Abstract
Bone age is a common indicator of children’s growth. However, traditional bone age assessment methods usually take a long time and are jeopardized by human error. To address the aforementioned problem, we propose an automatic bone age assessment system based on the convolutional neural network (CNN) framework. Generally, bone age assessment is utilized amongst 0–18-year-old children. In order to reduce its variation in terms of regression model building, our system consists of two steps. First, we build a maturity stage classifier to identify the maturity stage, and then build regression models for each maturity stage. In this way, assessing bone age through the use of several independent regression models will reduce the variation and make the assessment of bone age more accurate. Some bone sections are particularly useful for distinguishing certain maturity stages, but may not be effective for other stages, and thus we first perform a rough classification to generally distinguish the maturity stage, and then undertake fine classification. Because the skeleton is constantly growing during bone development, it is not easy to obtain a clear decision boundary between the various stages of maturation. Therefore, we propose a cross-stage class strategy for this problem. In addition, because fewer children undergo X-rays in the early and late stages, this causes an imbalance in the data. Under the cross-stage class strategy, this problem can also be alleviated. In our proposed framework, we utilize an MSCS-CNN (Multi-Step and Cross-Stage CNN). We experiment on our dataset, and the accuracy of the MSCS-CNN in identifying both female and male maturity stages is above 0.96. After determining maturity stage during bone age assessment, we obtain a 0.532 and 0.56 MAE (mean absolute error) for females and males, respectively.
1. Introduction
Bone age assessment is a method used to assess whether children are growing normally during development, and identify problems such as endocrine-system-related diseases. It judges the development of children by observing the growth of bones. Since the palm is a convenient place to photograph and observe the growth of bones, we usually use a palm X-ray to assess bone age. Traditionally, the two most widely used clinical methods for bone age assessment are the Greulich–Pyle (GP) atlas method [1] and the Tanner–Whitehouse (TW) method [2]. The GP method uses a template comparison method to give a bone age assessment value, and is undertaken by professionals. Therefore, the GP method relies on professional employees, and because it is a manual evaluation, there can be some human error. In addition, the TW method scores each bone block separately and then combines the scores of these bone blocks to give the bone age, and the fixed scoring method may be such that there are errors amongst different races. Races in different regions are unlikely to provide a more comprehensive evaluation than using a fixed set of parameters. According to the shortcomings of the two aforementioned widely used methods for bone age assessment, our purpose is to develop an automatic bone age assessment system that is automatic, simple to use, less labor-intensive, and for a single ethnic group (in this system, we use Taiwanese children as our experimental samples for our system). In addition, a CNN is currently the most powerful method of image recognition; thus, this system is built based on a CNN. A CNN can learn the correspondence between input and output after a large amount of parameter training. Due to the development of GPUs, a large amount of parameter training is not a problem. In recent years, CNNs have achieved considerable success in many computer vision applications. For example, many studies have researched the application of CNN technology in medical X-ray film recognition [3,4,5,6,7].
To solve the disadvantages of the two traditional methods (GP and TW) mentioned above, such as human error, labor intensity, and a lack of flexibility in scoring methods, in this paper we use a CNN to construct an automatic bone age assessment system. Because a CNN assesses bone age “automatically” via complex functions and weights, and since the weights of the network are unchanged after training, this can also reduce the human error problem. Finally, since the network weight is for a specific race (this article takes the Taiwanese race as an example), if other races are used as the training set, the network weight dedicated to a specific race can be easily reached; thus, the CNN is used to build a bone age assessment that can easily adjust the model parameters for different races. Due to the aforementioned advantages of a CNN, the use of a CNN can be very suitable for the needs of the aforementioned bone age assessment system.
In past research, Lee et al. [8] built a preprocessing engine to remove the background first, and then input the entire palm to CNN for classification, regarding each age as a class. With regard to the details of their CNN, under the leverage of parameters and accuracy, they chose GoogleNet as their classification model and used transfer learning and data augmentation to enhance the model performance. Spampinato et al. [9] constructed a bone age assessment system called BoNet, and determined their network architecture by experimenting with combinations of several parameters. Moreover, they introduced the deformation layer, as well as transfer learning, to improve the model performance. Unlike [9], they did not define the classification, but directly regressed the bone age in the output layer. Zhao et al. [10] constructed a segmentation system to remove the back-ground, and then used a generative adversarial network to enhance the image. They used Xception as their network architecture and proposed a transfer learning method called paced transfer learning. Furthermore, they used regression to assess bone age. Liu et al. [11] generated multi-scale images, used multiple VGG16 branches to extract multi-scale features and, finally, fused the features of each branch to regress bone age. Ren et al. [12] utilized attention maps to make the model more focused on the important parts of the palm, and then used Inception-v3 to regress bone age. Larson et al. [13] and Chen et al. [14] both used ResNet as their architecture and both utilized classification models, the former of which was classified in one-month intervals, while the latter was classified in one-year intervals.
The aforementioned studies all use the entire X-ray film as the input. In order to make the model more focused on key parts during bone age assessment, we will review several ROI-based studies. Son et al. [15] first assessed fingers and wrists as bROIs, and then used Faster R-CNN to extract 13 ROIs from the bROIs. Each ROI had an independent CNN model (VGG16). Finally, they averaged 13 ROI results for bone age, and each ROI CNN model classified the maturity levels of the ROI, rendering a total of nine maturity stages (A–I). Liang et al. [16] directly added the bone age regression network to Faster R-CNN, and their loss function contained three items: ROI class, coordinates, and bone age value. Bui et al.’s [17] method was similar to the aforementioned research, and used Faster R-CNN to extract eight ROIs. After extracting the ROIs, they trained Inception-v4 separately for different bone sections to classify ROI maturity. There were eight stages of maturity: A–I. Thereafter, they utilized Inception-v4 features to train support vector regression to regress bone age. Koitka et al. [18] used Faster R-CNN to extract the ROIs. Then, they used ResNet50 to regress bone age. For each gender, there were six regression networks, with the six models corresponding to six parts, respectively, i.e., distal phalanges, intermediate phalanges, proximal phalanges, carpal bones, radius bones, and ulna bones. He et al. [19] selected 14 ROIs based on the hands and wrists of the Chinese, and used these 14 ROIs to build a CNN model (AlexNet) to regress bone age. For each part, they took the first two highest probability stages to calculate a weighted score, and then the weighted scores of 14 parts to obtain the grade score, followed by mapping the grade score to the bone age comparison table to obtain the bone age. We chose 11 ROIs in our study, as shown in Figure 1.
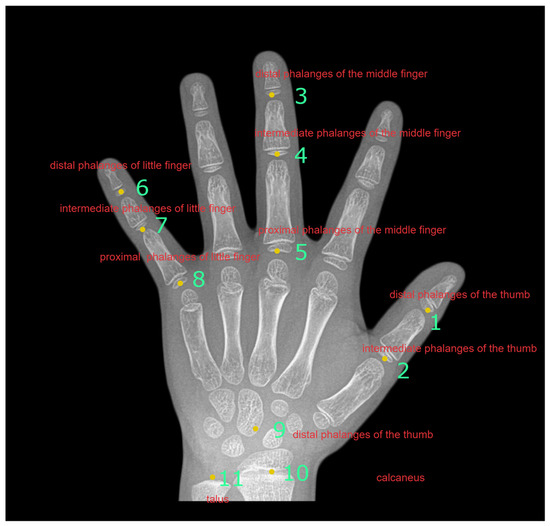
Figure 1.
Eleven bone ROIs used in this paper.
As mentioned above, using some specific blocks to assess bone age can eliminate unnecessary blocks before the CNN model makes predictions. In our system, we use 11 bones for bone age assessment: epiphyses in the distal phalanges of the thumb; epiphyses in the intermediate phalanges of the thumb; epiphyses in the distal phalanges of the middle finger; epiphyses in the intermediate phalanges of the middle finger; epiphyses in the proximal phalanges of the middle finger; epiphysis of the distal phalanx of the little finger; epiphysis of the middle phalanx of the little finger; epiphysis of the proximal phalanx of the little finger; carpal bones; ulna bones; and radius bones. For convenience, we add serial numbers to them in the order of B1–B11, as shown in Figure 1. In this article, we use the manual picking-up method. After the user clicks the center point of the block to be selected, the system will automatically cut out a fixed-size block; thus, this semi-automatic cutting method requires minimal labor, and avoids errors caused by automatic cutting. Incidentally, the selection of these 11 bones is based on the configuration of [20]; while [20] considers carpal bones, ulna bones, and radius bones as ROIs, we refer to [21] for carpal bones, ulna bones, and radius bones. These are divided into three ROIs to study the significance of these bones. There are two reasons for using the manual method. First, there are some errors in the automatic segmentation method. These errors will seriously affect the subsequent bone age calculation. Second, clicking is not a time-consuming task. In our method, only 11 mouse clicks are required, and can be completed in a few seconds at most.
Most of the aforementioned studies utilize either classification models or regression models. As for classification models, if the interval stage is too large, it will not be accurate enough. If the interval is too small, it will be more difficult to fit the model in model training, and more data will be required to train the model. Regarding regression models, a too-large bone age range may be more difficult to train. Therefore, we use a two-stage method comprising classification and then regression to build our bone age assessment system.
The remaining sections of this paper are as follows: We review CNN-related technologies and research on CNN-based automatic bone age assessment in Section 2, and detail the MSCS-CNN (Multi-Step and Cross-Stage CNN) in Section 3. Various experiments are compared in Section 4. Finally, we discuss our method and draw conclusions in Section 5.
2. Convolutional Neural Network Framework
2.1. Convolutional Neural Network Framework
In recent years, there have been reviews of several well-known convolutional neural network architectures [22,23,24,25,26]. Regardless of the architecture, layers are connected in different ways in order to increase the performance of the model. However, no matter how the architecture changes, there are two parts that will not change: first, using convolution kernels to extract features, and second, inputting features to the fully connected layer for various tasks.
2.1.1. Convolutional Layer
A convolutional layer mainly uses 2D convolutional operations on images (or any two-dimensional data) to extract features. Specifically, the convolution operation is as follows:
where is the -th pixel value on one of the feature maps on the layer, (.) is a non-linear activation function, is the number of kernels on layer , is the window size of kernels, and is the -th kernel’s weight on the -th kernel on layer .
2.1.2. Fully Connected Layer
After the convolutional layer extracts the features, it will input features into the fully connected layer to adapt to various tasks. The operation on the fully connected layer is as follows:
where is the -th neuron value on layer , (.) is a non-linear activation function, is the number of neurons on layer , and is the weight connecting the -th neuron on layer and the -th neuron on layer .
In summary, the basic architecture of the CNN comprises a convolutional layer, and a fully connected layer will be connected after the convolutional layer. A basic CNN architecture is shown in Figure 2.
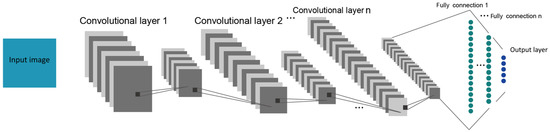
Figure 2.
CNN general architecture.
3. Materials and Methods
3.1. Dataset
The dataset used in this paper contains 2000 left-hand X-rays of children aged 0–18 years old. All X-ray films are provided by Cheng Hsin General Hospital. According to the maturity stages outlined in Ref. [27], the data for each stage are shown in Table 1. Ground truths were determined by 2 doctors using the GP method and were averaged.

Table 1.
Stage distribution of our dataset.
For the semi-automatic cutting of the B1–B8, B10, and B11 ROIs, we expand the manually clicked center point by 32 pixels on either side, so each block is 64×64 pixels, and B9 is the center point on both sides with 256 pixels facing outward. Thus, B6 is 512×512 pixels in size. In order to complete the ROI, including the bones, in B1 and B2 we use the connection between these two points as the axis of rotation and turn the connection to 90 degrees before cutting. Similarly, B3, B4 and B5 (B6, B7 and B8) use the connection between B3 (B6) and B5 (B8) to correct the image by turning the connection to 90 degrees, and B9, B10 and B11 use the connection between B10 and B11 to rotate to 0 degrees.
3.2. Framework of the MSCS-CNN
The first problem that we came across with our model was how to improve the accuracy of the CNN. Bone age assessment is generally used for humans aged 0–18 years. According to our experiment (see Section 4), we tried to fit all bone ages into one regression model. As bone age has large variation, this could have resulted in large error in the building of the regression model. In order to reduce the variation, we divide the X-ray film into several stages. A regression model will be built in each stage so that, in each regression model, only a small range of bone ages needs to be fitted.
At present, there needs to be a classifier that can accurately determine the maturity stage. Child development is a continuous process and there is no obvious decision boundary. Therefore, in this step, we propose a cross-stage class strategy for an intermediate stage which will exist in both the previous stage and the next stage. Ref. [27] shows that bone age can be divided into seven stages: infancy, toddlers, pre-puberty, early, mid-puberty, late puberty, and post-puberty. For convenience, we denote these stages as stages A to F. In our cross-stage class strategy mentioned above, we will convert the maturity stages into AB, BC, CD, DE, and EF. To illustrate this strategy, we take the B stage as an example for the AB and BC stages. The subsequent experiments will also show that the cross-stage strategy is not only helpful for bone age assessment, but also alleviates the problem of data imbalance.
The MSCS-CNN consists of three steps, as shown in Figure 3. Step 1 to step 3 involve coarse maturity stage classification, fine maturity stage classification and bone age regression. We will describe the details of each stage in the subsequent sections. In addition, the CNN model architectures that we used in the three steps are mostly the same. The only differences between the three-step models are the number of nodes and the activated function of the output layer.
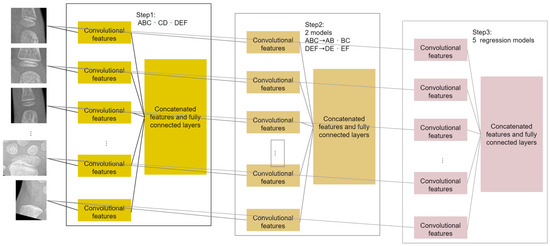
Figure 3.
Framework of the MSCS-CNN.
3.3. Step 1 of MSCS-CNN: Rough Maturity Stage Classification
If an error occurs in the classification step, it directly affects the accuracy of the subsequent bone age assessment and causes a larger bone age error. Therefore, we need to be as accurate as possible in the stage-classification step. Therefore, we use a two-step stage-classification strategy. In step 1, we perform rough maturity stage classification and divide the six stages of A–F into three rough maturity stages: ABC, CD and DEF.
In this step, we need to clearly separate the AB and EF stages, because as shown in Table 1, there are fewer data at these two ends. If directly using one-step classification will cause the CNN model to be poor in assisting the two end stages, and if the AB stage is misjudged to be the EF stage, the assessed bone age will be much larger than the ideal bone age value. On the contrary, if the EF stage is misjudged to be the class AB stage, the assessed bone age will be much smaller than the ideal bone age value. For these reasons, at this stage we need to make the AB and EF stages clear and distinct.
Therefore, we use the C stage and the D stage to augment data for the stages at both ends. As shown in Table 1, since the number of C and D stages is significantly greater than the number at both ends of the stage, we adopt this strategy. Taking the male data as an example, at this time there are 497 data in the ABC stage, 875 in the CD stage, and 574 in the DEF stage. This makes the trained CNN model distinguishable for the two end stages.
3.4. Step 2 of MSCS-CNN: Fine Maturity Stage Classification
We further subdivide the rough stages to reduce the variance between each stage in the regression model in the next step. In this step, we further divide the ABC stage into the AB and BC stages and divide the DEF stage into the DE and EF stages, the CD stage is not subdivided. After this step, we have a total of five fine stages: AB, BC, CD, DE and EF.
3.5. Step 3 of MSCS-CNN: REGRESSION MODEL
In this step, we will use the CNN model to assess the bone age value. Our bone age value is in years and includes a decimal point (e.g., 6.5 years denotes 6 years and 6 months). We build five independent models for the five fine stages in step 2. Before training the CNN regression model, we normalize the bone age value to make the range between 0 and 1. We use the following equation for normalization:
where represents the value of the original bone age after normalization, and represents belonging to the stages of step 2. In our data, the and values of each stage are shown in Table 2.

Table 2.
Bone age range of each stage.
After the regression model training is completed, for testing, we only need to return to Equation (3) to obtain x. We re-represent as the , and as the , and rewrite Equation (3) as follows:
3.6. CNN-based Automatic Bone Age Assessment
Our CNN architecture and its parameters are shown in Table 3. Our model is based on the residual convolution [24]. B1–B8 and blocks B10 and B11 use three layers of residual blocks and rescale the input to a 32×32 square image. Block 6 is relatively large, so we treat this block using a different configuration. We rescale block 6 to 256×256 and use five layers of residual blocks to extract features. After extracting 11 bone features, we concatenate them together. B1–B8 and B10 and B11 each extract 256 features. Block 9 extracts 512 features, and concatenating the 11 bone block features produces a total of 3072 features, which are then connected to the output by a fully connected layer. According to the target of the output layer, we use different numbers of neurons and different activation functions in the output. In our model, the softmax function is used for maturity stage classification (step 1 and step 2), and the sigmoid function is used for bone age regression (step 3).

Table 3.
Bone age range of each stage.
3.7. Using the Cross-Stage Strategy to Avoid Over-Training
In order to explain why we use a cross-stage strategy in the classification step, we first need to introduce the loss function used in stage identification (step 1 and step 2): categorical hinge loss (). is an extension of hinge loss (HL) [28]. While HL forces the classifier to learn the maximum margin, is a multi-class version of HL. Recently, many deep learning networks have used to learn models [29,30]. is mainly composed of classification correctness items and penalty items . Its definition is observed in Equations (5)–(7).
In Equations (5)–(7), denotes the number of classes, and both and represent the confidence in class , the former of which is ground truth, while the latter is that of the currently predicted results. For Equation (5), a higher value indicates that the sample has lower ambiguity; for Equation (6), a smaller value also makes the sample less ambiguous.
At this point, let us consider a case. Assuming that the prediction result of stage B at an epoch is that the confidence in both stages B and C is 0.5, as mentioned above, it is difficult to define a clear decision boundary for bone growth. Therefore, it may be acceptable to identify B as C at this time. At this time, if we continue to learn the model parameters, we may learn some unimportant features (such as noise) that easily lead to over-fitting, which will harm the accuracy during testing. Therefore, we hope that in this case, the loss function is as small as possible (the smaller the loss, the slighter the parameter adjustment). Referring to Equations (5)–(7), if the cross-stage strategy is not used, the will be 0.5 and the will be 0.5. The is 1, which will cause the model to continue to learn from these data. When adopting a cross-stage strategy, the will be 1 and the will be 0. The is 0; therefore, at this time, it will not continue to learn from these data, which will alleviate the over-fitting. We can be more general as long as B’s confidence is fully allocated to AB and BC, no matter how high AB and BC are. As long as the sum of the two is 1, the is 0. Because it is assumed in the pos that the data are stage B, we can rewrite Equation (5) as Equation (8). At this time, as long as the confidence is completely allocated to AB and BC, the is always equal to 1 and the is always 0, and the is also equal to 0.
In summary, our strategy is to create some stage data. As long as it is distinguished from the neighboring stage, we have acceptable results, and the aforementioned also shows that the cross-stage classification strategy can meet our needs when using .
Another advantage of the cross-stage classification strategy is that data such as X-rays often have data imbalances. We can use a large number of stages to augment data for a small number of stages. For example, stage C can help the learning of neurons connected to the BC stage, which we call neighbor stage data augmentation.
3.8. Evaluation Metrics
We evaluate the maturity stage classification effects of step 1 and step 2 using accuracy as an evaluation metric. The accuracy () is calculated using Equation (9), and the bone age assessment error is calculated using the mean absolute error () as Equation (10). A BA is the ground truth of bone age, while BA is the bone age assessed by means of our method.
4. Experiments
4.1. One-Step Bone Age Regression
In the MSCS-CNN, we first classify the maturity stage and then use the corresponding regression model to assess bone age according to the maturity stage classification results. In order to illustrate the use of maturity stage classification, we compare the results of a three-stage MSCS-CNN and only use step 3 to directly regress bone age, as shown in Table 4. The MAE is quite poor without classifying the maturity stage first. The biggest reason is that mentioned above. It is a more difficult task to fit a single network to a larger range of values. Based on this comparison, we illustrate the need for a maturity stage classification.

Table 4.
Comparing bone age assessment performance with and without maturity stage classification.
4.2. The Effect of Rough–Fine Classification
In order to reduce the bone age assessment error caused by classification errors, we present a two-step classification flow. To illustrate the effect of our approach, Table 5 compares the accuracy and bone age assessment difference between one-step stage classification and two-step stage classification.

Table 5.
Comparison of one- and two-stage classification performance.
It can be found that the effect of the two-step classification is much more accurate. This is because the difference between certain bone blocks in each stage is very small. If the classification is completed directly by one stage, it may cause a bone block that confuses the stage with other blocks. Moreover, Table 5 shows that if the maturity stage classification is wrong, the error of bone age will also be very large.
We believe that multi-stage classification is beneficial to the overall model performance and, in the following, will explain why two-stage classification is chosen in the MSCS-CNN. Therefore, we experiment with adding a stage before and after these two stages, which we call three-stage classification config. 1 and 2, respectively. In three-stage classification config. 1, ABCD and CDEF classifiers are added before step 1 of the MSCS-CNN. In three-stage classification config. 2, the A-F maturity stages are separated after step 2 of the MSCS-CNN. In classification config. 1, it is not difficult to classify ABCD and CDEF, and for step 1 of the MSCS-CNN, AB and EF can be effectively classified, so adding this step will not help the overall bone age assessment. For three-stage classification config. 2, as we will explain later, it is not an easy task to classify the various maturity stages perfectly, so adding this layer is harmful to the overall bone age assessment. Therefore, we choose two-step classification as the most appropriate. The classification effects of the above two configurations are shown in Table 6.

Table 6.
Two 3-stage classification accuracies.
4.3. The Effect of Cross-Stage Classification
As mentioned in Section 3.7, our cross-stage strategy can help us in stages. Here, we use experiments to test cross-stage strategy effects. We experiment on two configurations without cross-stages: config. 1 comprises AB, CD and EF, whereas config. 2 comprises A, B, C, D, E and F. Furthermore, we compare the staged classification accuracy and MAE, such as Equations (9) and (10), as shown in Table 7. It can be found that the classification accuracy under the non-cross-stage strategy is low, which means that it is difficult for us to clearly separate adjacent categories. An incorrect maturity stage classification result leads to the inaccurate assessment of bone age.

Table 7.
With and without cross-stage strategy performance comparison.
4.4. Maturity Stage Classification Configuration
As mentioned in Section 2, maturity stage classifications that are too rough or too fine have advantages and disadvantages. In this paper, the two maturity stages in Ref. [27] are merged into one maturity stage. In order to illustrate whether the fine class maturity stage is configured as in Ref. [27], we perform an experiment. We assume completely correct classification (100% accuracy) for these six stages, and then run the regression model independently. Its MAE is shown in the first row of Table 7, such an error is not much better than the performance of MSCS-CNN in this paper. Additionally, we have to consider that using a CNN model to perfectly separate the six mature stages for classification is a difficult task (as shown in Config 2 of Table 7, poor staging ability leads to the poor performance of bone age MAE), so we consider combining the two for one.
In addition, since the accuracy estimation of the MSCS-CNN in the maturity stage is relatively loose, for example, the C stage can be identified in the BC stage or the CD stage, we think it is correct, so it is unfair to use the accuracy comparison. Therefore, we perform another experiment to compare whether our cross-stage strategy is really helpful for the bone age assessment task, and we also assume that Config. 1 mentioned in Section 4.3 has completely correct staging, its MAE is as shown in the second row of Table 8, and its MAE is not much better than the MSCS-CNN. Thus, we refer to Table 5. If the cross-stage strategy is not used, it is difficult to separate them clearly to achieve good maturity stage classification performance, which also leads to the terrible MAE of Config. 1 in Table 7. Config. 2 assumes that the maturity stages A-F are individually separated, as mentioned in Section 4.3. We assume that the MAE results under the perfect classification of Config. 2 are shown in the row of Config. 2 in Table 7. Compared with Table 7 Config. 1, there is not much benefit. This result shows that we do not need to separate the two adjacent maturity stages clearly. Thus, we use a cross-stage strategy as a compromise.

Table 8.
MAE under perfect maturity stage classification.
4.5. Bone Age Assessment in Each Stagecs
In this section, we summarize the experimental results of bone age assessment under the framework that we have built. We have previously shown in Figure 4 that our frame-work, with respect to classification accuracy, demonstrates 1.0 and 0.96 for females and males, respectively, and provides 0.532 and 0.56 MAEs for females and males, respectively. Moreover, in this section, since the goal is to assess bone age, we will discuss in detail the performance of the framework that we have proposed. It can be observed that the overall MAE is good, and that our method obtains good MAE at all stages. This means that our system offers optimal prediction for children at all stages.
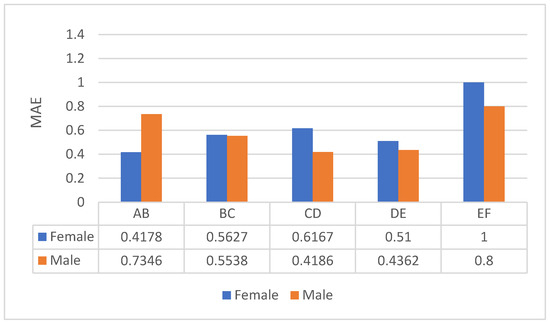
Figure 4.
Bone age assessment results for each stage on Stagecs.
4.6. Comparison with Other Bone Age Assessment Methods
Finally, we use the dataset in this article to compare several existing bone age assessment methods, and compare the entire image as the input for the model [8,10] and ROI-based methods [17,18]. The results are shown in Table 9, and it can be seen that our results are better than those in the existing literature. The main reason is that most of their models have better performance under data balance, but do not deal with data imbalance. The cross-stage strategy proposed in this article can indeed alleviate this problem.

Table 9.
MAE comparison with other bone age assessment methods.
5. Conclusions
Since the assessment of bone age is seen in many clinical applications amongst children, in order to make it easier to assess bone age, an automatic bone age assessment system is necessary. A CNN can meet these requirements. In order to make a CNN model more accurate for the assessment of bone age, this article proposes a multi-step framework. First, for the stage classifier, through a two-step method of rough stage classification and fine stage classification, stage identification can be as accurate as possible. In addition, due to the ambiguity between the stages, we also propose a cross-stage class strategy, combining cross-stage classes and cross-step strategies. Among females and males, the accuracy is 1.0 and 0.96, respectively, which makes the establishment of the subsequent bone age regression model more accurate. In order to prove that cross-stage classification has its advantages, we have also proven its effectiveness. This article is based on children in Taiwan. Generally speaking, children in different regions will have different developmental conditions due to factors such as race and diet. A simple solution is to build models for specific races. However, there are several shortcomings to this approach. First, it is time-consuming and labor-intensive, due to having to establish one’s own model and database. In addition, as the years go by, diets and living environments will change. This will cause the model to fail over time; therefore, building a universal bone age prediction model will be an important focus of future work.
Author Contributions
Software and Writing—original draft, C.-T.P.; Methodology, Supervision and Writing—review & editing, Y.-K.C. and S.-S.Y.; Resources, Validation, Y.-S.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Cheng Hsin General Hospital, Taipei City 112, Taiwan (CHGH109- (N)15).
Institutional Review Board Statement
This research was approved by the Institutional Review Board of the Cheng Hsin General Hospital, Taipei City 112 (IRB No. CHGH-IRB (735)108A-51).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data supporting this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Garn, S.M. Radiographic atlas of skeletal development of the hand and wrist. Am. J. Hum. Genet. 1959, 11, 282. [Google Scholar]
- Tanner, J.M.; Healy, M.J.R.; Cameron, N.; Goldstein, H. Assessment of Skeletal Maturity and Prediction of Adult Height (tw3 Method); W.B. Saunders: Philadelphia, PA, USA, 2001. [Google Scholar]
- Tanzi, L.; Vezzetti, E.; Moreno, R.; Aprato, A.; Audisio, A.; Massè, A. Hierarchical fracture classification of proximal femur X-ray images using a multistage deep learning approach. Eur. J. Radiol. 2020, 133, 109373. [Google Scholar] [CrossRef] [PubMed]
- Guan, B.; Zhang, G.; Yao, J.; Wang, X.; Wang, M. Arm fracture detection in X-rays based on improved deep convolutional neural network. Comput. Electr. Eng. 2020, 81, 106530. [Google Scholar] [CrossRef]
- Urban, G.; Porhemmat, S.; Stark, M.; Feeley, B.; Okada, K.; Baldi, P. Classifying shoulder implants in X-ray images using deep learning. Comput. Struct. Biotechnol. J. 2020, 18, 967–972. [Google Scholar] [CrossRef]
- Rahman, T.; Chowdhury, M.E.; Khandakar, A.; Islam, K.R.; Islam, K.F.; Mahbub, Z.B.; Kadir, M.A.; Kashem, S. Transfer learning with deep convolutional neural network (CNN) for pneumonia detection using chest X-ray. Appl. Sci. 2020, 10, 3233. [Google Scholar] [CrossRef]
- Dang, J.; Li, H.; Niu, K.; Xu, Z.; Lin, J.; He, Z. Kashin-beck disease diagnosis based on deep learning from hand X-ray images. Comput. Methods Programs Biomed. 2021, 200, 105919. [Google Scholar] [CrossRef]
- Lee, H.; Tajmir, S.; Lee, J.; Zissen, M.; Yeshiwas, B.A.; Alkasab, T.K.; Choy, G.; Do, S. Fully automated deep learning system for bone age assessment. J. Digit. Imaging 2017, 30, 427–441. [Google Scholar] [CrossRef]
- Spampinato, C.; Palazzo, S.; Giordano, D.; Aldinucci, M.; Leonardi, R. Deep learning for automated skeletal bone age assessment in X-ray images. Med. Image Anal. 2017, 36, 41–51. [Google Scholar] [CrossRef]
- Zhao, C.; Han, J.; Jia, Y.; Fan, L.; Gou, F. Versatile framework for medical image processing and analysis with application to automatic bone age assessment. J. Electr. Comput. Eng. 2018, 2018, 2187247. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, C.; Cheng, J.; Chen, X.; Wang, Z.J. A multi-scale data fusion framework for bone age assessment with convolutional neural networks. Comput. Biol. Med. 2019, 108, 161–173. [Google Scholar] [CrossRef]
- Ren, X.; Li, T.; Yang, X.; Wang, S.; Ahmad, S.; Xiang, L.; Stone, S.R.; Li, L.; Zhan, Y.; Shen, D.; et al. Regression convolutional neural network for automated pediatric bone age assessment from hand radiograph. IEEE J. Biomed. Health Inform. 2018, 23, 2030–2038. [Google Scholar] [CrossRef] [PubMed]
- Larson, D.B.; Chen, M.C.; Lungren, M.P.; Halabi, S.S.; Stence, N.V.; Langlotz, C.P. Performance of a deep-learning neural network model in assessing skeletal maturity on pediatric hand radiographs. Radiology 2018, 287, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, J.; Zhang, Y.; Lu, Y.; Liu, S. Automatic feature extraction in X-ray image based on deep learning approach for determination of bone age. Future Gener. Comput. Syst. 2020, 110, 795–801. [Google Scholar] [CrossRef]
- Son, S.J.; Song, Y.; Kim, N.; Do, Y.; Kwak, N.; Lee, M.S.; Lee, B.-D. Tw3-based fully automated bone age assessment system using deep neural networks. IEEE Access 2019, 7, 33346–33358. [Google Scholar] [CrossRef]
- Liang, B.; Zhai, Y.; Tong, C.; Zhao, J.; Li, J.; He, X.; Ma, Q. A deep automated skeletal bone age assessment model via region-based convolutional neural network. Future Gener. Comput. Syst. 2019, 98, 54–59. [Google Scholar] [CrossRef]
- Bui, T.D.; Lee, J.-J.; Shin, J. Incorporated region detection and classification using deep convolutional networks for bone age assessment. Artif. Intell. Med. 2019, 97, 1–8. [Google Scholar] [CrossRef]
- Koitka, S.; Kim, M.S.; Qu, M.; Fischer, A.; Friedrich, C.M.; Nensa, F. Mimicking the radiologists’ workflow: Estimating pediatric hand bone age with stacked deep neural networks. Med. Image Anal. 2020, 64, 101743. [Google Scholar] [CrossRef]
- He, M.; Zhao, X.; Lu, Y.; Hu, Y. An improved alexnet model for automated skeletal maturity assessment using hand X-ray images. Future Gener. Comput. Syst. 2021, 121, 106–113. [Google Scholar] [CrossRef]
- Giordano, D.; Spampinato, C.; Scarciofalo, G.; Leonardi, R. An automatic system for skeletal bone age measurement by robust processing of carpal and epiphysial/metaphysial bones. IEEE Trans. Instrum. Meas. 2010, 59, 2539–2553. [Google Scholar] [CrossRef]
- Guo, L.; Wang, J.; Teng, J.; Chen, Y. Bone age assessment based on deep convolutional features and fast extreme learning machine algorithm. Front. Energy Res. 2022, 9, 888. [Google Scholar] [CrossRef]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Processing Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A.A. Inception-v4, inception-resnet and the impact of residual connections on learning. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Gilsanz, V.; Ratib, O. Hand Bone Age: A Digital Atlas of Skeletal Maturity; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2005. [Google Scholar]
- Rosasco, L.; de Vito, E.; Caponnetto, A.; Piana, M.; Verri, A. Are loss functions all the same? Neural Comput. 2004, 16, 1063–1076. [Google Scholar] [CrossRef] [PubMed]
- Rajaraman, S.; Zamzmi, G.; Antani, S. Multi-loss ensemble deep learning for chest X-ray classification. arXiv 2021, arXiv:2109.14433. [Google Scholar]
- Díaz-Vico, D.; Fernández, A.; Dorronsoro, J.R. Companion losses for deep neural networks. In Proceedings of the International Conference on Hybrid Artificial Intelligence Systems, Bilbao, Spain, 22–24 September 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 538–549. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).