea Fisheries Division-Department of Primary Industry and Fisheries: Hobart, Australia, 1994; ISSN 1034-3288. [Google Scholar]
- Brooks, T.F.; Pope, D.S.; Marcolini, A.M. Airfoil Self-Noise and Prediction; Technical Report, NASA RP-1218; NASA: Hampton, VA, USA, 1989. [Google Scholar]
- Simonoff, J.S. Analyzing Categorical Data; Springer: New York, NY, USA, 2003. [Google Scholar]
- Simonoff, J.S. Smooting Methods in Statistics; Springer: New York, NY, USA, 1996. [Google Scholar]
- Yeh, I.C. Modeling of strength of high performance concrete using artificial neural networks. Cem. Concr. Res. 1998, 28, 1797–1808. [Google Scholar] [CrossRef]
- Harrison, D.; Rubinfeld, D.L. Hedonic prices and the demand for clean air. J. Environ. Econ. Manag. 1978, 5, 81–102. [Google Scholar] [CrossRef] [Green Version]
- Mackowiak, P.A.; Wasserman, S.S.; Levine, M.M. A critical appraisal of 98.6 degrees f, the upper limit of the normal body temperature, and other legacies of Carl Reinhold August Wunderlich. J. Am. Med. Assoc. 1992, 268, 1578–1580. [Google Scholar] [CrossRef]
- Ding, S.; Xu, L.; Su, C.; Jin, F. An optimizing method of RBF neural network based on genetic algorithm. Neural Comput. Appl. 2012, 21, 333–336. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).