Abstract
ICIs are a standard of care in several malignancies; however, according to overall response rate (ORR), only a subset of eligible patients benefits from ICIs. Thus, an ability to predict ORR could enable more rational use. In this study a ML-based ORR prediction model was built, with patient-reported symptom data and other clinical data as inputs, using the extreme gradient boosting technique (XGBoost). Prediction performance for unseen samples was evaluated using leave-one-out cross-validation (LOOCV), and the performance was evaluated with accuracy, AUC (area under curve), F1 score, and MCC (Matthew’s correlation coefficient). The ORR prediction model had a promising LOOCV performance with all four metrics: accuracy (75%), AUC (0.71), F1 score (0.58), and MCC (0.4). A rather good sensitivity (0.58) and high specificity (0.82) of the model were seen in the confusion matrix for all 63 LOOCV ORR predictions. The two most important symptoms for predicting the ORR were itching and fatigue. The results show that it is possible to predict ORR for patients with multiple advanced cancers undergoing ICI therapies with a ML model combining clinical, routine laboratory, and patient-reported data even with a limited size cohort.
1. Introduction
Immune checkpoint inhibitors (ICIs) are standard-of-care treatments in several malignancies, both in adjuvant and advanced settings [1,2,3,4,5,6,7,8,9,10,11,12]. However, treatment response assessment of the ICIs differs from traditional cancer therapies, with unique tumor response patterns such as pseudo- and hyperprogression [13]. Furthermore, the temporal association of radiological response to treatment may sometimes be obscure. While only a subset of patients responds to ICIs, novel tools to assess the treatment response are needed when aiming to improve patient care and the clinical value of ICIs.
Artificial intelligence (AI)-based analytics have gained growing interest in the field of cancer care. Machine learning models have been shown to predict responses to a variety of standard-of-care chemotherapy regimens from gene expression profiles of individual patients with high accuracy [14,15]. Furthermore, deep learning systems have shown promising results, especially in cancer diagnostics [16]. AI-based methods can be used to analyze vast data pools to create predictive and prognostic analytics for generating value-based healthcare assets. In addition, recent data show that machine learning (ML) algorithms could identify patients with cancer who are at risk of short-term mortality [17]. Tumor immunology is a very complex entity, and it is clear that none of the single factors known so far can predict benefit for ICI therapy with high accuracy. Therefore, it is likely that using multiple inputs would result in prediction models with higher sensitivity and specificity.
A comprehensive and timely assessment of patients’ symptoms is feasible via electronic (e) patient-reported outcomes (PROs) [18,19,20]. ePROs have been shown to improve quality of life (QoL) and survival and decrease emergency clinic visits in cancer patients receiving chemotherapy and in lung cancer follow-up [21,22]. Numerous studies have linked ICI treatment benefit to the presence of physician-assessed immune-related adverse events (irAEs), but the prognostic role of ePROs is an uninvestigated area [23,24,25]. While ML-based methodology can comprise numerous variable data sources to generate prediction models [26], the association of irAEs to ICI treatment benefit together with the complexity of tumor immunology generates an interesting landscape to investigate ML-based models.
We have previously shown that the real-world symptom data collected with the Kaiku Health ePRO tool from cancer patients receiving ICI therapy align with the data from clinical trials and that correlations between different symptoms occur, which might reflect therapeutic efficiency, side effects, or tumor progression [27,28]. We first explored the possibilities of ML-based prediction models on ePROs to create prediction models of symptom continuity of cancer patients receiving ICIs and showed that it is feasible [29]. Based on our previous work on ML modeling and the ePRO symptom correlations, we speculated that if symptoms can predict irAEs, symptoms could work as a surrogate to irAEs. That hypothesis was confirmed in our latest research showing that ML-based prediction models using ePRO and electronic health care record (EHR) data as an input can predict the presence and onset of irAEs with a high accuracy [30].
The aim of this study was to investigate whether it is possible to predict objective response rate (ORR) in patients undergoing ICIs for advanced cancers. Thus, pseudonymized and aggregated ePRO symptom data collected with the Kaiku Health ePRO tool, laboratory values, and demographics, in addition to prospectively collected clinician-assessed treatment responses and irAE data, were used to train and tune a prediction model built using an open-source Python library XBoost (extreme gradient boosting algorithm) to assess clinical response to ICI treatment.
2. Materials and Methods
The study subjects (n = 31) consisted of patients recruited to the prospective KISS trial investigating ePRO follow-up of cancer patients receiving ICIs at Oulu University Hospital. In brief, the trial included patients with advanced cancers (non-small cell lung cancer, melanoma, genito-urinary cancers, and head and neck cancers) treated with anti-PD-(L)1s in outpatient settings with the availability of internet access and email. At the initiation of the treatment phase (within 0–2 weeks from the first anti-PD-(L)1 infusion), the patients received an email notification to complete the baseline electronic symptom questionnaire of 18 symptoms and did so weekly thereafter until treatment discontinuation or six months of follow-up. The symptoms tracked by the Kaiku Health ePRO tool are potential signs and symptoms of immune-related adverse events, and symptom selection is based on the reported publications of the following clinical trials: CheckMate 017 (NCT01642004), CheckMate 026 (NCT02041533), CheckMate 057 (NCT01673867), CheckMate 066 (NCT01721772), CheckMate 067 (NCT01844505), KEYNOTE-010 (NCT01905657), and OAK (NCT02008227).
Besides recording the presence of a symptom, a severity algorithm of the ePRO tool grades the symptom according to the Common Terminology Criteria for Adverse Events (CTCAE) protocol, from 0 to 4, with no (0), mild (1), moderate (2), severe (3), and life-threatening (4) categories. In addition to ePRO-collected symptoms, data on demographics, treatment responses according to the Response Evaluation Criteria in Solid Tumors (RECIST 1.1), irAEs (nature of AE, date of onset and resolving, dates of change in AE severity, and the highest grade based on CTCAE classification), and laboratory values were prospectively collected prior and during the treatment period.
The KISS trial was approved by the Northern Ostrobothnia Health District ethics committee (number 9/2017), Valvira (number 361), and details of the study are publicly available at clinicaltrials.gov (NCT03928938). The study was conducted in accordance with the Declaration of Helsinki and Good Clinical Practice guidelines.
The ML-based prediction model was built using the extreme gradient boosting algorithm, implemented using an open-source Python library XGBoost, which is widely used for classification problems. Gradient boosting is an ensemble learning algorithm; thus, it is an ensemble of many decision trees—usually tens or hundreds—which are weak learners but, when combined using the gradient boosting approach, form a strong learner capable of capturing complex relationships in the training data.
The aim of this study was to create a ML-based model for predicting the presence of complete response (CR) or partial response (PR) based on evolving digitally collected patient-reported symptoms, the presence of physician-confirmed irAE, and laboratory values collected in a prospective manner from cancer patients receiving ICI therapies in the KISS trial [26]. The included data consisted of symptom data that were graded by the algorithm of the ePRO tool according to CTCAE, automatically via application programming interface (API)-fetched laboratory data (bilirubin, hemoglobin, ALP, ALT, platelets, leukocytes, creatinine, thyrotropin, and neutrophils) from the baseline (prior to the first drug infusion) throughout the treatment phase, demographics (age and sex), treatment responses, and the presence of irAEs at response assessment (yes/no), as well as the time (weeks) from therapy initiation.
ORR was defined as the proportion of patients in whom PR or CR responses were seen as the best overall response (BOR) according to RECIST 1.1. Stable disease (SD) was categorized as a non-response together with progressive disease (PD). Closest preceding laboratory values and reported symptoms, both as changes from the baseline, were linked to the treatment responses; thus, the timelines of ePROs, irAEs, and BORs were synchronized according to dates. In addition, the model accounted for whether the patient had had a diagnosed irAE prior to/at the time of response evaluation.
Treatment responses according to RECIST 1.1 were divided into binary categories. The output of the prediction model is a continuous value [0,1] depicting the probability for the positive event, i.e., objective response (CR or PR) versus no objective response (SD + PD) (Figure 1). With a classification threshold of 0.5, the continuous probabilities were converted into binary outcomes, i.e., when the predicted probability for the positive event is greater than 0.5, prediction is labeled positive (CR or PR as treatment response), and if less than 0.5, then negative (SD or PD as a treatment response). Thus, the modeling methodology used in this study follows a general framework of binary classification in ML.
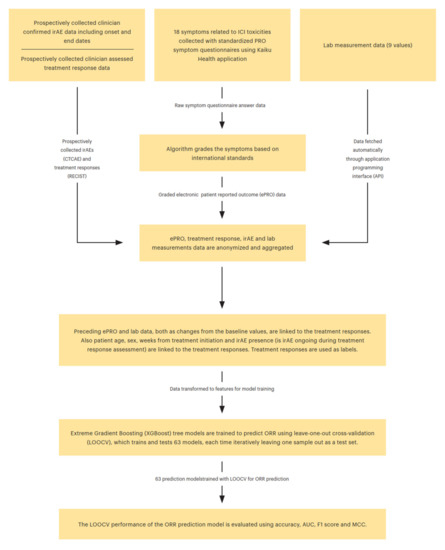
Figure 1.
Complete modeling framework for ORR prediction.
Prediction performance of the model for unseen samples was evaluated using leave-one-out cross-validation (LOOCV), which trained and tested 63 models, each time iteratively leaving one sample (related to one of the clinician-assessed treatment responses) out as a test set. Multiple response assessments across the same patients were used to create a timeline of best overall responses (BORs); however, in every time point analyzed, the parameters differ, comprising a new sample. Furthermore, the used gradient boosting trees-based algorithm (XGBoost) can handle intercorrelated observations or features, and, thus, correlated input parameters do not cause problems for the modeling.
The prediction performance of the model was evaluated with accuracy, AUC (area under curve), F1 score, and MCC (Matthew’s correlation coefficient). Accuracy describes how many predictions were correct as a percentage, and 100% indicates a perfect classification. AUC is a commonly used performance metric for binary classification ranging from 0 to 1, where 0.5 is random guessing and 1 is perfect classification. F1 score is the weighted average of precision (i.e., how many of the cases predicted as positive are positive) and recall (how many of the positive cases are detected), which attains values between 0 and 1, 1 indicating perfect precision and recall. MCC summarizes all possible cases for binary predictions: true and false positives, and true and false negatives. MCC can be considered as a correlation coefficient between the observed and the predicted classifications, and it attains values between −1 and 1, where 1 is perfect classification, 0 is random guessing, and −1 indicates completely contradictory classification.
3. Results
3.1. ML Prediction Model
The initial ePRO dataset included 992 filled symptom questionnaires from the 31 ICI-treated cancer patients in outpatient settings comprising 18 monitored symptoms collected weekly using the Kaiku Health ePRO tool (Table 1). The irAE data included physician-confirmed prospectively collected irAE (n = 26) data in the eCRFs of the KISS trial from those 31 patients, containing initiation and end dates, CTCAE class and severity, and nature (colitis, diarrhea, arthritis, rash, hyperglycemia, neutropenia, pneumonitis, itching, cholangitis, mucositis, hypothyreosis, and hepatitis). Prospectively assessed treatment responses (n = 63) by the study physicians were also retrieved from the eCRF. The patients with partial (PR) or complete (CR) responses (n = 19) were characterized as responders, while stable (SD) and progressive disease (PD) (n = 44) were categorized as a non-response.

Table 1.
The demographics of the study population.
The complete modeling framework for ORR prediction is illustrated in Figure 1. We also tested several other commonly used ML models, such as logistic regression, elastic-net regression, support vector machines, LightGBM, and random forests, but XGBoost had the best performance with the LOOCV evaluation, and, thus, it was chosen as the model for the study.
3.2. Performance Metrics for ORR Prediction
The model trained to predict ORR had a promising LOOCV performance with all four metrics: accuracy, AUC, F1 score, and MCC. The accuracy of predicting ORR was 75%. The AUC value (0.71) suggests a decent quality level of model performance. The F1 score (0.58) indicates that the model was feasible in predicting the treatment response, which was supported by the MCC value (0.40). In the confusion matrix for all 63 LOOCV ORR predictions can be seen a rather good sensitivity (0.58) and a high specificity (0.82) of the model (Figure 2). The false negatives (8/63 samples) were identified as the cases where the prediction model did not predict a presence of objective treatment response for a test dataset sample which was a true positive, i.e., CR or PR was present. The false positives (also 8/63 samples), on the other hand, were the cases where the model predicted the presence of CR or PR for the sample but the sample was a true negative, i.e., the response was SD or PD.
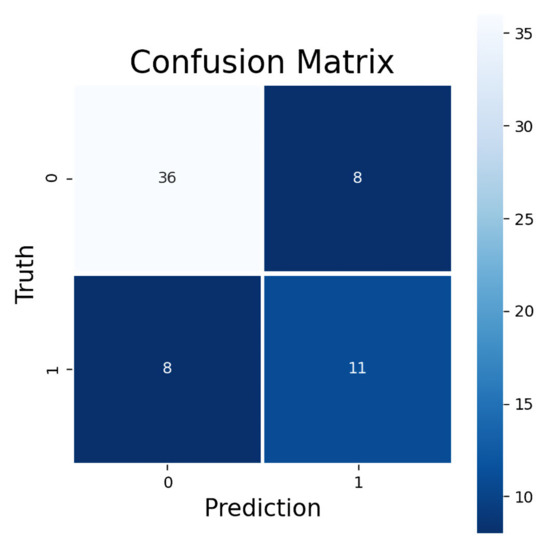
Figure 2.
Confusion matrix for predicted ORR. Upper-left corner shows correctly classified negative, lower-right corner correctly classified positive, upper-right corner false positive, and lower-left corner false negative samples. Negative samples consist of SD and PD responses and positive samples CR and PR responses.
3.3. Feature Importance Analysis
Figure 3 illustrates the feature importance from a model trained with all available samples (n = 63). The displayed importances depict the relative average improvement in prediction accuracy across all of the 100 decision trees in the model where a certain feature is utilized. The importance of each feature should be considered as relative to the others. As is presented in Figure 3, the two most important features for predicting the ORR were itching and fatigue. Figure 3 reveals that roughly half of the features contributed to the predictions. The features which do not contribute to the predictions could be removed from the prediction model using feature selection, but it does not impact the model performance, due to the tree structure of the used gradient boosting algorithm.
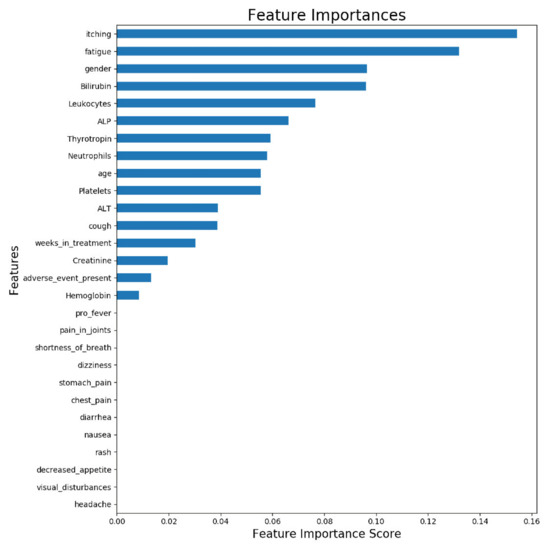
Figure 3.
Feature importance of ORR prediction model trained with all available samples. The displayed importances depict the relative average improvement in prediction accuracy across all 100 trees in the model where a certain feature is utilized. The importance of each feature should be considered as relative to the others.
4. Discussion
Digitalization is a global megatrend affecting the basic structures of human interaction. Digital transformation of healthcare aims to deliver the positive impact of technology in many forms, e.g., telemedicine, AI-aided medical devices, and vast data pools to create predictive analytics. Evolving data show that electronic health record-based predictive algorithms may improve clinicians´ prognostication and decision-making [31]. A recent study on the use of radiomics and machine learning revealed that the algorithm utilizing individual CT scans of advanced melanoma patients receiving single anti-PD-1 therapy outperformed traditional RECIST 1.1 criteria in predicting treatment response [32]. However, utilization of ePROs in creating ML algorithms is a novel approach.
In this study, we investigated ML modeling that combines prospectively collected data on ePROs, demographics, laboratory values, irAEs, and treatment responses. The aim of the study was to investigate whether these data inputs could be used to predict treatment benefit from ICI therapies in metastatic cancers. The results showed that it is possible to predict ORR with a high specificity, even using data from a patient cohort of multiple cancer types. The study highlights the possibilities of using pooled data from various sources for ML models and potentials of these models to improve the clinical value of cancer treatments.
Parallel to traditional follow-up of cancer patients, ePROs enable capturing of symptoms in a timely and comprehensive manner, and it integrates the patients’ perspective into the cancer care continuum [17,33]. Previous studies have provided evidence that ePRO follow-up can improve QoL, reduce emergency clinic visits, and, more importantly, improve survival in chemotherapy-treated patients with advanced cancers and in lung cancer [20,21]. We have previously shown that ePRO follow-up is also feasible for cancer patients receiving ICIs and that ePRO-collected symptom profiles mimic the AE results of ICI registration studies [26,27]. In addition, our earlier studies have highlighted the possibilities of ePRO data inputted ML models in facilitation of irAE detection, which could improve their treatment [29]. Furthermore, since irAEs often are linked to improved outcomes in patients treated with ICIs [23,24,25,26], we speculated that ePRO-collected symptom data could be used to predict treatment benefit. Compared to healthcare professional-collected symptoms, ePRO might provide additional value to the symptom assessment, especially with low-grade symptoms without external presentation such as itching. As far as we know, the present study is the first ever to combine ePRO-collected symptom data with ML modeling to predict treatment response with ICIs.
Even though this has been intensively studied for years, there are no known universal predictive factors for ICI benefit in cancer treatment suitable for clinical practice for multiple cancer types [34]. Our results indicate that multiple data points and sources collected over time can be used to generate an adaptive ML model able to predict treatment outcomes. Due to the complexity of cancer immunology, we speculate that no single universal marker for ICI benefit will be discovered, and more effort should be used in analyzing multidimensional data, combining not only tumor features but also clinical data, such as ePRO symptoms and routine laboratory values. Furthermore, our study suggests that ePRO data could be used as a non-invasive indicator for immune activation and, therefore, surrogate for ICI treatment benefit.
There are several limitations when interpreting our results. Our patient cohort is limited in size, which could decrease the generalizability of the results. Our model had high specificity for treatment benefit but only a moderate level of sensitivity, which might relate to the small cohort size or be a general feature of these models. Nevertheless, the used modeling methods and approaches were chosen to overcome the issues related to imbalanced datasets, and intercorrelated parameters were selected to minimize such bias. These methods and approaches included, e.g., utilization of sample weights (giving more emphasis to the rare positive samples in model training), utilization of F1 score and MCC as performance metrics, and using a regularized tree-based model, XGBoost. In addition, our cohort consisted of ICI-monotherapy-treated patients, and the results might not be applicable to patients treated with ICI combinatory therapies. Thus, our model inevitably requires validation in another, preferably larger, cohort. As far as we know, however, these types of datasets are currently unavailable.
In our opinion, ML models should be incorporated into the digital symptom follow-up of a cancer patient for optimal remote monitoring. The tool should include an interactive ePRO approach connecting the patient and care unit in a timely fashion and, preferably, also automatically integrate other clinical data such as laboratory values. When these datasets are available in a sole platform, adaptive ML models such as the one built in the present study can be used to bring additional important data, such as irAE and treatment benefit probabilities, for clinical decision-making. This digital tool could personalize cancer care and bring additional clinical value to the ICI treatments, especially considering the high costs and undefined predictive factors.
5. Conclusions
In healthcare, knowledge representation as part of the clinical decision support system is currently the most used AI approach. There are high hopes that AI could improve healthcare with early diagnostics and improved care in a more cost-effective manner compared to current measures. Yet the digital revolution in healthcare provides new ways to both collect clinically relevant data from each patient and connect it to large data pools of existing patient-level data for analysis with AI-based algorithms, aiming to personalize treatment schemas and follow-up based on individual risk assessment.
In conclusion, our study highlights the possibility of generating ML models for ICI treatment benefit. We used multiple inputs for the model, including ePRO symptom data, which could serve as a non-invasive surrogate for immune activation. The main results suggest that these models perform with a high specificity. Even though validation of the results in larger cohorts is required, the promising results favor digital approaches in ICI patient follow-up.
Author Contributions
Data curation, J.E. and H.V.; Formal analysis, S.I., J.E. and J.P.K.; Investigation, S.I. and J.P.K.; Methodology, J.E.; Project administration, H.V. and V.V.K.; Resources, S.I., V.V.K. and J.P.K.; Software, H.V.; Validation, S.I.; Writing—original draft, S.I., J.E. and J.P.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Oulu University, Emil Aaltonen Foundation, The Finnish Medical Foundation, and Finnish Cancer Society. Kaiku Health employees were involved in the data acquisition and analysis.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by Northern Ostrobothnia Health District Ethics Committee (9/2017) and registered at the Clinical Trials Register with identifier code NCT03928938 (26 April 2019).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The datasets generated and/or analyzed during the present study are not publicly available but are available from the corresponding author on reasonable request.
Conflicts of Interest
S.I. declares no competing interest. J.E., H.V., and V.K. are employees of Kaiku Health. J.P.K. is an advisor for Kaiku Health.
References
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef] [PubMed]
- Schachter, J.; Ribas, A.; Long, G.V.; Arance, A.; Grob, J.J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.; Lotem, M.; et al. Pembrolizumab versus ipilimumab for advanced melanoma: Final overall survival results of a multicentre, randomised, open-label phase 3 study (KEYNOTE-006). Lancet 2017, 390, 1853–1862. [Google Scholar] [CrossRef]
- Robert, C.; Schachter, J.; Long, G.V.; Arance, A.; Grob, J.J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.; Lotem, M.; et al. Pembrolizumab versus Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2015, 372, 2521–2532. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.S.; Hodi, F.S.; Wolchok, J.D.; Topalian, S.L.; Schadendorf, D.; Larkin, J.; Sznol, M.; Long, G.V.; Li, H.; Waxman, I.M.; et al. Safety Profile of Nivolumab Monotherapy: A Pooled Analysis of Patients with Advanced Melanoma. J. Clin. Oncol. 2017, 35, 785–792. [Google Scholar] [CrossRef] [PubMed]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 1627–1639. [Google Scholar] [CrossRef] [PubMed]
- Brahmer, J.; Reckamp, K.L.; Baas, P.; Crinò, L.; Eberhardt, W.E.; Poddubskaya, E.; Antonia, S.; Pluzanski, A.; Vokes, E.E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 123–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herbst, R.S.; Baas, P.; Kim, D.W.; Felip, E.; Pérez-Gracia, J.L.; Han, J.Y.; Molina, J.; Kim, J.H.; Arvis, C.D.; Ahn, M.J.; et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): A randomised controlled trial. Lancet 2016, 387, 1540–1550. [Google Scholar] [CrossRef]
- Reck, M.; Rodríguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csőszi, T.; Fülöp, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef] [Green Version]
- Rittmeyer, A.; Barlesi, F.; Waterkamp, D.; Park, K.; Ciardiello, F.; von Pawel, J.; Gadgeel, S.M.; Hida, T.; Kowalski, D.M.; Dols, M.C.; et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): A phase 3, open-label, multicentre randomised controlled trial. Lancet 2017, 389, 255–265. [Google Scholar] [CrossRef]
- Bellmunt, J.; de Wit, R.; Vaughn, D.J.; Fradet, Y.; Lee, J.L.; Fong, L.; Vogelzang, N.J.; Climent, M.A.; Petrylak, D.P.; Choueiri, T.K.; et al. Pembrolizumab as Second-Line Therapy for Advanced Urothelial Carcinoma. N. Engl. J. Med. 2017, 376, 1015–1026. [Google Scholar] [CrossRef] [Green Version]
- Tomita, Y.; Fukasawa, S.; Shinohara, N.; Kitamura, H.; Oya, M.; Eto, M.; Tanabe, K.; Kimura, G.; Yonese, J.; Yao, M.; et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2015, 373, 1803–1813. [Google Scholar]
- Balar, A.V.; Castellano, D.; O'Donnell, P.H.; Grivas, P.; Vuky, J.; Powles, T.; Plimack, E.R.; Hahn, N.M.; de Wit, R.; Pang, L.; et al. First-line pembrolizumab in cisplatin-ineligible patients with locally advanced and unresectable or metastatic urothelial cancer (KEYNOTE-052): A multicentre, single-arm, phase 2 study. Lancet Oncol. 2017, 18, 1483–1492. [Google Scholar] [CrossRef]
- Borcoman, E.; Kanjanapan, Y.; Champiat, S.; Kato, S.; Servois, V.; Kurzrock, R.; Goel, S.; Bedard, P.; Le Tourneau, C. Novel patterns of response under immunotherapy. Ann. Oncol. 2019, 30, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Clayton, E.A.; Matyunina, L.V.; McDonald, L.D.; Benigno, B.B.; Vannberg, F.; McDonald, J.F. Machine learning predicts individual cancer patient responses to therapeutic drugs with high accuracy. Sci. Rep. 2018, 8, 16444. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.C.; Naulaerts, S.; Bruna, A.; Ghislat, G.; Ballester, P.J. Predicting Cancer Drug Response In Vivo by Learning an Optimal Feature Selection of Tumour Molecular Profiles. Biomedicines 2021, 9, 1319. [Google Scholar] [CrossRef]
- McKinney, S.M.; Sieniek, M.; Godbole, V.; Godwin, J.; Antropova, N.; Ashrafian, H.; Back, T.; Chesus, M.; Corrado, G.S.; Darzi, A.; et al. International evaluation of an AI system for breast cancer screening. Nature 2020, 577, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Parikh, R.B.; Manz, C.; Chivers, C.; Regli, S.H.; Braun, J.; Draugelis, M.E.; Schuchter, L.M.; Shulman, L.N.; Navathe, A.S.; Patel, M.S.; et al. Machine Learning Approaches to Predict 6-Month Mortality Among Patients with Cancer. JAMA Netw. Open 2019, 2, e1915997. [Google Scholar] [CrossRef] [Green Version]
- Bennett, A.V.; Jensen, R.E.; Basch, E. Electronic patient-reported outcome systems in oncology clinical practice. CA Cancer J. Clin. 2012, 62, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Holch, P.; Warrington, L.; Bamforth, L.C.A.; Keding, A.; Ziegler, L.E.; Absolom, K.; Hector, C.; Harley, C.; Johnson, O.; Hall, G.; et al. Development of an integrated electronic platform for patient self-report and management of adverse events during cancer treatment. Ann. Oncol. 2017, 28, 2305–2311. [Google Scholar] [CrossRef]
- Basch, E.; Artz, D.; Dulko, D.; Scher, K.; Sabbatini, P.; Hensley, M.; Mitra, N.; Speakman, J.; McCabe, M.; Schrag, D. Patient online self-reporting of toxicity symptoms during chemotherapy. J. Clin. Oncol. 2005, 23, 3552–3561. [Google Scholar] [CrossRef]
- Basch, E.; Deal, A.M.; Dueck, A.C.; Scher, H.I.; Kris, M.G.; Hudis, C.; Schrag, D. Overall Survival Results of a Trial Assessing Patient-Reported Outcomes for Symptom Monitoring During Routine Cancer Treatment. JAMA 2017, 318, 197–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denis, F.; Yossi, S.; Septans, A.L.; Charron, A.; Voog, E.; Dupuis, O.; Ganem, G.; Pointreau, Y.; Letellier, C. Improving Survival in Patients Treated for a Lung Cancer Using Self-Evaluated Symptoms Reported Through a Web Application. Am. J. Clin. Oncol. 2017, 40, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Freeman-Keller, M.; Kim, Y.; Cronin, H.; Richards, A.; Gibney, G.; Weber, J.S. Nivolumab in Resected and Unresectable Metastatic Melanoma: Characteristics of Immune-Related Adverse Events and Association with Outcomes. Clin. Cancer Res. 2016, 22, 886–894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanlorenzo, M.; Vujic, I.; Daud, A.; Algazi, A.; Gubens, M.; Alcántara Luna, S.; Lin, K.; Quaglino, P.; Rappersberger, K.; Ortiz-Urda, S. Pembrolizumab Cutaneous Adverse Events and Their Association With Disease Progression. JAMA Dermatol. 2015, 151, 1206–1212. [Google Scholar] [CrossRef] [PubMed]
- Berner, F.; Bomze, D.; Diem, E.; Hasan Ali, O.; Fässler, M.; Ring, S.; Niederer, R.; Ackermann, C.J.; Baumgaertner, P.; Pikor, N. Association of Checkpoint Inhibitor-Induced Toxic Effects With Shared Cancer and Tissue Antigens in Non-Small Cell Lung Cancer. JAMA Oncol. 2019, 5, 1043–1047. [Google Scholar] [CrossRef]
- Scalable, Portable and Distributed Gradient Boosting (GBDT, GBRT or GBM) Library, for Python, R, Java, Scala, C++ and More. Runs on Single Machine, Hadoop, Spark, Flink and DataFlow: Dmlc/Xgboost. Distributed (Deep) Machine Learning Community. 2019. Available online: https://github.com/dmlc/xgboost (accessed on 22 May 2019).
- Iivanainen, S.; Alanko, T.; Peltola, K.; Konkola, T.; Ekström, J.; Virtanen, H.; Koivunen, J.P. ePROs in the follow-up of cancer patients treated with immune checkpoint inhibitors: A retrospective study. J. Cancer Res. Clin. Oncol. 2019, 145, 765–774. [Google Scholar] [CrossRef] [Green Version]
- Iivanainen, S.; Alanko, T.; Vihinen, P.; Konkola, T.; Ekstrom, J.; Virtanen, H.; Koivunen, J. Follow-Up of Cancer Patients Receiving Anti-PD-(L)1 Therapy Using an Electronic Patient-Reported Outcomes Tool (KISS): Prospective Feasibility Cohort Study. JMIR Form. Res. 2020, 4, e17898. [Google Scholar] [CrossRef]
- Iivanainen, S.; Ekstrom, J.; Virtanen, H.; Koivunen, J. Predicting Onset and Continuity of Patient-Reported Symptoms in Patients Receiving Immune Checkpoint Inhibitor (ICI) Therapies Using Machine Learning. Arch. Clin. Med. Case Rep. 2020, 4, 344–351. [Google Scholar] [CrossRef]
- Iivanainen, S.; Ekstrom, J.; Virtanen, H.; Kataja, V.V.; Koivunen, J.P. Electronic patient-reported outcomes and machine learning in predicting immune-related adverse events of immune checkpoint inhibitor therapies. BMC Med. Inform. Decis. Mak. 2021, 21, 205. [Google Scholar] [CrossRef]
- Bates, D.W.; Saria, S.; Ohno-Machado, L.; Shah, A.; Escobar, G. Big data in health care: Using analytics to identify and manage high-risk and high-cost patients. Health Aff. 2014, 33, 1123–1131. [Google Scholar] [CrossRef] [Green Version]
- Dercle, L.; Zhao, B.; Gönen, M.; Moskowitz, C.S.; Firas, A.; Beylergil, V.; Connors, D.E.; Yang, H.; Lu, L.; Fojo, T.; et al. Early Readout on Overall Survival of Patients with Melanoma Treated With Immunotherapy Using a Novel Imaging Analysis. JAMA Oncol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Kotronoulas, G.; Kearney, N.; Maguire, R.; Harrow, A.; Di Domenico, D.; Croy, S.; MacGillivray, S. What is the value of the routine use of patient-reported outcome measures toward improvement of patient outcomes, processes of care, and health service outcomes in cancer care? A systematic review of controlled trials. J. Clin. Oncol. 2014, 32, 1480–1501. [Google Scholar] [CrossRef] [PubMed]
- Shum, B.; Larkin, J.; Turajlic, S. Predictive biomarkers for response to immune checkpoint inhibition. Semin. Cancer Biol. 2021, 478, 31–44. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).