An Integrated Analysis of Metabolomics and Transcriptomics Reveals Significant Differences in Floral Scents and Related Gene Expression between Two Varieties of Dendrobium loddigesii
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Sample Collection
2.2. Gas Chromatography-Mass Spectrometry Analysis of Volatile Compounds in Flowers of Two Varieties
2.3. RNA Sequencing
2.4. Analysis of Differential Expressed Genes (DEGs)
2.5. Quantitative Real-Time PCR (qRT-PCR)
2.6. Combined Analysis of DEGs and DMs
3. Results
3.1. Analysis of Metabolites between Two Aromatic D. loddigesii
3.2. Illumina Sequencing and De Novo Transcriptome Assembly
3.3. Functional Annotation and Classification of Unigenes
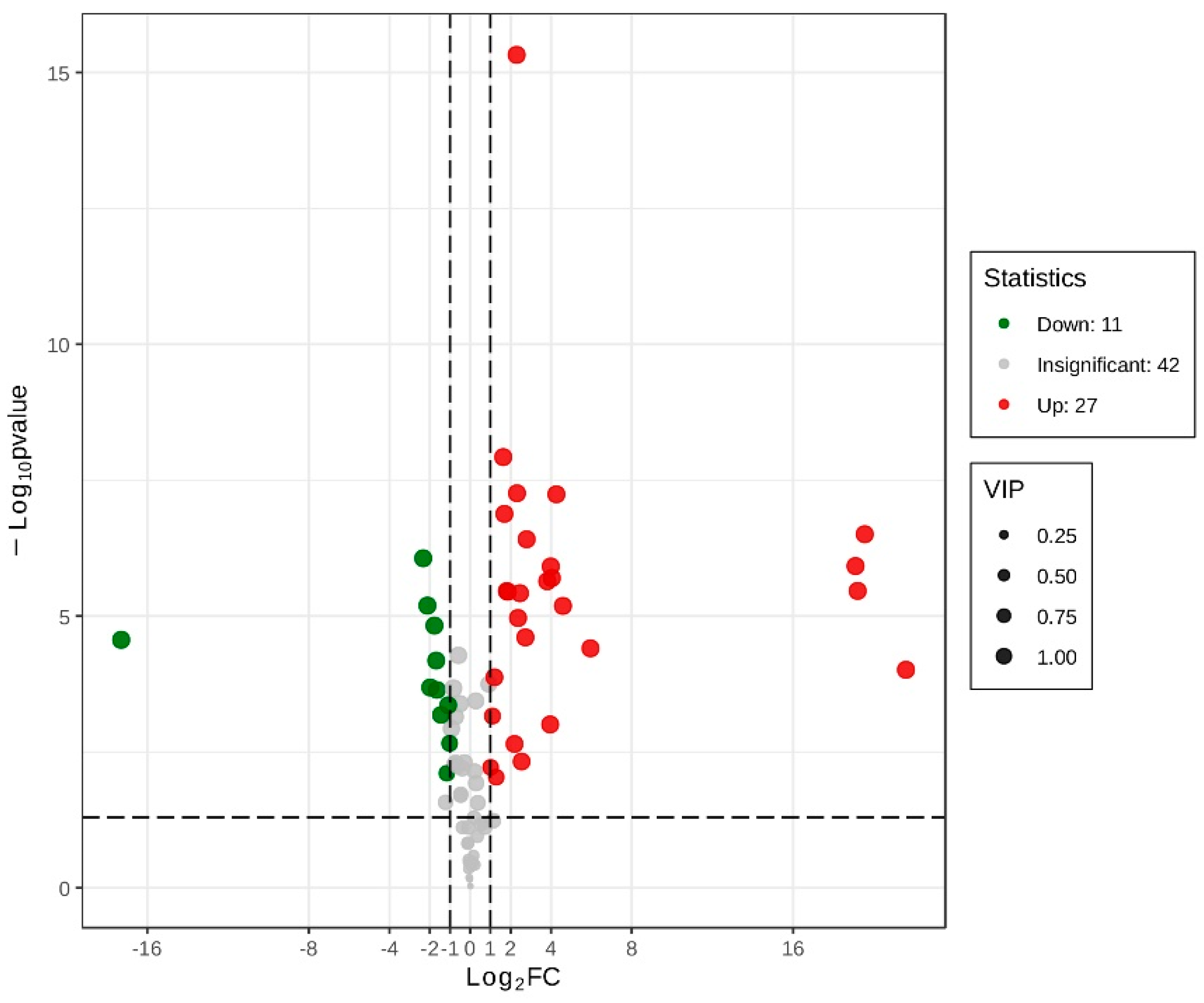
3.4. Functional Assessment and Verification of DEGs
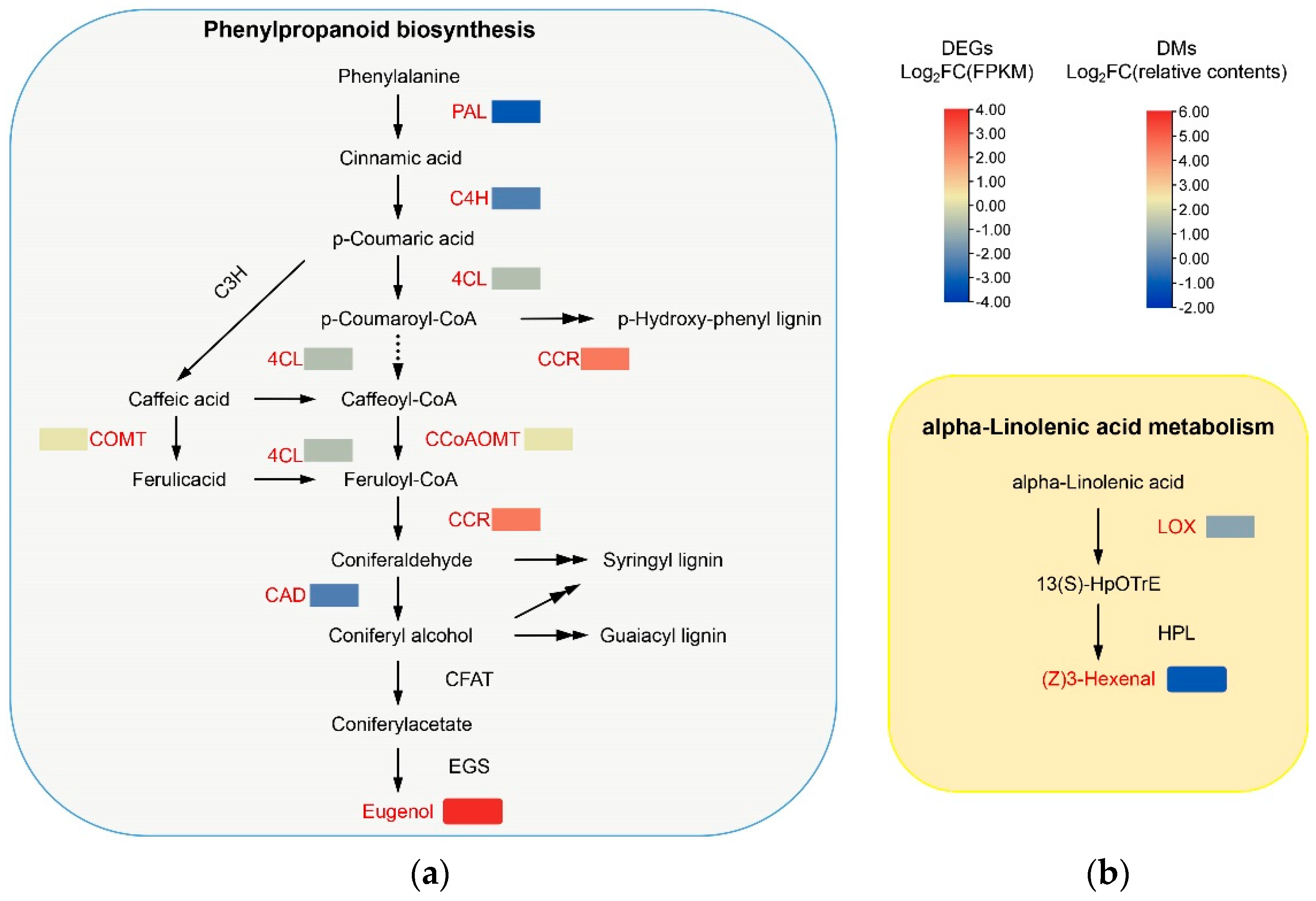
3.5. Combined Analysis of Transcriptome and Metabolome
4. Discussions
4.1. Identification and Comparison of Metabolites between Two Varieties
4.2. Association between Differential Metabolites and Differential Expressed Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chase, M.W.; Cameron, K.M.; Freudenstein, J.V.; Pridgeon, A.M.; Schuiteman, A. An updated classification of Orchidaceae. Bot. J. Linn. Soc. 2015, 177, 151–174. [Google Scholar] [CrossRef] [Green Version]
- Ho, C.K.; Chen, C.C. Moscatilin from the orchid Dendrobrium loddigesii is a potential anticancer agent. Cancer Investig. 2003, 21, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Matsuzaki, K.; Wang, J.; Daikonya, A.; Wang, N.L.; Yao, X.S.; Kitanaka, S. New phenanthrenes and stilbenes from Dendrobium loddigesii. Chem. Pharm. Bull. 2010, 58, 628–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grosso, V.; Farina, A.; Giorgi, D.; Nardi, L.; Diretto, G.; Lucretti, S. A high-throughput flow cytometry system for early screening of in vitro made polyploids in Dendrobium hybrids. Plant Cell Tissue Org. 2018, 132, 57–70. [Google Scholar] [CrossRef]
- Knudsen, J.T.; Eriksson, R.; StaHl, G.B. Diversity and distribution of floral scent. Bot. Rev. 2006, 72, 1. [Google Scholar] [CrossRef]
- Ramya, M.; Jang, S.; An, H.-R.; Lee, S.-Y.; Park, P.-M.; Park, P.H. Volatile Organic Compounds from Orchids: From Synthesis and Function to Gene Regulation. BMC Plant Biol. 2020, 19, 337. [Google Scholar] [CrossRef] [Green Version]
- Pichersky, E.; Dudareva, N. Scent engineering: Toward the goal of controlling how flowers smell. Trends Biotechnol. 2007, 25, 105–110. [Google Scholar] [CrossRef] [Green Version]
- Vainstein, A.; Lewinsohn, E.; Pichersky, E.; Weiss, D. Floral fragrance. New inroads into an old commodity. Plant Physiol. 2001, 127, 1383–1389. [Google Scholar] [CrossRef]
- Huang, X.L.; Zheng, B.Q.; Wang, Y. Study of Aroma Compounds in Flowers of Dendrobium chrysotoxum in Different Florescence Stages and Diurnal Variation of Full Blooming Stage(Article). Forest Res. 2018, 31, 142–149. [Google Scholar] [CrossRef]
- Dudareva, N.; Klempien, A.; Muhlemann, J.K.; Kaplan, I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 2013, 198, 16–32. [Google Scholar] [CrossRef]
- Hendel-Rahmanim, K.; Masci, T.; Vainstein, A.; Weiss, D. Diurnal regulation of scent emission in rose flowers. Planta 2007, 226, 1491–1499. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Yuan, M.; Li, B.; Li, C.; Shi, Q. Variation of floral volatiles and fragrance reveals the phylogenetic relationship among nine wild tree peony species. Flavour Frag. J. 2020, 35, 227–241. [Google Scholar] [CrossRef]
- Qiu, S.; Zheng, W.J.; Xia, K.; Tang, F.L.; Zhao, J.; Ding, L.; Zhao, Z.G. Volatile components in flowers of Dendrobium moniliforme. Guihaia 2019, 39, 1482–1495. [Google Scholar] [CrossRef]
- Cao, H.; Xu, F.; Lu, L.; Ji, Y.L.; Chen, X.; Zhang, B.Q.; Li, H. GC-MS Analysis of Volatile Components in Flowers of Four Kinds of Fragrant Dendrobium Species. Chin. Agric. Sci. Bull. 2021, 37, 56–62. [Google Scholar]
- Yuan, M.Y.; Wang, Y.Q.; Li, Y.Z.; Xie, Y.; Hu, J.M.; Yang, L. Analysis of Aroma Components in Dendrobium loddigesii Rolfe Flower by SPME-GC-MS FLAVOUR FRAGR. Cosmetics 2018, 4, 23–25+69. [Google Scholar]
- Li, N.H.; Dong, Y.X.; Lv, M.; Qian, L.; Sun, X.; Liu, L.; Cai, Y.P.; Fan, H.H. Combined Analysis of Volatile Terpenoid Metabolism and Transcriptome Reveals Transcription Factors Related to Terpene Synthase in Two Cultivars of Dendrobium officinale Flowers. Front. Genet. 2021, 12, 661296. [Google Scholar] [CrossRef]
- Zhao, C.H.; Yu, Z.M.; Silva, J.; He, C.M.; Wang, H.B.; Si, C.; Zhang, M.; Zeng, D.Q.; Duan, J. Functional Characterization of a Dendrobium officinale Geraniol Synthase DoGES1 Involved in Floral Scent Formation. Int. J. Mol. Sci. 2020, 21, 7005. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Mortazavi, A.; Williams, B.A.; Mccue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Dinesh, A.N.P.G. Advances in biosynthesis, regulation, and metabolic engineering of plant specialized terpenoids. plant Sci. 2020, 294, 110457. [Google Scholar] [CrossRef]
- Li, C.H.; Huang, M.Z.; Mo, S.H.; Yin, J.M. Volatile Components in Flowers of Four Dendrobium Species. J.Trop. Subtrop. Bot. 2015, 23, 454–462. [Google Scholar] [CrossRef]
- Krist, S.; Unterweger, H.; Bandion, F.; Buchbauer, G. Volatile compound analysis of SPME headspace and extract samples from roasted Italian chestnuts (Castanea sativa Mill.) using GC-MS. Eur. Food Res. Technol. 2004, 219, 470–473. [Google Scholar] [CrossRef]
- Baek, Y.S.; Ramya, M.; An, H.R.; Park, P.M.; Lee, S.Y.; Baek, N.I.; Park, P.H. Volatiles Profile of the Floral Organs of a New Hybrid Cymbidium, ‘Sunny Bell’ Using Headspace Solid-Phase Microextraction Gas Chromatography-Mass Spectrometry Analysis. Plants 2019, 8, 251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuna, F.S.R.d.; Calevo, J.; Bari, E.; Giovannini, A.; Boselli, C.; Tava, A. Characterization and Antioxidant Activity of Essential Oil of Four Sympatric Orchid Species. Molecules 2019, 24, 3878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feussner, I.; Wasternack, C. The lipoxygenase pathway. Annu. Rev. Plant Biol. 2002, 53, 275–297. [Google Scholar] [CrossRef] [PubMed]
- Gigot, C.; Ongena, M.; Fauconnier, M.L.; Wathelet, J.P.; Thonart, P. The lipoxygenase metabolic pathway in plants: Potential for industrial production of natural green leaf volatiles. Biotechnol. Agron. Soc. 2010, 14, 451–460. [Google Scholar] [CrossRef] [Green Version]
- Dexter, R.; Qualley, A.; Kish, C.M.; Ma, C.J.; Koeduka, T.; Nagegowda, D.A.; Dudareva, N.; Pichersky, E.; Clark, D. Characterization of a petunia acetyltransferase involved in the biosynthesis of the floral volatile isoeugenol. Plant J. 2007, 49, 265–275. [Google Scholar] [CrossRef] [Green Version]






| Sample ID | TH | TL |
|---|---|---|
| Raw reads | 134,806,660 | 136,435,192 |
| Clean reads | 127,791,342 | 130,146,106 |
| Clean bases (Gb) | 19.17 | 19.53 |
| Clean reads rate (%) | 92.28 | 95.23 |
| Clean Q20 bases rate (%) | 97.79 | 97.72 |
| Clean Q30 bases rate (%) | 93.36 | 93.23 |
| Item | Total Number | N50 (bp) | Mean Length (bp) | Total Bases (bp) |
|---|---|---|---|---|
| Transcripts | 230,951 | 1627 | 952 | 219,753,857 |
| Unigenes | 175,089 | 1745 | 1168 | 204,522,529 |
| Database | NR | SwissProt | KEGG | KOG | GO | Total Annotation | Total Unigene |
|---|---|---|---|---|---|---|---|
| Number of annotated unigenes | 120,597 | 70,272 | 86,544 | 64,171 | 71,288 | 121,096 | 175,089 |
| Percentage (%) | 68.88 | 40.14 | 49.43 | 36.65 | 40.72 | 69.16 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Du, Z.; Yang, X.; Wang, L.; Xia, K.; Chen, Z. An Integrated Analysis of Metabolomics and Transcriptomics Reveals Significant Differences in Floral Scents and Related Gene Expression between Two Varieties of Dendrobium loddigesii. Appl. Sci. 2022, 12, 1262. https://doi.org/10.3390/app12031262
Wang S, Du Z, Yang X, Wang L, Xia K, Chen Z. An Integrated Analysis of Metabolomics and Transcriptomics Reveals Significant Differences in Floral Scents and Related Gene Expression between Two Varieties of Dendrobium loddigesii. Applied Sciences. 2022; 12(3):1262. https://doi.org/10.3390/app12031262
Chicago/Turabian StyleWang, Sha, Zhihui Du, Xiyu Yang, Lanlan Wang, Kuaifei Xia, and Zhilin Chen. 2022. "An Integrated Analysis of Metabolomics and Transcriptomics Reveals Significant Differences in Floral Scents and Related Gene Expression between Two Varieties of Dendrobium loddigesii" Applied Sciences 12, no. 3: 1262. https://doi.org/10.3390/app12031262
APA StyleWang, S., Du, Z., Yang, X., Wang, L., Xia, K., & Chen, Z. (2022). An Integrated Analysis of Metabolomics and Transcriptomics Reveals Significant Differences in Floral Scents and Related Gene Expression between Two Varieties of Dendrobium loddigesii. Applied Sciences, 12(3), 1262. https://doi.org/10.3390/app12031262






