Off-Axis Holographic Interferometer with Ensemble Deep Learning for Biological Tissues Identification
Abstract
1. Introduction
2. Method
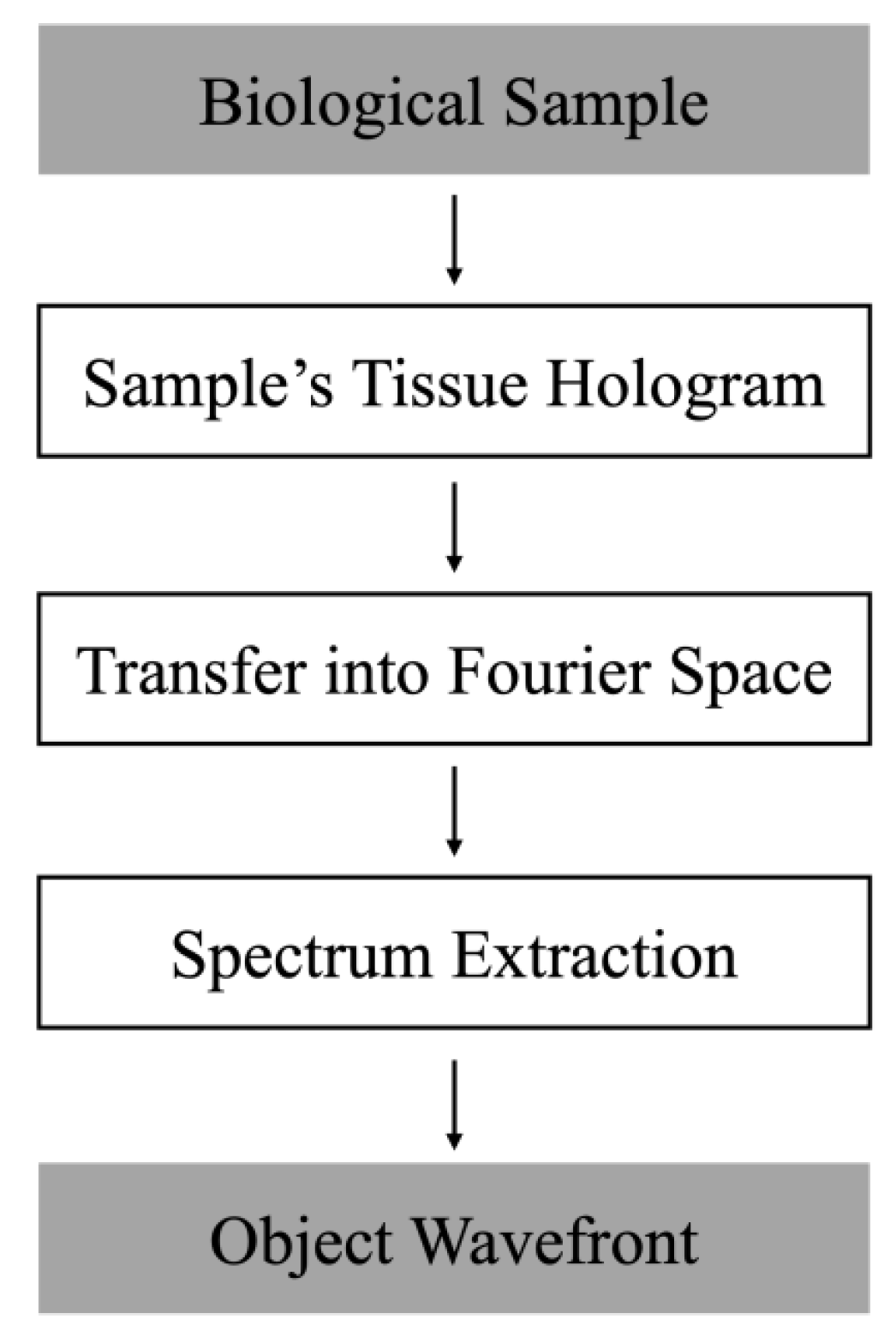
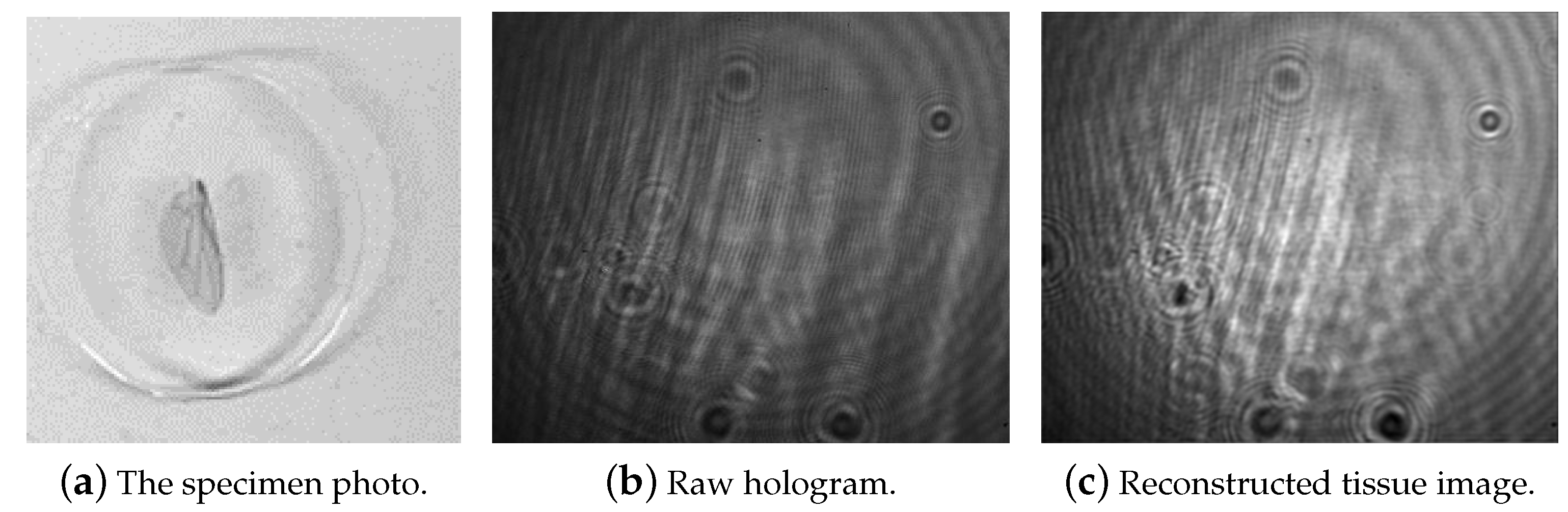
2.1. Single-Shot Off-Axis Digital Holographic Interferometer
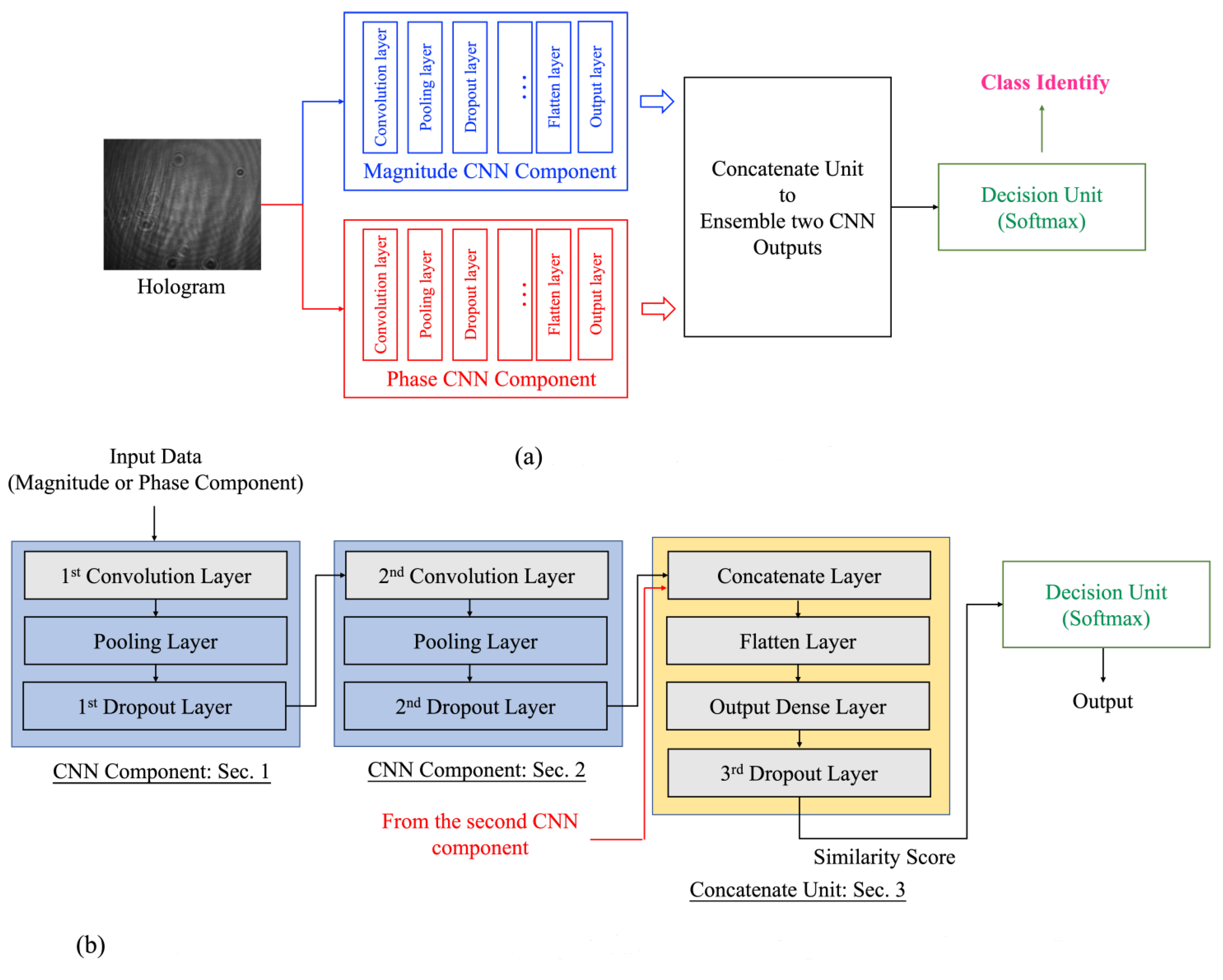
2.2. Ensemble Deep-Learning Network
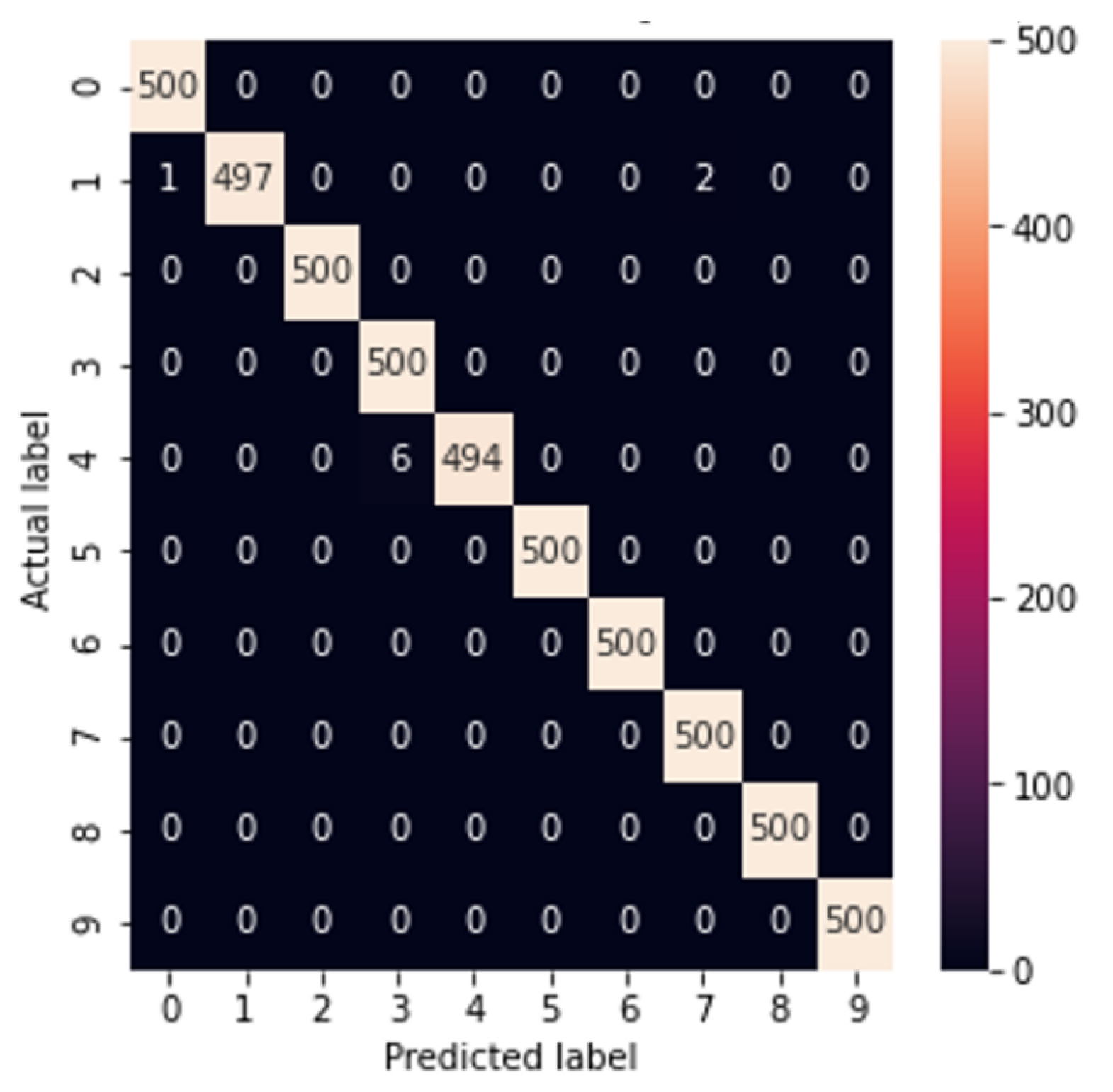
3. Experiments
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| I-EDL | Interferometer and an ensemble deep learning |
| 3D | Three-dimensional |
| EDL-IOHC | Ensemble deep learning invariant hologram classification |
| CGH | Computer generated hologram |
| DFT | Discrete Fourier transform |
| IDFT | Inverse discrete Fourier transform |
| CMOS | Complementary metal–oxide–semiconductor |
| MO | Microscope objective |
| NDF | Neural density filter |
| BS | Beam splitter |
| CNN | Convolutional neural network |
| GPU | Graphics processing unit |
References
- Lugt, A.V. Signal detection by complex spatial filtering. IEEE Trans. Inf. Theory 1964, 10, 139–145. [Google Scholar] [CrossRef]
- Poon, T.C.; Kim, T. Optical image recognition of three-dimensional objects. Appl. Opt. 1999, 38, 370–381. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Poon, T.C. Extraction of 3-D location of matched 3-D object using power fringe-adjusted filtering and Wigner analysis. Opt. Eng. 1999, 38, 2176–2183. [Google Scholar] [CrossRef]
- Kim, T.; Poon, T.C. Three-dimensional matching by use of phase-only holographic information and the Wigner distribution. JOSA A 2000, 17, 2520–2528. [Google Scholar] [CrossRef] [PubMed]
- Park, S.C.; Lee, K.J.; Lee, S.H.; Kim, S.C.; Kim, E.S. Robust recognition of partially occluded 3-D objects from computationally reconstructed hologram by using a spatial filtering scheme. In Proceedings of the Practical Holography XXIII: Materials and Applications; SPIE: Bellingham, WA, USA, 2009; Volume 7233, pp. 254–262. [Google Scholar]
- Lam, H.; Tsang, P.; Poon, T.C. Hologram classification of occluded and deformable objects with speckle noise contamination by deep learning. JOSA A 2022, 39, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Gabor, D. A new microscopic principle. Nature 1948, 161, 777–778. [Google Scholar] [CrossRef] [PubMed]
- Sahin, E.; Stoykova, E.; Mäkinen, J.; Gotchev, A. Computer-generated holograms for 3D imaging: A survey. ACM Comput. Surv. (CSUR) 2020, 53, 1–35. [Google Scholar] [CrossRef]
- Goodman, J.W.; Lawrence, R. Digital image formation from electronically detected holograms. Appl. Phys. Lett. 1967, 11, 77–79. [Google Scholar] [CrossRef]
- Leal-León, N.; Medina-Melendrez, M.; Flores-Moreno, J.; Álvarez-Lares, J. Object wave field extraction in off-axis holography by clipping its frequency components. Appl. Opt. 2020, 59, D43–D53. [Google Scholar] [CrossRef] [PubMed]
- Cuche, E.; Marquet, P.; Depeursinge, C. Spatial filtering for zero-order and twin-image elimination in digital off-axis holography. Appl. Opt. 2000, 39, 4070–4075. [Google Scholar] [CrossRef] [PubMed]
- Kreis, T. Handbook of Holographic Interferometry: Optical and Digital Methods; John Wiley & Sons: Heppenheim, German, 2006. [Google Scholar]
- Sirico, D.; Cavalletti, E.; Miccio, L.; Bianco, V.; Pirone, D.; Memmolo, P.; Sardo, A.; Ferraro, P. Holographic tracking and imaging of free-swimming Tetraselmis by off-axis holographic microscopy. In Proceedings of the 2021 International Workshop on Metrology for the Sea, Learning to Measure Sea Health Parameters (MetroSea). Reggio Calabria, Italy, 4–6 October 2021; pp. 234–238. [Google Scholar]
- Thorlabs, I. HeNe Lasers: Red. Available online: https://www.thorlabs.com/newgrouppage9.cfm?objectgroup_id=1516 (accessed on 7 December 2022).
- Thorlabs, I. Laser Safety Curtain System Kits. 2021. Available online: https://www.thorlabs.com/newgrouppage9.cfm?objectgroup_id=10720 (accessed on 7 December 2022).
- Microscope Central. Available online: https://microscopecentral.com/products/nikon-be-plan-40x-microscope-objective (accessed on 7 December 2022).
- Zhu, Y.; Yeung, C.H.; Lam, E.Y. Digital holography with polarization multiplexing for underwater imaging and descattering. In Proceedings of the 2021 International Workshop on Metrology for the Sea, Learning to Measure Sea Health Parameters (MetroSea). Reggio Calabria, Italy, 4–6 October 2021; pp. 219–223. [Google Scholar]
- Lam, H.; Tsang, P.W. Invariant classification of holograms of deformable objects based on deep learning. In Proceedings of the 2019 IEEE 28th International Symposium on Industrial Electronics (ISIE), Vancouver, BC, Canada, 12–14 June 2019; pp. 2392–2396. [Google Scholar]
- Lam, H.; Tsang, P.; Poon, T.C. Ensemble convolutional neural network for classifying holograms of deformable objects. Opt. Express 2019, 27, 34050–34055. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.H.; Stojkovic, I.; Obradovic, Z. A robust data scaling algorithm to improve classification accuracies in biomedical data. BMC Bioinform. 2016, 17, 359. [Google Scholar] [CrossRef] [PubMed]
- Mach-Zehnder Interferometers. 2021. Available online: https://www.sciencedirect.com/topics/engineering/mach-zehnder-interferometer (accessed on 7 December 2022).




| Specimen Class | Class Label |
|---|---|
| Cucurbita Stem | 0 |
| Pine Stem | 1 |
| Corn (Zea mays) Seed | 2 |
| House Fly Wing | 3 |
| Honeybee Wing | 4 |
| Bird Feather | 5 |
| Corpus Ventriculi | 6 |
| Liver Section | 7 |
| Lymph Node | 8 |
| Human Chromosome | 9 |
| Optical Parameters | Values |
|---|---|
| Wavelength of light | nm |
| Pixel size | 3.45 m |
| Size of hologram | 2056 rows × 2546 columns |
| Off-axis angle | 1.5 degrees |
| Dataset | Success Rate |
|---|---|
| Test set | 99.60% |
| Complete set (both out- and in-training sets) | 99.82% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lam, H.; Zhu, Y.; Buranasiri, P. Off-Axis Holographic Interferometer with Ensemble Deep Learning for Biological Tissues Identification. Appl. Sci. 2022, 12, 12674. https://doi.org/10.3390/app122412674
Lam H, Zhu Y, Buranasiri P. Off-Axis Holographic Interferometer with Ensemble Deep Learning for Biological Tissues Identification. Applied Sciences. 2022; 12(24):12674. https://doi.org/10.3390/app122412674
Chicago/Turabian StyleLam, Hoson, Yanmin Zhu, and Prathan Buranasiri. 2022. "Off-Axis Holographic Interferometer with Ensemble Deep Learning for Biological Tissues Identification" Applied Sciences 12, no. 24: 12674. https://doi.org/10.3390/app122412674
APA StyleLam, H., Zhu, Y., & Buranasiri, P. (2022). Off-Axis Holographic Interferometer with Ensemble Deep Learning for Biological Tissues Identification. Applied Sciences, 12(24), 12674. https://doi.org/10.3390/app122412674







