Comparison of Selection Criteria for Model Selection of Support Vector Machine on Physiological Data with Inter-Subject Variance
Abstract
1. Introduction
2. Materials and Methods
2.1. MS of SVM
2.2. LSSC
| Algorithm 1: Pseudocode used for LSSC in MS |
Input: - C and values to be tested - Data of N subjects that can be used for MS Output: evaluated performance (P) for the tested model P = 0 Normalization for the data of N subjects For = Dataset of subject i = {Dataset of subject , } Train SVM using C, , and = classification accuracy of the SVM for P = End For ReturnP |
2.3. Classification Problems on Physiological Data
3. Results
4. Discussion
4.1. Statistical Analysis on Classification Accuracy
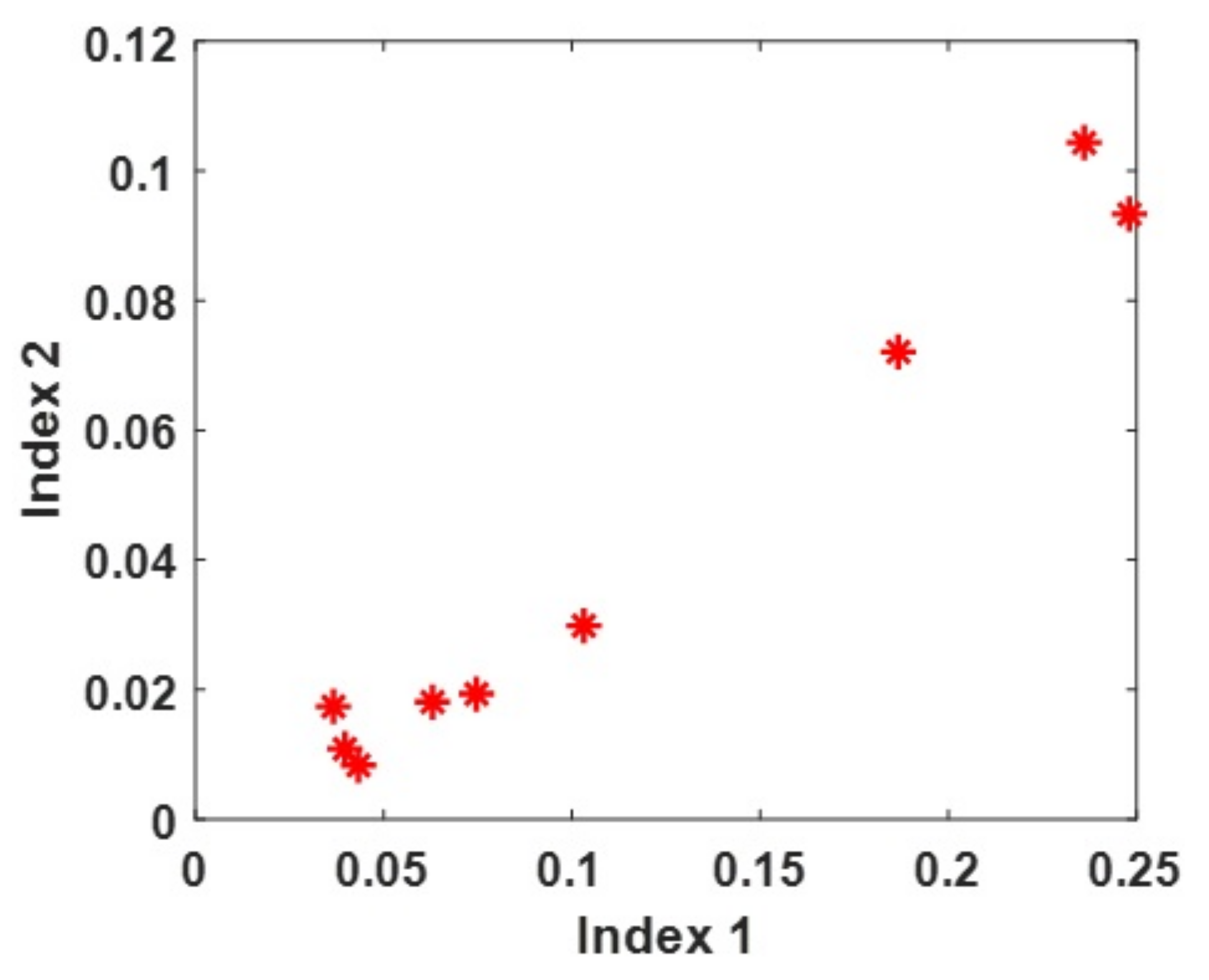
4.2. Relationship between ISV and LSSC
4.3. Comparison with the Results of Previous Studies
4.4. Contributions of This Study
- Comparison outcomes of selection criteria on classification accuracy and computational time were presented and analyzed. The results can be used when a selection criterion is selected for a classification problem on physiological data. The LSSC yielded the highest classification accuracy, but its computational time was the longest. DBTC was lower than the LSSC in classification accuracy, but it was associated with a relatively short computational time.
- The effectiveness of LSSC in classification accuracy was verified experimentally. It outperformed the classification accuracies of others, and the difference was statistically significant. In addition, the advantages were more pronounced for data that had larger ISV.
- The change in relative superiority was investigated when selection criteria were used for the data that contained ISV. The relative effectiveness of DBTC was increased more than KSC when the selection criterion was utilized for physiological data compared with data without ISV.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Description on Datasets and Extracted Features
Appendix A.1. Problem 1
| Signal | Features |
|---|---|
| ECG | Normalized mean and variance of RR intervals, ratio of the low (0–0.08 Hz) to the high-frequency (0.15–0.5 Hz) spectral power for RR intervals |
| EDA (hand) | Normalized mean and variance, the number of orienting responses, sum of the startle magnitudes, sum of the response duration, sum of the areas under responses |
| EDA (foot) | Same as EDA (hand) |
| EMG | Normalized mean |
| Respiration | Normalized mean and variance, spectral power (0–0.1, 0.1–0.2, 0.2–0.3, and 0.3–0.4 Hz) |
Appendix A.2. Problem 2
| Classification | Features |
|---|---|
| Time domain | Mean and median of RR intervals, root mean square of the successive RR interval differences (RMSSD), standard deviation of RR interval differences (SDSD), previous four features for relative RR intervals, standard deviation, skewness, and kurtosis of RR intervals, ratio of SDRR over RMSSD, percentage of RR interval differences more than 25 and 50 ms |
| Spectral feature | Spectral power of RR intervals for very low (≤0.04 Hz), low (0.04–0.15 Hz), and high (0.15–0.4 Hz) frequency bands, ratio of the low to the high-frequency spectral power for RR intervals |
| Nonlinear measure | Two Poincaré plot descriptors |
Appendix A.3. Problems 3–5
| Problem | Signal | Features |
|---|---|---|
| 3 | PPG | Standard deviation of pulse amplitudes, DC amplitude, standard deviation of amplitude differences between pulses, signal energy, bandwidth |
| 4 | PPG | Mean of pulse intervals, normalized power in low-frequency range (0.04–0.15 Hz) of HRV, DC amplitude, mean rise time (time from valley to peak), mean ratio of rise to fall time (time from peak to valley), bandwidth |
| EDA | Mean | |
| 5 | PPG | Mean of pulse intervals, normalized power in low-frequency range of HRV, mean amplitude of pulse amplitudes, DC amplitude, mean rise time, signal energy, bandwidth |
| EDA | Mean |
Appendix A.4. Problem 6
Appendix A.5. Problems 7 and 8
| Classification | Features |
|---|---|
| Time domain | Mean, median, standard deviation, skewness, and kurtosis of RR intervals, root mean square of the successive RR interval differences (RMSSD), standard deviation of RR interval differences (SDSD), ratio of SDRR over RMSSD, previous eight features for relative RR intervals, percentage of RR interval differences more than 25 and 50 ms, HR, sample entropy of RR intervals |
| Spectral features | Spectral power of RR intervals for very low (≤0.04 Hz), low (0.04–0.15 Hz), high (0.15–0.4 Hz), and whole frequency bands, ratio of the low to the high-frequency spectral power for RR intervals, ratio of the high to the low-frequency spectral power for RR intervals |
| Nonlinear measure | Higuchi fractal dimension, two Poincaré plot descriptors |
Appendix A.6. Problem 9
| Classification | Features |
|---|---|
| RR interval | Distance between current and previous beats, distance between current and next beats, average of previous ten-RR intervals, average of previous RR intervals in the last 20 min, normalized version of previous four features |
| Higher order statistics | Kurtosis and skewness for five segments of each beat |
| Wavelet transform | 23-dimensional descriptors using Daubechies wavelet function with three levels of decomposition |
| Morphological descriptor | Maximum values for the first and last segments and minimum values for the second and third segments when each beat is divided into four segments |
References
- Lee, J.S.; Lee, S.J.; Choi, M.; Seo, M.; Kim, S.W. QRS detection method based on fully convolutional networks for capacitive electrocardiogram. Expert Syst. Appl. 2019, 134, 66–78. [Google Scholar] [CrossRef]
- Lynn, H.M.; Kim, P.; Pan, S.B. Data Independent Acquisition Based Bi-Directional Deep Networks for Biometric ECG Authentication. Appl. Sci. 2021, 11, 1125. [Google Scholar] [CrossRef]
- Tirado-Martin, P.; Sanchez-Reillo, R. BioECG: Improving ECG Biometrics with Deep Learning and Enhanced Datasets. Appl. Sci. 2021, 11, 5880. [Google Scholar] [CrossRef]
- Zhang, P.; Li, F.; Zhao, R.; Zhou, R.; Du, L.; Zhao, Z.; Chen, X.; Fang, Z. Real-Time Psychological Stress Detection According to ECG Using Deep Learning. Appl. Sci. 2021, 11, 3838. [Google Scholar] [CrossRef]
- Ayoobi, N.; Sharifrazi, D.; Alizadehsani, R.; Shoeibi, A.; Gorriz, J.M.; Moosaei, H.; Khosravi, A.; Nahavandi, S.; Chofreh, A.G.; Goni, F.A.; et al. Time Series Forecasting of New Cases and New Deaths Rate for COVID-19 using Deep Learning Methods. arXiv 2021, arXiv:2104.15007. [Google Scholar] [CrossRef]
- Moosaei, H.; Ketabchi, S.; Razzaghi, M.; Tanveer, M. Generalized Twin Support Vector Machines. Neural Process. Lett. 2021, 53, 1545–1564. [Google Scholar] [CrossRef]
- Mangasarian, O.L. Data mining via support vector machines. In Proceedings of the IFIP Conference on System Modeling and Optimization, Trier, Germany, 23–27 July 2001; pp. 91–112. [Google Scholar]
- Lee, Y.J.; Mangasarian, O.L. SSVM: A smooth support vector machine for classification. Comput. Optim. Appl. 2001, 20, 5–22. [Google Scholar] [CrossRef]
- Choi, M.; Koo, G.; Seo, M.; Kim, S.W. Wearable Device-Based System to Monitor a Driver’s Stress, Fatigue, and Drowsiness. IEEE Trans. Instrum. Meas. 2018, 67, 634–645. [Google Scholar] [CrossRef]
- Ortega, S.; Fabelo, H.; Halicek, M.; Camacho, R.; Plaza, M.d.l.L.; Callicó, G.M.; Fei, B. Hyperspectral superpixel-wise glioblastoma tumor detection in histological samples. Appl. Sci. 2020, 10, 4448. [Google Scholar] [CrossRef]
- Setiowati, S.; Franita, E.L.; Ardiyanto, I. A review of optimization method in face recognition: Comparison deep learning and non-deep learning methods. In Proceedings of the 9th International Conference on Information Technology and Electrical Engineering (ICITEE), Phuket, Thailand, 12–13 October 2017; pp. 1–6. [Google Scholar]
- Pandit, R.; Kolios, A. SCADA data-based support vector machine wind turbine power curve uncertainty estimation and its comparative studies. Appl. Sci. 2020, 10, 8685. [Google Scholar] [CrossRef]
- Rizwan, A.; Iqbal, N.; Ahmad, R.; Kim, D.H. WR-SVM Model Based on the Margin Radius Approach for Solving the Minimum Enclosing Ball Problem in Support Vector Machine Classification. Appl. Sci. 2021, 11, 4657. [Google Scholar] [CrossRef]
- Ayat, N.E.; Cheriet, M.; Suen, C.Y. Automatic model selection for the optimization of SVM kernels. Pattern Recogn. 2005, 38, 1733–1745. [Google Scholar] [CrossRef]
- Adankon, M.M.; Cheriet, M. Optimizing resources in model selection for support vector machine. Pattern Recogn. 2007, 40, 953–963. [Google Scholar] [CrossRef]
- Zhang, X.; Qiu, D.; Chen, F. Support vector machine with parameter optimization by a novel hybrid method and its application to fault diagnosis. Neurocomputing 2015, 149, 641–651. [Google Scholar] [CrossRef]
- Kapp, M.N.; Sabourin, R.; Maupin, P. A dynamic model selection strategy for support vector machine classifiers. Appl. Soft Comput. 2012, 12, 2550–2565. [Google Scholar] [CrossRef]
- Li, W.; Liu, L.; Gong, W. Multi-objective uniform design as a SVM model selection tool for face recognition. Expert Syst. Appl. 2011, 38, 6689–6695. [Google Scholar] [CrossRef]
- Huang, C.M.; Lee, Y.J.; Lin, D.K.; Huang, S.Y. Model selection for support vector machines via uniform design. Comput. Stat. Data Anal. 2007, 52, 335–346. [Google Scholar] [CrossRef]
- Wu, C.H.; Tzeng, G.H.; Goo, Y.J.; Fang, W.C. A real-valued genetic algorithm to optimize the parameters of support vector machine for predicting bankruptcy. Expert Syst. Appl. 2007, 32, 397–408. [Google Scholar] [CrossRef]
- Namdeo, A.; Singh, D. Challenges in evolutionary algorithm to find optimal parameters of SVM: A review. Mater. Today-Proc. 2021. [Google Scholar] [CrossRef]
- Vapnik, V.; Chapelle, O. Bounds on error expectation for support vector machines. Neural Comput. 2000, 12, 2013–2036. [Google Scholar] [CrossRef]
- Anguita, D.; Ridella, S.; Rivieccio, F.; Zunino, R. Hyperparameter design criteria for support vector classifiers. Neurocomputing 2003, 55, 109–134. [Google Scholar] [CrossRef]
- Sun, J.; Zheng, C.; Li, X.; Zhou, Y. Analysis of the distance between two classes for tuning SVM hyperparameters. IEEE Trans. Neural Netw. 2010, 21, 305–318. [Google Scholar] [PubMed]
- Yin, S.; Yin, J. Tuning kernel parameters for SVM based on expected square distance ratio. Inform. Sci. 2016, 370, 92–102. [Google Scholar] [CrossRef]
- Duan, K.; Keerthi, S.S.; Poo, A.N. Evaluation of simple performance measures for tuning SVM hyperparameters. Neurocomputing 2003, 51, 41–59. [Google Scholar] [CrossRef]
- Duarte, E.; Wainer, J. Empirical comparison of cross-validation and internal metrics for tuning SVM hyperparameters. Pattern Recogn. Lett. 2017, 88, 6–11. [Google Scholar] [CrossRef]
- Choi, M.; Seo, M.; Lee, J.S.; Kim, S.W. Fuzzy support vector machine-based personalizing method to address the inter-subject variance problem of physiological signals in a driver monitoring system. Artif. Intell. Med. 2020, 105, 101843. [Google Scholar] [CrossRef]
- Gholamiangonabadi, D.; Kiselov, N.; Grolinger, K. Deep Neural Networks for Human Activity Recognition with Wearable Sensors: Leave-one-subject-out Cross-validation for Model Selection. IEEE Access 2020, 8, 133982–133994. [Google Scholar] [CrossRef]
- Rojas-Domínguez, A.; Padierna, L.C.; Valadez, J.M.C.; Puga-Soberanes, H.J.; Fraire, H.J. Optimal hyper-parameter tuning of SVM classifiers with application to medical diagnosis. IEEE Access 2017, 6, 7164–7176. [Google Scholar] [CrossRef]
- Kumar, S. Neural Networks: A Classroom Approach; Tata McGraw-Hill Education: New York, NY, USA, 2004. [Google Scholar]
- Diosan, L.; Rogozan, A.; Pecuchet, J.P. Improving classification performance of support vector machine by genetically optimising kernel shape and hyper-parameters. Appl. Intell. 2012, 36, 280–294. [Google Scholar] [CrossRef]
- Healey, J.A.; Picard, R.W. Detecting stress during real-world driving tasks using physiological sensors. IEEE Trans. Intell. Transp. Syst. 2005, 6, 156–166. [Google Scholar] [CrossRef]
- Goldberger, A.L.; Amaral, L.A.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. Physiobank, physiotoolkit, and physionet. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [PubMed]
- Koldijk, S.; Sappelli, M.; Verberne, S.; Neerincx, M.A.; Kraaij, W. The swell knowledge work dataset for stress and user modeling research. In Proceedings of the 16th International Conference on Multimodal Interaction, Istanbul, Turkey, 12–16 November 2014; pp. 291–298. [Google Scholar]
- Nkurikiyeyezu, K.; Yokokubo, A.; Lopez, G. The Effect of Person-Specific Biometrics in Improving Generic Stress Predictive Models. arXiv 2019, arXiv:1910.01770. [Google Scholar] [CrossRef]
- Leeb, R.; Brunner, C.; Müller-Putz, G.; Schlögl, A.; Pfurtscheller, G. BCI Competition 2008–Graz Data Set B; Graz University of Technology: Graz, Austria, 2008. [Google Scholar]
- Lopez, G.; Kawahara, Y.; Suzuki, Y.; Takahashi, M.; Takahashi, H.; Wada, M. Effect of direct neck cooling on psychological and physiological state in summer heat environment. Mech. Eng. J. 2016, 3, 15-00537. [Google Scholar] [CrossRef][Green Version]
- Moody, G.B.; Mark, R.G. The impact of the MIT-BIH arrhythmia database. IEEE Eng. Med. Biol. 2001, 20, 45–50. [Google Scholar] [CrossRef]
- Mondéjar-Guerra, V.; Novo, J.; Rouco, J.; Penedo, M.G.; Ortega, M. Heartbeat classification fusing temporal and morphological information of ECGs via ensemble of classifiers. Biomed. Signal Proces. 2019, 47, 41–48. [Google Scholar] [CrossRef]
- Ang, K.K.; Chin, Z.Y.; Wang, C.; Guan, C.; Zhang, H. Filter bank common spatial pattern algorithm on BCI competition IV datasets 2a and 2b. Front. Neurosci. 2012, 6, 39. [Google Scholar] [CrossRef]
- Nkurikiyeyezu, K.; Yokokubo, A.; Lopez, G. Affect-aware thermal comfort provision in intelligent buildings. In Proceedings of the 8th International Conference on Affective Computing and Intelligent Interaction Workshops and Demos (ACIIW), Cambridge, UK, 3–6 September 2019; pp. 331–336. [Google Scholar]
- Son, Y.; Kim, W. Missing Value Imputation in Stature Estimation by Learning Algorithms Using Anthropometric Data: A Comparative Study. Appl. Sci. 2020, 10, 5020. [Google Scholar] [CrossRef]
- Le, N.Q.K.; Hung, T.N.K.; Do, D.T.; Lam, L.H.T.; Dang, L.H.; Huynh, T.T. Radiomics-based machine learning model for efficiently classifying transcriptome subtypes in glioblastoma patients from MRI. Comput. Biol. Med. 2021, 132, 104320. [Google Scholar] [CrossRef]
- Do, D.T.; Le, N.Q.K. Using extreme gradient boosting to identify origin of replication in Saccharomyces cerevisiae via hybrid features. Genomics 2020, 112, 2445–2451. [Google Scholar] [CrossRef]
| Problem | #S | #F | Data (Sample Size) |
|---|---|---|---|
| 1. Stress detection 1 | 10 | 22 | low (166) / medium + high stress (217) |
| 2. Stress detection 2 | 18 | 20 | normal (1800)/stress (1800) |
| 3. Stress detection 3 | 28 | 5 | normal (1692)/stress (1548) |
| 4. Fatigue detection | 28 | 7 | normal (1692)/fatigue (3073) |
| 5. Drowsiness detection | 28 | 8 | normal (1692)/drowsiness (3110) |
| 6. Motor imaginary | 9 | 18 | left (1706)/right hand (1717) |
| 7. Thermal comfort 1 | 11 | 29 | VH with cooler (1100)/VH (1100) |
| 8. Thermal comfort 2 | 11 | 29 | VH with cooler (1100)/hot (1100) |
| 9. VEB detection | 16 | 45 | normal (1600)/VEB (1600) |
| Problem | KSC | KSC2 | DBTC | ESDR | XAB | GACV | LSSC | Optimal |
|---|---|---|---|---|---|---|---|---|
| 1 | 81.53 | 82.24 | 87.29 | 79.47 | 76.30 | 83.83 | 83.96 | 96.23 |
| 2 | 74.00 | 73.97 | 72.97 | 73.42 | 71.19 | 64.19 | 79.33 | 89.22 |
| 3 | 82.06 | 81.94 | 81.95 | 82.17 | 82.85 | 83.36 | 83.49 | 91.17 |
| 4 | 88.90 | 88.85 | 89.10 | 89.02 | 90.42 | 90.67 | 90.51 | 96.57 |
| 5 | 90.84 | 90.42 | 90.99 | 90.73 | 92.85 | 92.44 | 91.82 | 97.37 |
| 6 | 72.80 | 72.80 | 72.95 | 72.74 | 70.22 | 50.65 | 73.41 | 76.21 |
| 7 | 63.32 | 63.36 | 63.64 | 60.77 | 59.05 | 59.68 | 69.23 | 77.00 |
| 8 | 64.05 | 64.00 | 64.55 | 61.77 | 59.91 | 65.00 | 70.73 | 79.50 |
| 9 | 85.72 | 85.22 | 84.97 | 86.44 | 84.16 | 86.06 | 87.38 | 94.34 |
| Mean | 78.13 | 78.09 | 78.71 | 77.39 | 76.33 | 75.10 | 81.09 | 88.62 |
| SD | 10.16 | 10.07 | 10.47 | 11.07 | 12.28 | 15.26 | 8.42 | 8.73 |
| Problem | KSC | KSC2 | DBTC | ESDR | XAB | GACV | LSSC |
|---|---|---|---|---|---|---|---|
| 1 | 5 | 4 | 1 | 6 | 7 | 3 | 2 |
| 2 | 2 | 3 | 5 | 4 | 6 | 7 | 1 |
| 3 | 5 | 7 | 6 | 4 | 3 | 2 | 1 |
| 4 | 6 | 7 | 4 | 5 | 3 | 1 | 2 |
| 5 | 5 | 7 | 4 | 6 | 1 | 2 | 3 |
| 6 | 3 | 3 | 2 | 5 | 6 | 7 | 1 |
| 7 | 4 | 3 | 2 | 5 | 7 | 6 | 1 |
| 8 | 4 | 5 | 3 | 6 | 7 | 2 | 1 |
| 9 | 4 | 5 | 6 | 2 | 7 | 3 | 1 |
| Mean | 4.22 | 4.89 | 3.67 | 4.78 | 5.22 | 3.67 | 1.44 |
| SD | 1.20 | 1.76 | 1.80 | 1.30 | 2.28 | 2.35 | 0.73 |
| Problem | KSC | KSC2 | DBTC | ESDR | XAB | GACV | LSSC |
|---|---|---|---|---|---|---|---|
| 1 | 2.21 | 2.22 | 0.20 | 0.21 | 2.12 | 1.31 | 4.91 |
| 2 | 181.85 | 181.47 | 12.22 | 15.80 | 141.15 | 89.84 | 738.46 |
| 3 | 205.72 | 207.95 | 38.16 | 9.90 | 104.05 | 82.10 | 1309.69 |
| 4 | 203.88 | 205.48 | 14.84 | 19.58 | 142.74 | 95.23 | 1467.00 |
| 5 | 209.10 | 210.07 | 13.35 | 20.86 | 150.53 | 98.61 | 1529.19 |
| 6 | 313.58 | 316.27 | 42.36 | 16.06 | 169.13 | 129.10 | 577.19 |
| 7 | 102.80 | 103.30 | 10.85 | 16.87 | 75.34 | 49.74 | 224.57 |
| 8 | 90.65 | 90.68 | 8.18 | 9.35 | 69.58 | 44.45 | 199.50 |
| 9 | 191.61 | 191.53 | 11.51 | 15.64 | 483.72 | 203.65 | 654.59 |
| Mean | 166.82 | 167.66 | 16.85 | 13.81 | 148.71 | 88.23 | 745.01 |
| KSC | KSC2 | DBTC | ESDR | XAB | GACV | ||
|---|---|---|---|---|---|---|---|
| value | 1.2 × | 9.6 × | 6.5 × | 1.9 × | 9.0 × | 2.7 × |
| Problem | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|
| 0.103 | 0.187 | 0.037 | 0.063 | 0.040 | 0.043 | 0.248 | 0.236 | 0.075 | |
| 0.030 | 0.072 | 0.017 | 0.018 | 0.011 | 0.008 | 0.093 | 0.104 | 0.019 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, M.; Jeong, J.J. Comparison of Selection Criteria for Model Selection of Support Vector Machine on Physiological Data with Inter-Subject Variance. Appl. Sci. 2022, 12, 1749. https://doi.org/10.3390/app12031749
Choi M, Jeong JJ. Comparison of Selection Criteria for Model Selection of Support Vector Machine on Physiological Data with Inter-Subject Variance. Applied Sciences. 2022; 12(3):1749. https://doi.org/10.3390/app12031749
Chicago/Turabian StyleChoi, Minho, and Jae Jin Jeong. 2022. "Comparison of Selection Criteria for Model Selection of Support Vector Machine on Physiological Data with Inter-Subject Variance" Applied Sciences 12, no. 3: 1749. https://doi.org/10.3390/app12031749
APA StyleChoi, M., & Jeong, J. J. (2022). Comparison of Selection Criteria for Model Selection of Support Vector Machine on Physiological Data with Inter-Subject Variance. Applied Sciences, 12(3), 1749. https://doi.org/10.3390/app12031749






