Featured Application
The clustering implementation being presented can be used to discover clusters and identify outliers in a dataset. This implementation provides a fast prediction feature that makes it a compelling choice for applications, such as a streaming clustering service.
Abstract
An implementation of the HDBSCAN* clustering algorithm, Tribuo Hdbscan, is presented in this work. The implementation is developed as a new feature of the Java machine learning library Tribuo. This implementation leverages concurrency and achieves better performance than the reference Java implementation. Tribuo Hdbscan provides prediction functionality, which is a novel technique to make fast predictions for unseen data points using an HDBSCAN* clustering model. Tribuo Hdbscan cluster results and performance measurements are also compared with the state-of-the-art HDBSCAN* implementation, the Python module hdbscan.
1. Introduction
Cluster analysis is the task of arranging a set of similar data points into groups. It is an unsupervised machine learning task that can discover patterns or identify related groups from a dataset. There are many different algorithms proposed for cluster analysis [1,2]. The K-Means algorithm is commonly used for clustering since it is fast and easy to understand. However, there are some potential problems with this algorithm. First, the number of clusters, or partitions, need to be provided as input to the algorithm. The number of clusters that may be present in a new dataset is, however, not always known. Another problem is that K-means is more of a partitioning algorithm than a clustering algorithm, since it partitions all the data into groups by minimizing distances between data points. These problems are also exist in other clustering algorithms. For example, the affinity propagation and spectral clustering algorithms also partition all the data into groups, so even outliers or noisy data points are included in the clusters. Another well-known algorithm is agglomerative clustering, which like K-Means, requires the number of clusters to be provided as input to the algorithm when a threshold parameter is not used.
The HDBSCAN* clustering algorithm [3] is a density-based algorithm. Unlike K-Means, it does not require that every data point is assigned to a cluster, since it identifies dense clusters. Points not assigned to a cluster are considered as outliers, or noise. An algorithm that can effectively find distinct groups in a dataset and identify outliers is a valuable technique. There is an established Python implementation of the HDBSCAN* algorithm [4] which is included as a scikit-learn compatible project. It is well known that Python is a very popular language for data mining and machine learning tasks, such as clustering. Java, on the other hand, is not as popular for these types of tasks, even though there are numerous Java enterprise applications running in production currently. Machine learning libraries offer the benefit of providing a consistent API to variety of learning algorithms and utilities. Tribuo [5] is a recent open-sourced Java machine learning library. Tribuo is proven to be robust and performant, but currently it only supports the K-Means clustering algorithm. Therefore, it is valuable to build an implementation of the HDBSCAN* algorithm in the Tribuo library.
This work offers a new and optimized implementation of the HDBSCAN* algorithm in Java. A full tutorial about this implementation is available at [6]. The proposed implementation leverages Tribuo’s infrastructure to provide a train/predict interface consistent with the library’s other learning algorithms and features. This gives the ability to train models which discover clusters and identify outliers. This work introduces a novel prediction technique which uses a trained model to make predictions for unseen data points natively in Java. Functionality such as this provides the foundation for applications, such as a streaming clustering service. Furthermore, the methodology presented in this work illustrates a generic development process which can be reused for the implementation of an existing learning algorithm in an existing machine learning library.
The remainder of this article is structured as follows. A background and literature review are presented in Section 2. Section 3 provides a description of the methodology of this work. Section 4 discusses the results of the comparisons between the HDBSCAN* implementations. Finally, Section 5 presents a summary discussion of the work.
2. Background and Literature Review
2.1. DBSCAN
One of the first, and perhaps the most well-known density-based clustering algorithm is DBSCAN [7]. It was first proposed in 1996 and remains a relevant technique for clustering tasks today. DBSCAN stands for density-based spatial clustering of applications with noise. The algorithm requires a rather obscure distance parameter as input, which is used as a threshold for determining the data points that will be assigned to clusters, and those marked as outliers. There is no obvious intuition for establishing the best value for this parameter. As the authors of [3] point out, DBSCAN may have issues when the density of individual clusters in a dataset varies.
2.2. HDBSCAN*
More recently, the HDBSCAN* algorithm [3] was introduced. It stands for Hierarchical DBSCAN*. The asterisk suffix indicates an improvement the same authors of [3] make to DBSCAN. These authors extend their work in [8] by introducing a complete framework for cluster analysis and outlier detection which includes an enhanced explanation of the HDBSCAN* algorithm. HDBSCAN* improves on DBSCAN by establishing a hierarchical representation of the clusters. The hierarchy derived from the execution of the algorithm can be used very effectively for cluster extraction and outlier detection. HDBSCAN* overcomes the limitations of DBSCAN by identifying clusters of any density. Although there are several benefits provided by the HDBSCAN* algorithm, there is one drawback that should be considered. The algorithm has an overall asymptotic complexity of . Furthermore, there are multiple sub-steps of the algorithm, which themselves have a complexity of . This complexity can have a significant impact on the algorithm execution time for large datasets. There are implementations of the K-Means algorithm mentioned earlier, which have a complexity of . Table 1 summarizes the strengths and weaknesses of the DBSCAN and HDBSCAN* algorithms.

Table 1.
Summary of the strengths and weaknesses of DBSCAN and HDBSCAN*.
2.2.1. Other Methods
There are other clustering algorithms which can be used to perform clustering when the number of clusters are unknown. Perhaps the most similar to DBSCAN and HDBSCAN* is OPTICS, which stands for ordering points to identify clustering structure [9]. OPTICS can be used for cluster analysis, although the original algorithm does not produce an explicit clustering of the data. Instead, it produces an ordered representation of the data density-based clustering structure. It overcomes the limitation of DBSCAN by identifying cluster structures of varying density, but is not as comprehensive as HDBSCAN* to identify clusters while excluding outliers.
Hierarchical clustering methods are another form of clustering analysis which can work without the need to specify the number of clusters. These methods construct clusters by recursively partitioning the data points in either a top–down or bottom–up approach [10]. Divisive clustering starts with a single cluster containing all points and is divided into sub-clusters based on some criteria, which are successively divided into further sub-clusters. There are different strategies to determine when the process should be terminated. Agglomerative hierarchical clustering starts with each data point in its own cluster and merges clusters sharing some criteria, and continues to successively merge clusters. Again, there are different strategies which can be employed to terminate the process. Hierarchical clustering methods can provide high interpretability since the results can be presented in a dendrogram, and the choices for cluster splits or merges are well defined. These methods do not provide any mechanism for identifying outliers. They also tend to have high asymptotic complexity and large memory requirements.
In the previous section, it was mentioned that partitioning algorithms, such as K-Means require the number of clusters, k, to be defined as input to the algorithm. In some cases, the value for k is unknown. There are techniques which can be used to determine the optimal number of clusters from a dataset, such as the frequently mentioned elbow method. The elbow method is somewhat subjective, and, therefore, unreliable. There are also more analytical techniques that leverage silhouette analysis to estimate the optimal number of clusters from a dataset. One such example is an algorithm called k-SCC [11]. Combining the k-SCC algorithm with K-Means effectively achieves a natural discovery of clusters from a dataset. However, this combination is not able to distinguish outlier points from the clusters as can be done using HDBSCAN*.
2.2.2. The HDBSCAN* Algorithm
This section describes the HDBSCAN* algorithm presented in [8]. There are two required input parameters to this algorithm in addition to the dataset targeted for clustering. The first parameter is the minimum number of points s to be used in the distance calculation, as it will be shown later in this paper. The second required parameter is the minimum cluster size p that defines a lower bound on the number of data points required to form a cluster. The dataset is a set of homogeneous points where a point is a record of numerical features. Figure 1 shows a simple dataset of 39 two-dimensional points that will be used as an example throughout this section.
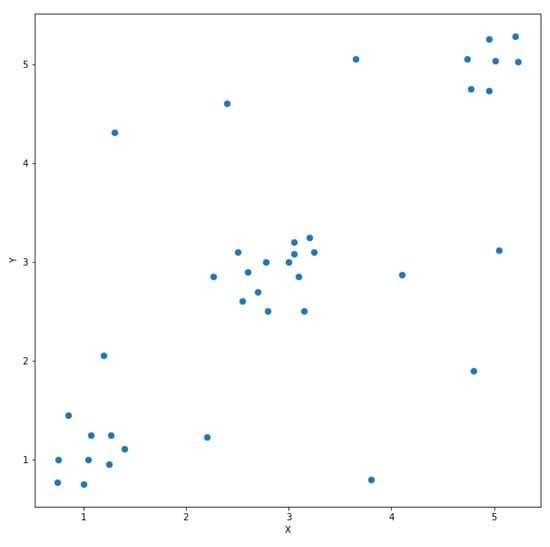
Figure 1.
A simple dataset of two-dimensional points.
The first step in the algorithm is to compute the core distances for each point in the dataset. The core distance of a point is the distance to the kth nearest neighbor. The value of k includes the point itself. This is a technique to obtain an approximate density for each point. Figure 2 shows the core distances for the points and for , from the simple dataset. Note that both points’ 5th nearest neighbor is common. The point has a smaller core distance value than the point because its neighboring points are closer, which implies it has a higher approximate density.
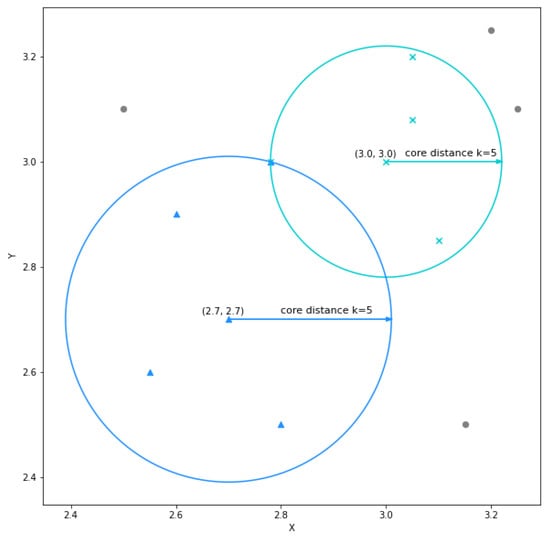
Figure 2.
The core distances for the points and for , using a zoomed in view on a particular area of the simple dataset.
Using the core distances, a new distance metric is computed which is called the mutual reachability distance. The mutual reachability distance between two points x and y, is the maximum value of: the core distance of x, the core distance of y, or the distance between x and y. The points and shown in Figure 2 have a mutual reachability distance equal to the distance between the two points. Concretely, for , the core distance of is , the core distance of is and the distance between the two points is . Therefore, the mutual reachability distance between these points is .
Next, the mutual reachability distances between the points can be used to establish a weighted graph, where the data points are vertices and an edge between any two points has the weight of the mutual reachability distance of those points. This complete graph is only a conceptual artifact in the algorithm, since it is the graph’s minimum spanning tree which needs to be computed. A minimum spanning tree is a spanning tree whose sum of edge weights is as small as possible. That is, it has all vertices connected with a subset of the edges from the complete graph, without any cycles and with the minimum possible total edge weight. The resulting minimum spanning tree is modified by adding a self-edge to each vertex with the point’s core distance as the weight. This gives a graph called the extended minimum spanning tree. Figure 3 shows the minimum spanning tree of the simple dataset.
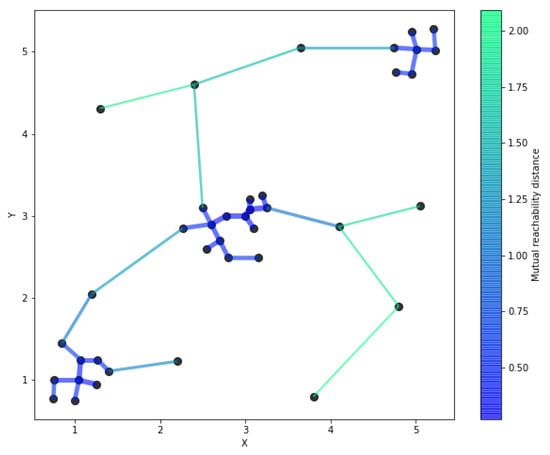
Figure 3.
The minimum spanning tree of the simple dataset. An extended minimum spanning tree would contain self-edges for each point, with the weight of the self-edge being the point’s core distance value.
Now, the graph can be used to build the HDBSCAN* hierarchy. To begin with, there is one cluster label, and all the points are assigned to this cluster. This cluster is added to a cluster list. The graph is then sorted by edge weight in ascending order. Starting from the bottom of the graph, edges are iteratively removed from the extended minimum spanning tree. Edges with equal weights must be removed simultaneously. The weight value of the edge(s) being removed is used to denote the current hierarchical level. As an edge is removed, the cluster containing the removed edge is explored. Clusters that have become disconnected and contain fewer points than s (minimum cluster size) are all assigned with the noise label. A cluster that has become disconnected, but has more than s points, is assigned a new cluster label. This is called a cluster split. Additionally, the new cluster is added to the list of clusters. A new hierarchy level is created when an edge removal has resulted in new clusters due to cluster splits. By the end of the process, in the last level of the hierarchy, all the points in the dataset will have been assigned to noise. The hierarchy produced by this process is the HDBSCAN* hierarchy. Figure 4 shows a table representation of the HDBSCAN* hierarchy of the simple dataset. Each column of the table represents a data point, and the rows contain the cluster assignments as the algorithm runs. In the top row, all the points are assigned to the same, single cluster. In the second row, some points are set to zero because they have been split off and are marked as outliers. In the third row, there are now two distinct clusters and one additional outlier. By the last row, all the points are assigned as outliers or noise points. During the construction of the HDBSCAN* hierarchy, a list of the clusters is also maintained, where each cluster holds a reference to its parent.

Figure 4.
A tablular representation of the HDBSCAN* hierarchy of the simple dataset.
With the HDBSCAN* hierarchy and the list of clusters computed, the next step is to identify the prominent clusters from this hierarchy. To proceed, a new measure needs to be established that can be used towards determining the stability of a cluster. This is called lambda, such that = 1/edge weight. Further, for a cluster we also define values and to be the lambda value when a new cluster was created from a cluster split, and the lambda value when that same cluster was itself split, respectively. For each point in a cluster, we can define the value as the lambda value at which that point dropped out of the cluster. The stability for a cluster can be computed, as shown in Equation (1).
The stability needs to be propagated through the clusters. Leaf clusters are clusters with no children, and they can be identified from the list of clusters. Starting with these leaf clusters, traverse up using the reference to the cluster’s parent. Leaf clusters always propagate their stability to their parents and add themselves as a propagated descendent in the parent cluster. For non-leaf clusters one of two things will occur. If the cluster being processed has a higher stability than the cumulative stability of its descendants, it alone will be propagated to the parent cluster. Otherwise, the cumulative stability of all the current cluster’s descendants will be propagated to the parent cluster. Clearly, no propagation occurs for the root cluster since it has no parent. When this process is finished, the root cluster will contain the references to descendent clusters with the highest stabilities. The clusters with the highest stabilities are the most prominent clusters. Using the HDBSCAN* hierarchy together with the details of the most prominent clusters, the list of cluster assignments for each data point can be quickly generated. This is the most significant artifact generated as output of the HDBSCAN* algorithm described in [8]. The prominent clusters identified using the simple dataset are shown in Figure 5. Three clusters are identified, and the five yellow points are determined to be outliers.
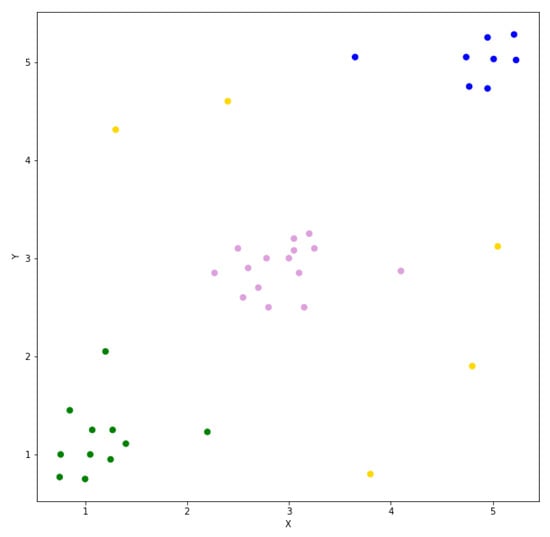
Figure 5.
The clustering result produced by HDBSCAN* using the simple dataset.
Another important artifact, that can be obtained from the execution of the HDBSCAN* algorithm, are the outlier scores for each data point. The literature [8] names this as a point’s GLOSH which stands for Global–Local Outlier Score from Hierarchies. Fortunately, computing a data point’s GLOSH is not too complex, but requires that some additional bookkeeping was done during the construction of the HDBSCAN* hierarchy. Earlier in this section, it was mentioned that the weight value of the edge being removed is used to denote the current hierarchical level. This weight value, and the last cluster label must be noted for every point, at the moment the point is marked as noise, or assigned the noise label, to use the same wording as before. A data point, x, has the values and , which are: the weight value of the point right before it was marked as noise, and the lowest propagated child death level from its last labeled cluster, respectively. The value of the outlier score for a point x can be obtained using Equation (2).
This section is summarized by Algorithm 1 which shows the main steps of the HDBSCAN* algorithm.
| Algorithm 1: HDBSCAN* main steps. |
|
Compute the core distance for the k nearest neighbors for all points in the dataset; Compute the extended minimum spanning tree from a weighted graph, where the mutual reachability distances are the edges; Build the HDBSCAN* hierarchy from the extended minimum spanning tree; Find the prominent clusters from the hierarchy; Calculate the outlier scores; |
2.3. HDBSCAN* Implementations
In this paper, two existing implementations of HDBSCAN* are being reviewed: Python and Java implementations.
2.3.1. Python HDBSCAN* Implementation
As mentioned in Section 1, there is a well-established Python implementation of the HDBSCAN* algorithm called hdbscan [4]. Researchers in [12] indicate that this is currently the best performing implementation available. In addition, the current version of this Python module provides some innovative features beyond what is provided by the framework described in [8]. One example is the ability to predict the cluster assignment for unseen data points using a trained model. Another example of an innovative feature from hdbscan is the soft clustering functionality it provides. The version of hdbscan being reviewed and used in this work is 0.8.27 with Python 3.9.1.
To understand the details of this particular implementation of HDBSCAN*, the source code is reviewed. The hdbscan project has a dependency on scikit-learn and uses many of its features. It uses the KDTree and BallTree classes which are nearest neighbor algorithms. The main class, HDBSCAN, inherits from scikit-learn’s BaseEstimator and ClusterMixin classes. This provides a lot of functionality out of the box and gives the module a standardized API. This code also uses numpy extensively and takes advantage of its optimized numerical array operations while providing support for multithreading. Leveraging proven libraries, such as scikit-learn and numpy, certainly contribute to the good performance of this implementation. In addition to this, the hdbscan module has implemented several variations of the algorithms which calculate the core distances and construct the minimum spanning tree. Considering all these points, it is easy to understand why this HDBSCAN* implementation is the best performing implementation available. Furthermore, this module is being actively maintained and improved by the development community. The hdbscan Python module will be consulted as a reference throughout this paper.
2.3.2. Java HDBSCAN* Implementation
There is an existing Java Implementation of the HDBSCAN* algorithm. It was developed as part of the framework for cluster analysis and outlier detection, based on the HDBSCAN* algorithm, introduced in [8]. The code for this implementation is carefully reviewed and several tests are performed using different input datasets. In general, the code is well-written, and logically organized. This Java implementation of the HDBSCAN* algorithm will be referred to as the reference implementation throughout this work. Interestingly, this same reference implementation is also used as the reference implementation by the authors of [12] and in the documentation of the hdbscan Python module’s GitHub repository. The reference implementation is a good basis for a new Java implementation. However, there are some changes needed and issues which should be addressed. The most significant among them are as follows:
- There are cases where important results are written to disk and subsequently read in again using customized file offset logic. Machine learning libraries do not commonly require access to the file system to persist results. Further, I/O can lead to performance issues;
- No unit tests. The reference implementation has no self-contained unit tests to verify the algorithm;
- There is some constraints functionality present in the reference implementation which adds complexity to several steps of the algorithm. This feature is not required;
- None of the logic leverages parallelization. There are several blocks of logic with high asymptotic complexity which could cause performance issues.
Beyond these issues, it should be noted that this reference implementation is not being actively maintained and can not be easily integrated with an existing Java application. This highlights the value of developing a new HDBSCAN* implementation as a feature of an existing machine learning library for Java.
2.4. Tribuo Machine Learning Library
Tribuo [5] was developed by the Oracle Labs Machine Learning Research Group, a team within Oracle Corporation. They realized there is need for a single node machine learning framework on the JVM. Initially it was only available internally to Oracle but has been recently open sourced to help build the machine learning ecosystem on the Java platform. The version of Tribuo being reviewed, and at which it has been forked, is 4.2.0-SNAPSHOT. This version is currently compiled with Java 8.
Some initial work has been done to evaluate the Tribuo library. When comparing machine learning tasks, such as classification, regression, and clustering, results from these experiments [13] show that Tribuo and scikit-learn achieve very similar performance measurements. This indicates that Tribuo is an interesting subject of further study.
Tribuo Project Source Code
To implement a new algorithm in the Tribuo project, a detailed review of the source code is conducted. Overall, the code is very well organized, and has a very modular design. The first thing to mention is that Tribuo uses a maven build system. At the root of the project there are Classification, Regression, and Clustering maven modules, among others. The Clustering module is the parent of the KMeans module. It seems logical that a new clustering algorithm should be added here, adjacent to the KMeans module.
Looking more closely at the source code shows there are several frequently used classes which provide interesting functionality. There is a DenseVector class which is backed by an array of doubles. This class provides methods for many vector operations, such as addition, subtraction, and dot product. Most notably, the class provides methods which can be used to compute the distance between two vectors, including Euclidean and cosine distances. There is also a SparseVector class which implements the same interface as DenseVector but is optimized for, as the name suggests, sparse array data.
In general, to perform a machine learning task with Tribuo, there is a standard workflow that should be followed. At a high-level, the first step is to instantiate the appropriate Trainer object. Calling a trainer’s train method (and providing a dataset), returns a trained model to the user. For example, calling the train method on an instantiated KMeansTrainer object returns a KMeansModel object. A model may have attributes or methods specific to its task. Continuing with the example, a KMeansModel has a getCentroids method which returns the points which are the cluster centroids. Every model implements the Model interface, and one of the methods in this interface is predict. This indicates that every model should provide some functionality to make predictions on unseen data points.
The Tribuo K-Means implementation leverages concurrency to perform portions of the algorithm in parallel. Specifically, Java parallel streams are used, and are assigned to a custom ForkJoinPool that controls the number of threads in the thread pool. This concurrency technique is considered for the new HDBSCAN* implementation.
Tribuo provides a feature called Provenance which is ubiquitous throughout the library’s code. This feature captures the details on how any model, dataset, and evaluation were created in Tribuo. The main benefit it offers is that any of these objects can be regenerated from scratch, which is valuable for reproducibility. Implementing a new feature in Tribuo requires some additional considerations to cooperate with the Provenance system.
3. Materials and Methods
This section presents the methodology followed in this work.
3.1. Study the HDBSCAN* Algorithm
3.1.1. The Theoretical Approach
The first step of the methodology is to thoroughly understand the HDBSCAN* algorithm from a theoretical point of view. To achieve this, the pseudo-code for the algorithms, and their accompanying explanations, described in [8] need to be carefully studied. This is a non-trivial task.
3.1.2. The Practical Approach
Next, the source code for the two existing implementations of the HDBSCAN* algorithms are examined. The best way to do this is by opening the code in an IDE such as IntelliJ IDEA. Using an integrated development environment greatly simplifies navigation, and provides facilities to identify call hierarchies, and quickly explore different flows through the code. Using the debugger to step through a live execution of the code is extremely helpful. The ability to inspect variables which are complex data structures and confirm conditional branch logic and loop termination conditions provides valuable insight.
The Python implementation of the HDBSCAN* algorithm is widely used, so it is important to focus on the features it provides, and the way it is used. This includes observing both the required and optional parameters the algorithm accepts. The outputs generated by the algorithm and their format should also be noted. The machine learning community is comfortable with the hdbscan module and its API, therefore, a new Java implementation should not be significantly different.
The reference Java implementation is analyzed more critically. There is a potential that some of its code can be reused in the new implementation being proposed. This requires a very rigorous examination of the existing logic, to benefit from its strengths and overcome its weaknesses. In Section 2, some issues with the reference implementation were described, which have been identified during this review. These issues will be addressed at various steps throughout the remainder of the process.
3.2. Initial Development Phase
The next step in the methodology of this work is to perform some code changes directly to a copy of the reference implementation’s source code. The objective of this step is to reduce the reference implementation down to a minimal HDBSCAN* algorithm. The major tasks will be described in this section. During the course of performing these tasks, several other minor code changes are made.
3.2.1. Add Unit Tests
The HDBSCAN* algorithm consists of complex logic. It follows that a test-driven development (TDD) approach is needed to facilitate an optimal development process. Therefore, the first thing to do is develop some unit tests. The scope of these tests is broad, that is, the algorithm in its entirety is tested as a unit. Currently there is no requirement to test individual sub-components of the algorithm, for example the extended minimum spanning tree, or the outlier score calculations. This is because they are not individual units and will not be used by any other code. If, in the future, a sub-component was going to be called by other code, for a different purpose, that would be the appropriate time to add finer grained tests.
Adding initial unit tests is straightforward. Datasets can be generated using scikit-learn [14], to produce isotropic Gaussian blobs, which are simply sets of points normally distributed around a defined number of centroids. This makes it easy to vary the number of samples, the number of features, and the number of centroids. The generated datasets are written to .csv files. The reference implementation is executed using a .csv file as input, and the resulting outputs are used to make assertions in the unit tests. Several different unit tests are created each with a unique dataset. Once unit tests are established, changes can be made safely since they can be verified quickly by executing the tests.
3.2.2. Remove Unneeded File Input and Output
The reference implementation currently produces several output artifacts during the execution of the algorithm. In one case, there are important results written to disk which are subsequently read in again using customized file offset logic. The output artifacts are not required by the new implementation being presented. Rather than persist computed data structures to disk which are referenced again, efficient in-memory representations are developed. The appropriate structure in this case is a Map with an Integer key, and an array of integers as the value. The Integer map key is the level of the hierarchy. A value of zero indicates the top level of the hierarchy. The map value is an array of integers that represents the current cluster labels at the current level of the hierarchy. As mentioned above, there is a list of cluster objects maintained for the duration of the algorithm. Each cluster has a hierarchy level attribute, which corresponds to the key in the hierarchy map structure. This level attribute indicates the hierarchy level when the cluster was first created.
Additionally, the reference implementation contains code that is generating custom output files to be used with a separate visualization application included with the framework described in [8]. All this logic is also removed.
3.2.3. Remove Constraints Functionality
The reference implementation contains some interesting functionality which enables the algorithm to apply certain constraints to the clustering result. This constraints input provides a mechanism to instruct the algorithm about relationships defined between data points. Specifically, this could be used to indicate that a pair of points must be clustered together, or conversely, that a pair of points must be clustered separately. Although this could be useful, it is observed that a similar constraint feature is not present in the popular Python HDBSCAN* implementation, nor is it mentioned in any of the other reviewed literature. As a result, a similar feature is not planned for the new implementation, Tribuo Hdbscan. Therefore, the constraints functionality code is removed from the initial working implementation. This clean-up needs to be performed carefully, since many methods and several classes need to be changed. The removal of the constraints functionality has the benefit of reducing the logical complexity of the working implementation.
3.3. Review the Tribuo Project
Now that a working, minimal HDBSCAN* implementation has been derived from the reference implementation, there is one more step in the methodology to be completed before coding in the Tribuo project can be started. The latest version of the Tribuo project needs to be carefully analyzed to see how a new HDBSCAN* implementation will fit in. The best way to do this is by opening the code project in an IDE, such as IntelliJ IDEA. As described earlier, using an integrated development environment greatly facilitates the process of performing an initial analysis of a code base. The debugger can be used to step through live executions of the project’s unit tests. This helps to establish a thorough understanding of how some of the framework classes should be used, and how interfaces should be implemented.
The Tribuo project uses a maven build system, so it is straightforward to build the project’s artifacts locally. Local access to these jar files provides the ability to build test applications or Java notebooks to further explore the functionality of the Tribuo library. It is expected that the proposed HDBSCAN* implementation will leverage concurrency in one or more steps of the algorithm. Therefore, areas of the Tribuo code where concurrency has been implemented, are identified and analyzed as part of this review. The current version of Tribuo provides an implementation of K-Means clustering. This specific part of the project should be carefully reviewed since the new clustering algorithm being added will likely be quite similar. The results of the analysis of the Tribuo project are captured back in Section 2, in the Background and Literature Review, since they should be considered as a component of the background for this work.
3.4. Main Development Phase
This step of the methodology of this work involves coding the HDBSCAN* algorithm in the Tribuo project. The objective of this step is to have a complete and optimized implementation of the HDBSCAN* algorithm, Tribuo Hdbscan. The coding work is broken down into four major tasks which are described in this section.
3.4.1. Add the Tribuo Hdbscan Module
The first thing that needs to be completed, is to fork the Tribuo GitHub repository. With the code checked out locally, a full build should be performed to ensure that the code compiles and all the unit tests complete successfully. This also indicates that the local environment is correctly configured to work with the Tribuo project.
During the review of the Tribuo project, it was observed that there is a Clustering module present at the root of the project. A new module, Hdbscan is added under the Clustering module. The adjacent KMeans module serves as a good example to follow when creating the pom.xml file and initial Java package and class structures. Like the KMeansTrainer class implements the Trainer interface, and the KMeansModel class extends the Model class, the new HdbscanTrainer and HdbscanModel classes will do the same, respectively. Initially, only skeletons of these classes are created, without any actual HDBSCAN* logic. This task is complete when a successful build can be executed, and the hdbscan artifacts are installed to the appropriate location in the local maven repository.
3.4.2. Implement the HDBSCAN* Core Logic
This is perhaps the most significant task in this work. To implement HDBSCAN* in Tribuo, the existing code from the working, minimal HDBSCAN* implementation is used as a basis for the code being added into the skeletons of the classes prepared in the previous task. As blocks of code are added, changes are made as required. For example, the input to the algorithm is a Tribuo specific Dataset <ClusterID> object, and the data should be transformed into an array of DenseVector objects which were described in Section 2. Differences such as this require several code modifications to achieve an implementation that will compile, at the very least. The changes must also be made carefully to avoid introducing subtle bugs before the unit tests can be integrated and executed. During the process, the code being added needs to be meticulously scrutinized, and detailed notes are kept about potential issues or improvements that should be made. These types of changes should only be attempted after the unit tests are integrated. The train method of the HdbscanTrainer class is the main entry point to the algorithm and an HdbscanModel object must be returned which provides access to the cluster assignments, and outlier scores. No intermediate state can be stored in an instance of an HdbscanTrainer object. The trainer could be invoked several times, possibly in parallel, with different datasets and must return a distinct, correct model object in every case. Furthermore, it is possible that a model object could be shared among client codes. The attributes of an HdbscanModel instance need to be immutable, so copies of the cluster assignments and outlier scores must be made before being returned. Additionally, there is some code required to generate the details of the clustering which is a requirement for the Provenance feature.
Adding the unit tests, established as part of the working minimal implementation, to the new HDBSCAN* implementation in Tribuo is a critical aspect of this task. Once the code for the new implementation compiles, adding the unit tests immediately follows. Although the datasets for the unit tests already exist as .csv files, some initial effort is required to figure out how to load these as Dataset <ClusterID> objects. Recall that a Dataset <ClusterID> object is provided as input to an HdbscanTrainer instance’s train method. It is a major milestone when the first unit test in the Tribuo Hdbscan module is successfully executed which makes the same assertions as those from the working, minimal HDBSCAN* implementation. This indicates that for a specific dataset, the reference implementation and the Tribuo Hdbscan implementation produce the identical cluster assignments and outlier scores. Further comparisons between these two implementations are performed in the upcoming Compare the HDBSCAN* implementations section.
3.4.3. Develop a Novel Prediction Technique
As described in Section 2, the Tribuo library has a standard workflow that should be followed when using the API. Instantiating a Trainer object and calling the Trainer’s train method with a dataset, returns a trained model to the user. A model can be used to make predictions on unseen data points, by calling the model’s predict method. This implies that the new HdbscanModel class must also implement the predict method. However, the reference implementation does not provide any prediction functionality, and a technique for making predictions is not found in any of the reviewed literature.
The Python implementation of the HDBSCAN* algorithm called hdbscan [4], which was described earlier, provides the functionality to predict the cluster assignments and outlier scores. A careful review of the code implementing this prediction functionality is performed. The hdbscan module’s implementation relies on several computed structures not currently available in Tribuo Hdbscan, accompanied by logic specific to those data structures. Unfortunately, implementing prediction logic similar to what is done in the hdbscan Python module is not feasible at this time. To achieve the goal of providing prediction functionality as part of Tribuo Hdbscan, a novel prediction technique for HDBSCAN* clusters is being proposed. Conceptually, the technique is straight-forward. First, determine a good set of cluster exemplars which are representative of the HDBSCAN* clustering result. The use of exemplar points for clustering data is described by the authors of [15] which supports this approach. Then, for a new data point, it can be approximated with the cluster label and outlier score of its nearest exemplar point. The cluster exemplars are computed during training and are stored as an attribute of the HdbscanModel. This makes prediction calls very fast. However, how are a “good” set of cluster exemplars determined? What is the appropriate number of exemplars given a dataset and its computed clusters? An algorithm is developed which provides the answers to these questions. Algorithm 2 shows the high-level steps of the algorithm.
| Algorithm 2: Compute cluster exemplars. |
 |
The input to the algorithm is the dataset, and a list of cluster assignments. First, the cluster assignments are used to create a map, where the key of this map is the cluster label, and the value is a collection of the points assigned to this cluster label. The collection of points is sorted by a point’s outlier score. Next, the number of exemplars must be established. Let l be the size of the dataset and let c represent the number of identified clusters, for a given dataset and its corresponding HDBSCAN* result. The exemplar number formula is shown in Equation (3).
There are some important details to mention about this formula. The square root of half of l allows the number of exemplars to grow as the size of the dataset increases, but the growth slows as size of the dataset increases. Adding the value of c ensures there are always at least as many exemplars as there are cluster assignments. Lastly, the floor function is applied to return the greatest integer less than or equal to the computed value. The exemplar formula is not rigorously proven by induction or through formal methods. Instead, it is empirically shown to produce values for the exemplar number which result in accurate predictions, without growing too large, which would reduce prediction speed. It is certainly possible that there exists a different formula that provides a better balance between prediction accuracy and prediction speed. Additionally, the constant of 2 in the formula could be a configurable input parameter to the algorithm to give the user a means to influence this behavior based on their requirements.
To provide some intuition for the values produced by this formula, Table 2 shows some sample “exemplar numbers” calculated for different configurations.

Table 2.
Examples of exemplar numbers calculated for different scenarios.
With the total number of exemplars established, the map of cluster assignments is iterated. The number of exemplar points to be drawn from each cluster is proportional to the size of the dataset. A cluster with more assigned points, will have more exemplars drawn from it. Out of all the points assigned to a cluster, the points selected as exemplars are those with the lowest outlier scores. With one exception: the cluster of noise points. Recall that any outlier points are assigned to the noise cluster label. Points selected as exemplars from the noise points are those with the highest outlier score. This process determines a set of cluster exemplars which are used for predicting the cluster assignments and outlier scores for new data points. This novel technique, which uses carefully computed cluster exemplars to estimate cluster assignments, has an asymptotic complexity of . To make a prediction for a new data point, the point is compared with each cluster exemplar, and there are approximately exemplar points.
This prediction functionality is useful to approximate how a point would fit into an existing HDBSCAN* cluster model. New points do not change or contribute to the existing cluster model. A Tribuo Hdbscan clustering model used for predictions should be regularly retrained using up-to-date data to ensure the model provides the most accurate clustering representation.
3.4.4. Add Concurrency
During the review of the Java reference implementation, several blocks of logic with high asymptotic complexity were identified. For example, the first step in the algorithm to compute the core distances has a time complexity of . The second step in the algorithm is to compute the extended minimum spanning tree graph. This also takes . These findings are consistent with the descriptions the authors of [8] provide in their detailed complexity analysis of the HDBSCAN* algorithm. Rather than repeat all the details from this complexity analysis here, a more practical approach to understanding the performance of the algorithm is taken. The execution times of each major step of the algorithm are collected when training models using a variety of datasets. These are not meant to be rigorous experiments. Only a general idea of where the algorithm spends most of its time is needed at this point. In fact, the first step in the algorithm which computes the core distances consistently takes the longest out of all the steps. It is the next step that computes the extended minimum spanning tree graph that takes the second longest.
Therefore, it makes sense to focus on parallelizing these two steps of the algorithm, starting with the core distance calculations. In Java 8, there are several APIs which can be used for implementing concurrency. An initial attempt is made to use a ForkJoinPool which is designed for work that can be broken into smaller pieces recursively. This implementation does not perform well and ended up taking longer in all cases than the sequential version of the code. Next, the core distance calculations were implemented using the ExecutorService API. For datasets larger than 5000 records, executing this logic in parallel with multiple threads is always faster than the sequential implementation. For small datasets, the overhead of starting and shutting down the ExecutorService appears to degrade performance. For this reason, when the HdbscanTrainer is instantiated with the variable numThreads equal to one, this condition will be detected, and the original sequential core distance calculation will be executed. This provides a mechanism to achieve optimal performance results for smaller datasets. The use of Java’s Parallel Streams API is not explored here since some cumbersome steps would have been required to transform the existing data structures into streams-compatible datatypes.
Exact measurements comparing various core distance calculation execution times are not observed or presented here. This analysis is too fine grained. It is more important to focus on measuring the time it takes to execute the complete algorithm, since that is how the code will be used in practice. Such measurements will be presented and discussed in the following section.
The next step of this task is to parallelize the block of logic that computes the extended minimum spanning tree graph. Similar to the core distance calculation, there is a loop nested within a loop which causes the time complexity to be . Carefully reviewing this logic, again with the intention of executing it in parallel, reveals that each execution of the outer loop cannot be safely performed in separate threads. Each subsequent execution of the outer loop depends on a result from the previous executions of the loop. This indicates each iteration of the outer loop needs to be made sequentially. Instead, an implementation which again uses the ExecutorService API is developed which submits only the code within the inner loop to a thread pool executor. This solution still required some synchronization on a specific block of code to maintain the safety of the logic. Unfortunately, this multithreaded implementation of the logic that computes the extended minimum spanning tree graph did not perform well with any of datasets used for testing. At this time, this step of the HDBSCAN* algorithm will not be parallelized.
3.5. Compare the HDBSCAN* Implementations
The last step in the methodology of this work is to compare Tribuo Hdbscan to the other HDBSCAN* implementations reviewed in this work. These comparisons are described in this section, and the results are presented in the next section. The comparisons are captured using Jupyter notebooks and they are available for review online [16]. It is well known that Jupyter includes a Python kernel by default. However, Jupyter does not natively support Java. Fortunately, a Java kernel can be added to Jupyter using a project called IJava [17].
3.5.1. Cluster Assignments and Outlier Scores
The first comparison to make is between the reference implementation and Tribuo Hdbscan. A model is trained for both implementations using the same dataset, and the resulting cluster assignments and outlier scores are compared. This process is repeated with three different datasets. The first and second datasets both use Gaussians, or generated isotropic Gaussian blobs, which are simply sets of points normally distributed around a defined number of centroids. The first dataset uses Gaussians with four centroids and 2000 points, where each point has three features. The second dataset uses gaussians with three centroids and 4000 points, where each point has seven features. The third dataset is more of a real-world dataset, which is the usage behavior of credit card holders over a six-month period [18]. After data preprocessing, this dataset contains 8949 records where each record has seventeen features.
A similar comparison is made between Tribuo Hdbscan and the Python module hdbscan [4]. A model is trained for both implementations using the same dataset, and the resulting cluster assignments and outlier scores are compared. The process is repeated using the same three different datasets just described in the previous paragraph. The HDBSCAN* algorithm is deterministic, which means that the algorithm produces the same cluster assignments and outlier scores when the input remains fixed. Therefore, there is no need to execute each individual test multiple times.
3.5.2. Predictions
Next, a comparison is made using the predictions made by Tribuo Hdbscan and the Python module hdbscan. Recall that the reference implementation does not provide prediction functionality. A model is trained for both implementations using the same training dataset, and then each model is used to make predictions on a separate test dataset. This process is repeated with three different datasets. All three datasets use Gaussians because artificial datasets such as this provide the point’s assigned cluster, which is useful for evaluating the quality of the predictions. Note that each of the three datasets is split into separate training and test files in advance, to avoid any subtle differences that may occur in the way each library splits the data after it has been loaded. The first dataset uses Gaussians with four centroids and 2000 points, where each point has three features. In this case the data are split with 99% for training and only 1% for test. Note that for density-based clustering algorithms such as HDBSCAN*, splitting the data like this does not result in over fitting. In fact, this should improve the quality of the predictions. The second dataset uses Gaussians with three centroids and 5000 points, where each point has four features. The third dataset uses Gaussians with five centroids and 5000 points, where each point has four features. For both the second and third datasets, the data are split with 80% for training and 20% for test.
3.5.3. Performance
Additionally, the performance of the Tribuo Hdbscan implementation is compared to the other HDBSCAN* implementations reviewed in this work. The first aspect of performance to compare is the model training times. The approach is simple, a model is trained for each implementation using the same dataset, and the training times are recorded. Each test is performed three times and the average training times are calculated. This process is repeated with three different datasets. The first dataset is the same credit card usage data already described above. The second dataset uses Gaussians with six centroids and 50,000 points, where each point has seven features. The third dataset uses Gaussians with six centroids and 100,000 points, where each point has seven features.
The second aspect of performance to compare is the model prediction times, thus Tribuo Hdbscan is compared to the Python module hdbscan [4]. A model is trained for both implementations using the same training dataset. Then, each model is used to make predictions on a separate test dataset and the prediction times are recorded. Again, each test is performed three times and the average training times are calculated. This process is repeated with two different datasets. Each dataset is split into separate training and test files in advance, to avoid any subtle differences that may occur in the way each library splits the data after it has been loaded. The first dataset uses Gaussians with six centroids and 50,000 points, where each point has seven features. The second dataset uses Gaussians with six centroids and 100,000 points, where each point has seven features. For both datasets, the data are split with 60% for training and 40% for predictions. For accurate predictions, the data should not be split in this way. The best practice would be to allocate a much higher percentage for the training set. For these tests however, it is more interesting to provide a larger volume of data to the prediction call.
4. Results and Discussion
This section presents the results of the comparisons made between Tribuo Hdbscan and the other HDBSCAN* implementations reviewed in this work.
4.1. Cluster Assignments and Outlier Scores
Tribuo Hdbscan and the reference Java implementation produce identical cluster assignments and outlier scores for each of the three datasets. A JUnit assertion in the notebook is used to validate these results. This confirms that the new HDBSCAN* implementation, Tribuo Hdbscan, produces correct results.
When comparing Tribuo Hdbscan with the Python module hdbscan, there are some differences in the cluster assignments. Adjusted mutual information scores are an effective way to compare the assignments when the labeling technique is different. For example, Tribuo Hdbscan uses the label “0” to indicate an outlier where hdbscan uses “−1”. Interestingly, for the first dataset of Gaussians with four centroids and 2000 points, both models obtained an adjusted mutual information score close to 0.80 when comparing the computed cluster assignments to the actual cluster assignments. Recall that an adjusted mutual information value of one indicates perfect correlation between results. Computing the adjusted mutual information between the Tribuo Hdbscan and the hdbscan cluster assignments gives a score of 0.98. For the second dataset of Gaussians with three centroids and 5000 points, both models achieve an adjusted mutual information score of 1.0 when comparing the computed cluster assignments to the actual cluster assignments. The third dataset of credit card usage data produces the least similar cluster assignments. There are no “ground truth” cluster assignments to use for comparison for this dataset. Computing the adjusted mutual information between the Tribuo Hdbscan and the hdbscan cluster assignments gives a score of 0.82. This dataset effectively demonstrates that subtle differences between the way these two HDBSCAN* implementations have been developed can produce different clustering results. Table 3 shows the adjusted mutual information scores from the cluster assignments for trained models of each clustering implementation for the described datasets. Scores of N/A are shown for the Credit Card dataset since there are no “ground truth” cluster labels that can be used to calculate an adjusted mutual information score.

Table 3.
The adjusted mutual information scores of the cluster assignments for trained models.
When comparing the outlier scores between Tribuo Hdbscan with the Python module hdbscan, the results were quite different for every dataset. Because these are decimal values between zero and one, some investigation was made to determine if the differences were very small fraction values. Even with difference tolerances of 0.01, the results are still significantly different.
4.2. Predictions
Tribuo Hdbscan and the Python module hdbscan produce identical cluster assignment predictions for each of the three datasets. This confirms that the novel technique to make fast predictions provided with Tribuo Hdbscan produces high-quality results. Table 4 shows the adjusted mutual information scores for the predicted cluster assignments of these two clustering implementations for the described datasets. Recall again that the reference implementation does not provide prediction functionality.

Table 4.
The adjusted mutual information scores for the predicted cluster assignments.
It is interesting to note that both implementations make the identical prediction errors for the dataset of Gaussians with four centroids and 2000 points. The adjusted mutual information score for the predicted cluster assignments for this test case is 0.87. Both models identify three clusters and assign several points as outliers or noise, even though this synthetic dataset is defined to have four clusters. Recall that a similar observation is made above when reviewing the cluster assignments for training step of the dataset. Fortunately, this particular dataset was defined with only three features, so it is possible to plot and visualize. The resulting visualization does show that two of the clusters appear nearly combined, which explains this result.
Lastly, since the cluster models trained by these two HDBSCAN* implementations demonstrated significantly different outlier scores, the predicted outlier scores are not compared.
4.3. Performance
The results from comparing the model training times are best visualized. Figure 6 shows a clustered column chart comparing the model training times for the three datasets. Tribuo Hdbscan performs better than the reference implementation. Even though there is some overhead from the Tribuo framework, and some extra steps to compute the cluster exemplars, the improved training times are a direct result of parallelizing the core distance calculations. On the test hardware, instantiating the HdbscanTrainer class with the variable numThreads set to eight produced the best results, as can be seen in the relevant notebooks [16].
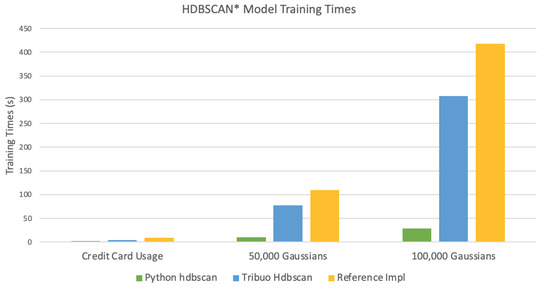
Figure 6.
A clustered column chart comparing the model training times for the three datasets.
The Python module hdbscan is faster than Tribuo Hdbscan by an order of magnitude in the time taken to train the models. This implementation takes advantage of scikit-learn’s KDTree and BallTree nearest neighbor algorithms to compute the core distances. It also uses numpy extensively and leverages its optimized numerical array operations.
The results from comparing the model prediction times are also best visualized. Figure 7 shows a clustered column chart comparing the model prediction times for the two datasets.
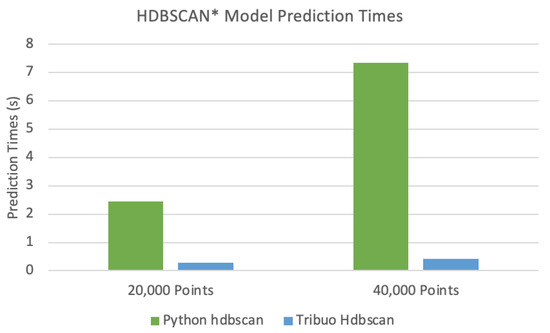
Figure 7.
A clustered column chart comparing the model prediction times for the two datasets.
The novel prediction technique provided with Tribuo Hdbscan is able to make predictions much faster than the Python module hdbscan. This technique, which uses computed cluster exemplars to estimate cluster assignments, has an asymptotic complexity of as discussed above. The Python module hdbscan closely follows the HDBSCAN* algorithm to make predictions which has a theoretical complexity of . In practice, the hdbscan module leverages pre-computed structures stored in memory, so it should be somewhat faster, but it is still slower than the technique proposed in this work.
5. Conclusions and Future Work
An implementation of the HDBSCAN* clustering algorithm, Tribuo Hdbscan, is developed and presented in this work. This implementation is proven to produce the same cluster assignments and outlier scores as the reference Java implementation. This implementation leverages concurrency and improves on the performance of this reference implementation for model training by 25%. The Python module hdbscan is the state-of-the-art HDBSCAN* implementation, which is well-known and widely used. It significantly outperforms Tribuo Hdbscan at training clustering models. Tribuo Hdbscan provides prediction functionality, which is a novel technique to make fast predictions for unseen data points using an HDBSCAN* clustering model. This functionality is demonstrated to be correct since it produces the same cluster predictions as the Python module hdbscan given the same input data. Surprisingly, Tribuo Hdbscan makes predictions ten times faster than hdbscan.
There are numerous Java enterprise applications running in production today. Being able to generate density-based clustering models, using the Tribuo machine learning library, natively in a Java project is an important point. The fast prediction capabilities make Tribuo Hdbscan a compelling choice for applications such as a streaming clustering service.
There are some suggestions for future work on this HDBSCAN* implementation. Improving the performance of model training needs to be investigated to achieve model training times more comparable to the Python module hdbscan. The current analysis of Tribuo Hdbscan shows that the first step of the algorithm which computes the core distances, although improved, still takes about 35% of the total processing time. It is the next step in the algorithm which computes the extended minimum spanning tree graph that takes the largest percentage of the processing time, at 45%. Carefully examining the Python module hdbscan shows that there are some advances made in these steps of the algorithm. To compute the core distances, it leverages either the KDTree or BallTree [14] classes which are optimized nearest neighbor algorithms. To compute the minimum spanning tree, a technique described as a Dual tree Boruvka minimum spanning tree computation can be employed. The authors of both [19,20] describe this technique in their work. Researching these areas and reimplementing the first two steps of the HDBSCAN* algorithm in Tribuo Hdbscan should significantly improve the performance of model training.
Author Contributions
Conceptualization, G.S. and M.A.-K.; methodology, G.S. and M.A.-K.; software, G.S.; validation, G.S. and M.A.-K.; formal analysis, G.S.; investigation, G.S. and M.A.-K.; resources, G.S. and M.A.-K.; data curation, G.S.; writing—original draft preparation, G.S.; writing—review and editing, G.S. and M.A.-K.; visualization, G.S. and M.A.-K.; supervision, M.A.-K.; project administration, G.S. and M.A.-K.; funding acquisition, G.S. and M.A.-K. All authors have read and agreed to the published version of the manuscript.
Funding
This work is partially funded by Lewis University.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
A publicly available dataset was analyzed in this study. These data can be found here: https://www.kaggle.com/arjunbhasin2013/ccdata, accessed on 10 September 2021.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Al-sharoa, E.; Al-khassaweneh, M.A.; Aviyente, S. Detecting and tracking community structure in temporal networks: A low-rank+ sparse estimation based evolutionary clustering approach. IEEE Trans. Signal Inf. Process. Over Netw. 2019, 5, 723–738. [Google Scholar] [CrossRef]
- Al-Sharoa, E.; Al-khassaweneh, M.; Aviyente, S. Low-rank estimation based evolutionary clustering for community detection in temporal networks. In Proceedings of the ICASSP 2019–2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 5381–5385. [Google Scholar]
- Campello, R.J.; Moulavi, D.; Sander, J. Density-based Clustering Based on Hierarchical Density Estimates. In Advances in Knowledge Discovery and Data Mining, Proceedings of the Pacific-Asia Conference on Knowledge Discovery and Data Mining; Pei, J., Tseng, V.S., Cao, L., Motoda, H., Xu, G., Eds.; Springer: Berlin, Germany, 2013; pp. 160–172. [Google Scholar]
- McInnes, L.; Healy, J.; Astels, S. hdbscan: Hierarchical density based clustering. J. Open Source Softw. 2017, 2, 205. [Google Scholar] [CrossRef]
- Machine Learning in Java—Tribuo. Available online: https://tribuo.org/ (accessed on 24 July 2021).
- HDBSCAN* Clustering Tutorial. Available online: https://tribuo.org/learn/4.2/tutorials/clustering-hdbscan-tribuo-v4.html (accessed on 22 January 2022).
- Ester, M.; Kriegel, H.; Sander, J. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. In Proceedings of the Second International Conference on Knowledge Discovery and Data Mining, Portland, OR, USA, 2 August 1996; pp. 226–231. [Google Scholar]
- Campello, R.J.; Moulavi, D.; Zimek, A.; Sander, J. Hierarchical Density Estimates for Data Clustering, Visualization, and Outlier Detection. ACM Trans. Knowl. Discov. Data (TKDD) 2015, 10, 5–56. [Google Scholar] [CrossRef]
- Ankerst, M.; Breunig, M.; Kriegel, H.; Sander, J. OPTICS: Ordering Points To Identify the Clustering Structure. In Proceedings of the 1999 ACM SIGMOD International Conference on Management of Data, United States, Philadelphia, PA, USA, 1–3 June 1999; ACM: New York, NY, USA, 1999; pp. 49–60. [Google Scholar]
- Maimon, O.; Rokach, L. Clustering methods. In Data Mining and Knowledge Discovery Handbook, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 321–352. [Google Scholar]
- Dinh, D.T.; Fujinami, T.; Huynh, V.N. Estimating the Optimal Number of Clusters in Categorical Data Clustering by Silhouette Coefficient. In KSS 2019: Knowledge and Systems Sciences; Chen, J., Huynh, V., Nguyen, G.N., Tang, X., Eds.; Communications in Computer and Information Science; Springer: Singapore, 2019; Volume 1103. [Google Scholar]
- McInnes, L.; Healy, J. Accelerated Hierarchical Density Based Clustering. In Proceedings of the 2017 IEEE International Conference on Data Mining Workshops (ICDMW), New Orleans, LA, USA, 18–21 November 2017; pp. 33–42. [Google Scholar]
- JavaVsPythonMLlibs. Available online: https://github.com/geoffreydstewart/JavaVsPythonMLlibs (accessed on 15 September 2021).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Sia, W.; Lazarescu, M. Clustering Large Dynamic Datasets Using Exemplar Points. In Machine Learning and Data Mining in Pattern Recognition, Proceedings of the 4th International Conference on Machine Learning and Data Mining in Pattern Recognition, Leipzig, Germany, 18–20 July 2005; Springer: Berlin/Heidelberg, Germany, 2005; pp. 163–173. [Google Scholar]
- TribuoHdbscan. Available online: https://github.com/geoffreydstewart/TribuoHdbscan (accessed on 10 November 2021).
- IJava. Available online: https://github.com/SpencerPark/IJava (accessed on 22 September 2021).
- Credit Card Dataset for Clustering. Available online: https://www.kaggle.com/arjunbhasin2013/ccdata (accessed on 10 September 2021).
- Curtin, R.; March, W.; Ram, P.; Anderson, D.; Gray, A.; Isbell, C. Tree-Independent Dual-Tree Algorithms. In Proceedings of the 30th International Conference on Machine Learning, Atlanta, GA, USA, 17 June 2013; pp. 1435–1443. [Google Scholar]
- March, W.; Ram, P.; Gray, A. Fast Euclidean Minimum Spanning tree: Algorithm, Analysis, and Applications. In ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Proceedings of the 16th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Washington, DC, USA, 24–27 July 2010; Association for Computing Machinery: New York, NY, USA, 2010; pp. 603–612. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).