A Deep Learning-Based Model for Tree Species Identification Using Pollen Grain Images
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Image Processing and Data Augmentation
2.3. Learning Models, Learning Algorithms, and Learning Environment
2.4. Simulation Conditions and Performance Evaluation
3. Results
3.1. Tree Species Classification Accuracy for Test Data with Varying Focal Points
3.2. Tree Species Classification Accuracy for Images at Different Focal Points when Training Data Were Used as Test Data
3.3. Tree Species Classification Accuracy according to Focal Points
3.4. Misclassification Patterns according to Tree Species and Pollen Type
4. Discussion
5. Conclusions and Future Work
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CNN | convolutional neural network |
| MCC | Matthews correlation coefficient |
| TP | true positive |
| TN | true negative |
| FP | false positive |
| FN | false negative |
| A.j. | Acer japonicum |
| B.p | Betula platyphylla |
| C.co. | Cornus controversa |
| C.cr. | Castanea crenata |
| C.cu. | Castanopsis cuspidata |
| C.si. | Celtis sinensis |
| C.t. | Carpinus tschonoskii |
| Cam.j. | |
| Car.j. | Carpinus japonica |
| F.j | Fagus japonica |
| F.c | Fagus crenata |
| F.s. | Fraxinus sieboldiana |
| G.i. | Gamblea innovans |
| J.m. | Juglans mandshurica |
| P.a. | Phellodendron amurense |
| Q.c | Quercus crispula |
| Q.ser | Quercus serrata |
| Q.ses | Quercus sessilifolia |
| U.d. | Ulmus davidiana var. japonica |
| Z.s. | Zelkova serrata |
References
- Nakamura, J. Pollen Analysis; Kokonsyoin: Tokyo, Japan, 1967. [Google Scholar]
- Stillman, E.; Flenley, J.R. The needs and prospects for automation in palynology. Quat. Sci. Rev. 1996, 15, 1–5. [Google Scholar] [CrossRef]
- Holt, K.A.; Bennett, K.D. Principles and methods for automated palynology. New Phytol. 2014, 203, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Nakae, M.; Hosoya, T. Status survey of digitization of natural history collections in Japan. Jpn. Soc. Degit. Arch. 2019, 3, 345–349. [Google Scholar]
- GBIF Survey. 2015. Available online: https://science-net.kahaku.go.jp/contents/resource/GBIF_20151005_questionnaire.pdf (accessed on 21 April 2022).
- Li, P.; Flenley, J.R. Pollen texture identification using neural networks. Grana 1999, 38, 59–64. [Google Scholar] [CrossRef]
- Marcos, J.V.; Nava, R.; Cristóbal, G.; Redondo, R.; Escalante-Ramírez, B.; Bueno, G.; Dénizc, Ó.; González-Porto, A.; Pardoe, C.; Chung, F.; et al. Automated pollen identification using microscopic imaging and texture analysis. Micron 2015, 68, 36–46. [Google Scholar] [CrossRef] [Green Version]
- Daood, A.; Ribeiro, E.; Bush, M. Pollen recognition using a multi-layer hierarchical classifier. In Proceedings of the 2016 23rd International Conference on Pattern Recognition (ICPR), Cancun, Mexico, 4–8 December 2016; pp. 3091–3096. [Google Scholar]
- Gonçalves, A.B.; Souza, J.S.; da Silva, G.G.; Cereda, M.P.; Pott, A.; Naka, M.H.; Pistori, H. Feature extraction and machine learning for the classification of Brazilian Savannah pollen grains. PLoS ONE 2016, 11, e0157044. [Google Scholar] [CrossRef] [Green Version]
- France, I.; Duller, A.W.G.; Duller, G.A.T.; Lamb, H.F. A new approach to automated pollen analysis. Quat. Sci. Rev. 2000, 19, 537–546. [Google Scholar] [CrossRef]
- Takagi, M.; Shimoda, H. Handbook of Image Analysis—Revised Edition; University of Tokyo Press: Tokyo, Japan, 2004. [Google Scholar]
- Ghazi, M.M.; Yanikoglu, B.; Aptoula, E. Plant identification using deep neural networks via optimization of transfer learning parameters. Neurocomputing 2017, 235, 228–235. [Google Scholar] [CrossRef]
- Yamashita, T. An Illustrated Guide to Deep learning; Kodansha: Tokyo, Japan, 2016. [Google Scholar]
- Makino, K.; Nishizaki, H. Deep Learning to Begin with the Arithmetic and Raspberry Pi; CQ Shuppansha: Tokyo, Japan, 2018. [Google Scholar]
- Sevillano, V.; Aznarte, J.L. Improving classification of pollen grain images of the POLEN23E dataset through three different applications of deep learning convolutional neural networks. PLoS ONE 2018, 13, e0201807. [Google Scholar] [CrossRef] [Green Version]
- Gallardo-Caballero, R.; García-Orellana, C.J.; García-Manso, A.; González-Velasco, H.M.; Tormo-Molina, R.; Macías-Macías, M. Precise pollen grain detection in bright field microscopy using deep learning techniques. Sensors 2019, 19, 3583. [Google Scholar] [CrossRef] [Green Version]
- Mahbod, A.; Schaefer, G.; Exker, R.; Ellinger, I. Pollen grain microscopic image classification using an ensemble of fine-tuned deep convolutional neural networks. In Proceedings of the ICPR 2021: Pattern Recognition. ICPR International Workshops and Challenges, Virtual Event, 10–15 January 2021. [Google Scholar]
- Sevillano, V.; Holt, K.; Aznarte, J.L. Precise automatic classification of 46 different pollen types with convolutional neural networks. PLoS ONE 2020, 15, e0229751. [Google Scholar] [CrossRef] [PubMed]
- Kubera, E.; Kubik-Komar, A.; Piotrowska-Weryszko, K.; Skrzypiec, M. Deep learning methods for improving pollen monitoring. Sensors 2021, 21, 3526. [Google Scholar] [CrossRef] [PubMed]
- Boldeanu, M.; Cucu, H.; Burileanu, C.; Mărmureanu, L. Multi-input convolutional neural networks for automatic pollen classification. Appl. Sci. 2021, 11, 11707. [Google Scholar] [CrossRef]
- Ortega-Cisneros, S.; Ruiz-Varela, J.M.; Rivera-Acosta, M.A.; Rivera-Dominguez, J.; Moreno-Villalobos, P. Pollen grains classification with a deep learning system GPU-trained. IEEE Lat. Am. Trans. 2022, 20, 22–31. [Google Scholar] [CrossRef]
- Chen, X.; Ju, F. Automatic classification of pollen grain microscope images using a multi-scale classifier with SRGAN deblurring. Appl. Sci. 2022, 12, 7126. [Google Scholar] [CrossRef]
- POLEN23E. 2013. Available online: https://www.quantitative-plant.org/dataset/polen23e (accessed on 3 June 2022).
- Battiato, S.; Ortis, A.; Trenta, F.; Ascari, L.; Politi, M.; Siniscalco, N. POLLEN13K: A large scale microscope pollen grain image dataset. In Proceedings of the 2020 IEEE International Conference on Image Processing (ICIP), Abu Dhabi, United Arab Emirates, 25–28 October 2020. [Google Scholar]
- Minowa, Y.; Nagasaki, Y. Convolutional neural network applied to tree species identification based on leaf images. J. For. Plan. 2020, 26, 1–11. [Google Scholar] [CrossRef]
- Morita, Y. Classification, Morphological Categories, General Structure and Names of Pollen and Spores—Encyclopedia of Pollen Science; Asakurashoten: Tokyo, Japan, 1994. [Google Scholar]
- Shavlik, J. Transfer Learning. 2009. Available online: https://ftp.cs.wisc.edu/machine-learning/shavlik-group/torrey.handbook09.pdf (accessed on 25 August 2021).
- Perez, L.; Wang, J. The effectiveness of data augmentation in image classification using deep learning. arXiv 2017, arXiv:1712.04621. [Google Scholar]
- Faegri, K.; Iversen, J. Textbook of Pollen Analysis, 4th ed.; Faegri, K., Kaland, P.E., Krzywinski, K., Eds.; John Wiley & Sons Ltd.: New York, NY, USA, 1989. [Google Scholar]
- NIH. ImageJ. 2004. Available online: https://imagej.nih.gov/ij/ (accessed on 5 January 2022).
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef] [Green Version]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7 June 2015; pp. 1–12. [Google Scholar]
- NVIDIA. 2016. Available online: https://developer.nvidia.com/cuda-toolkit/ (accessed on 5 January 2022).
- Jia, Y.; Shelhamer, E.; Donahue, J.; Karayev, S.; Long, J.; Girshick, R.; Guadarrama, S.; Darrell, T. Caffe: Convolutional architecture for fast feature embedding. In Proceedings of the 22nd ACM International Conference on Multimedia, Orlando, FL, USA, 3–7 November 2014; pp. 675–678. [Google Scholar]
- Matthews, B.W. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Et Bio-phys. Acta (BBA)—Protein Struct. 1975, 405, 442–451. [Google Scholar] [CrossRef]
- Motoda, H.; Tsumoto, S.; Yamaguchi, T.; Numao, M. Fundamentals of Data Mining; Ohmsha: Tokyo, Japan, 2006. [Google Scholar]
- Witten, I.H.; Frank, E.; Hall, M.A. Data Mining, Practical Machine Learning Tools and Techniques, 3rd ed.; Morgan Kaufmann Publishers: Burlington, NJ, USA, 2011; pp. 163–180. [Google Scholar]
- Scikit-Learn. 2007. Available online: https://scikit-learn.org/stable/modules/model_evaluation.html#matthews-corrcoef (accessed on 5 February 2022).
- Minowa, Y.; Kubota, Y. Identification of broad-leaf trees using deep learning based on field photographs of multiple leaves. J. For. Res. 2022, 27, 246–254. [Google Scholar] [CrossRef]
- Minowa, Y.; Kubota, Y.; Nakatsukasa, S. Verification of a deep learning-based tree species identification model using images of broadleaf and coniferous tree leaves. Forests. 2022, 13, 943. [Google Scholar] [CrossRef]
- Goëau, H.; Bonnet, P.; Joly, A. LifeCLEF plant identification task 2015. CLEF (Working Notes). In Proceedings of the Working Notes for CLEF 2014 Conference, Sheffield, UK, 15–18 September 2014. [Google Scholar]
- Raschka, S.; Mirjalili, V. Python Machine Learning Programming; Impress: Tokyo, Japan, 2018. (In Japanese) [Google Scholar]
- Li, Z.; Hoiem, D. Learning without forgetting. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 2935–2947. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhang, Z.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Huang, G.; Liu, Z.; Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Okatani, T. On deep learning. J. Robotics. Soc. Jpn. 2015, 33, 92–96. [Google Scholar] [CrossRef]
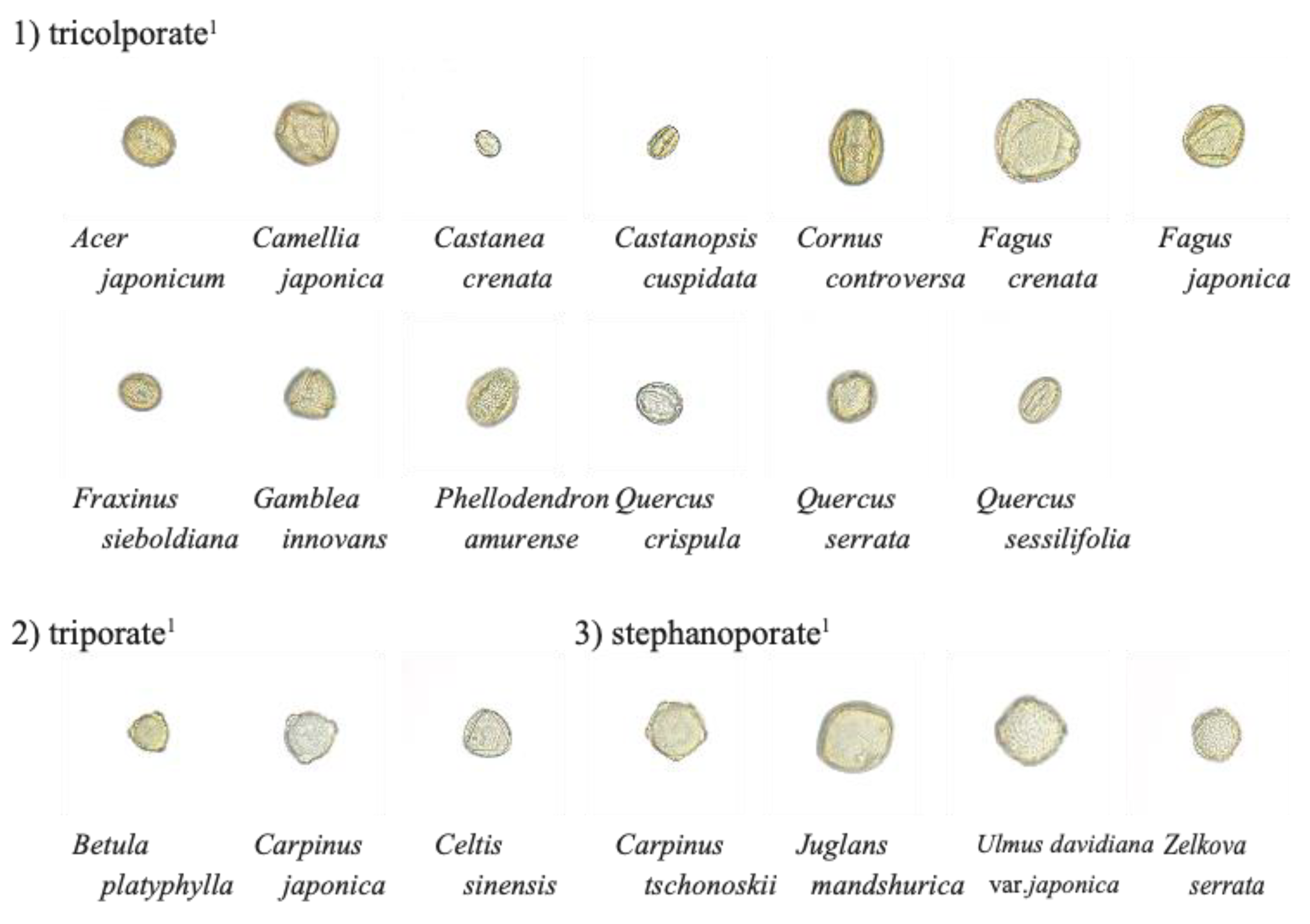
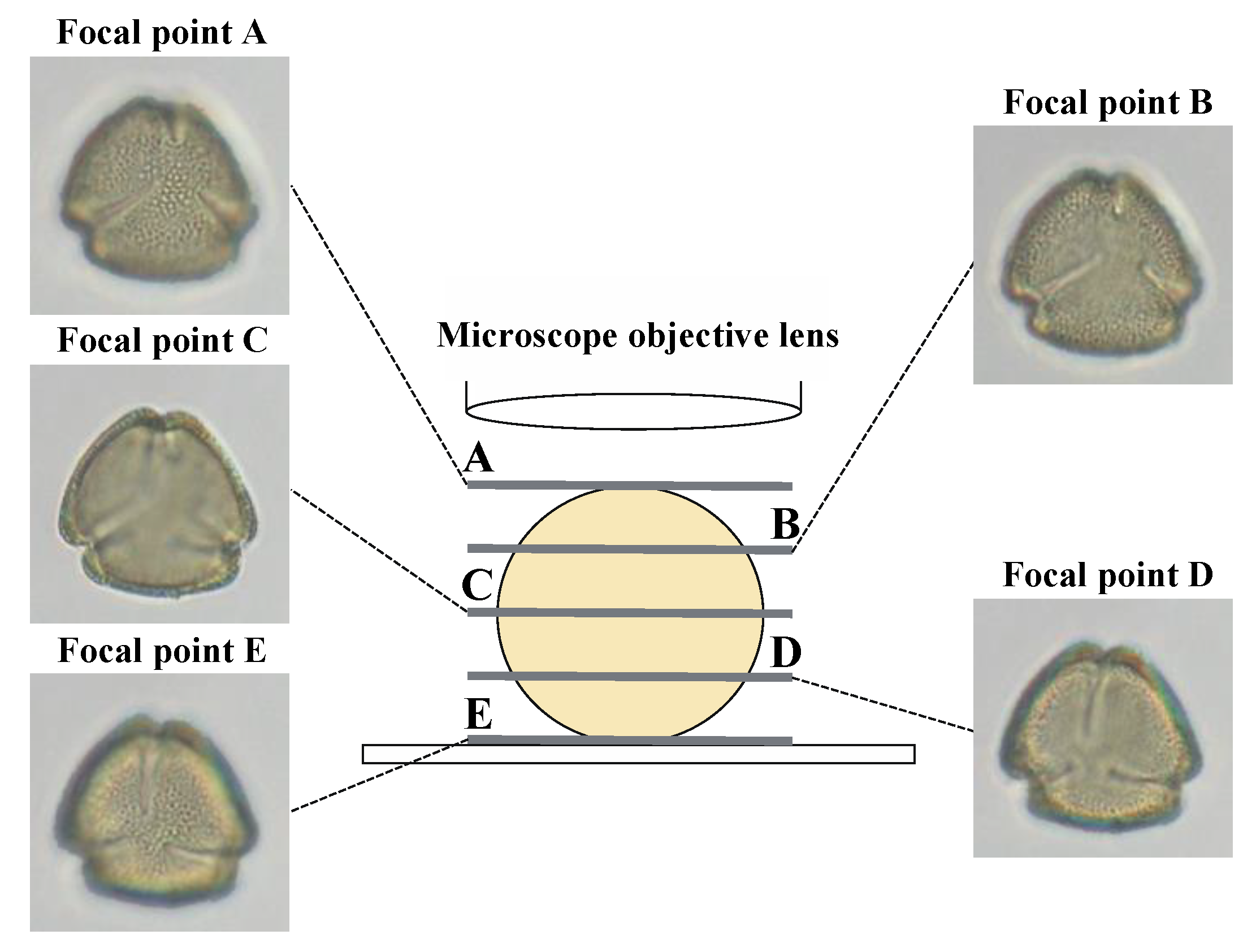
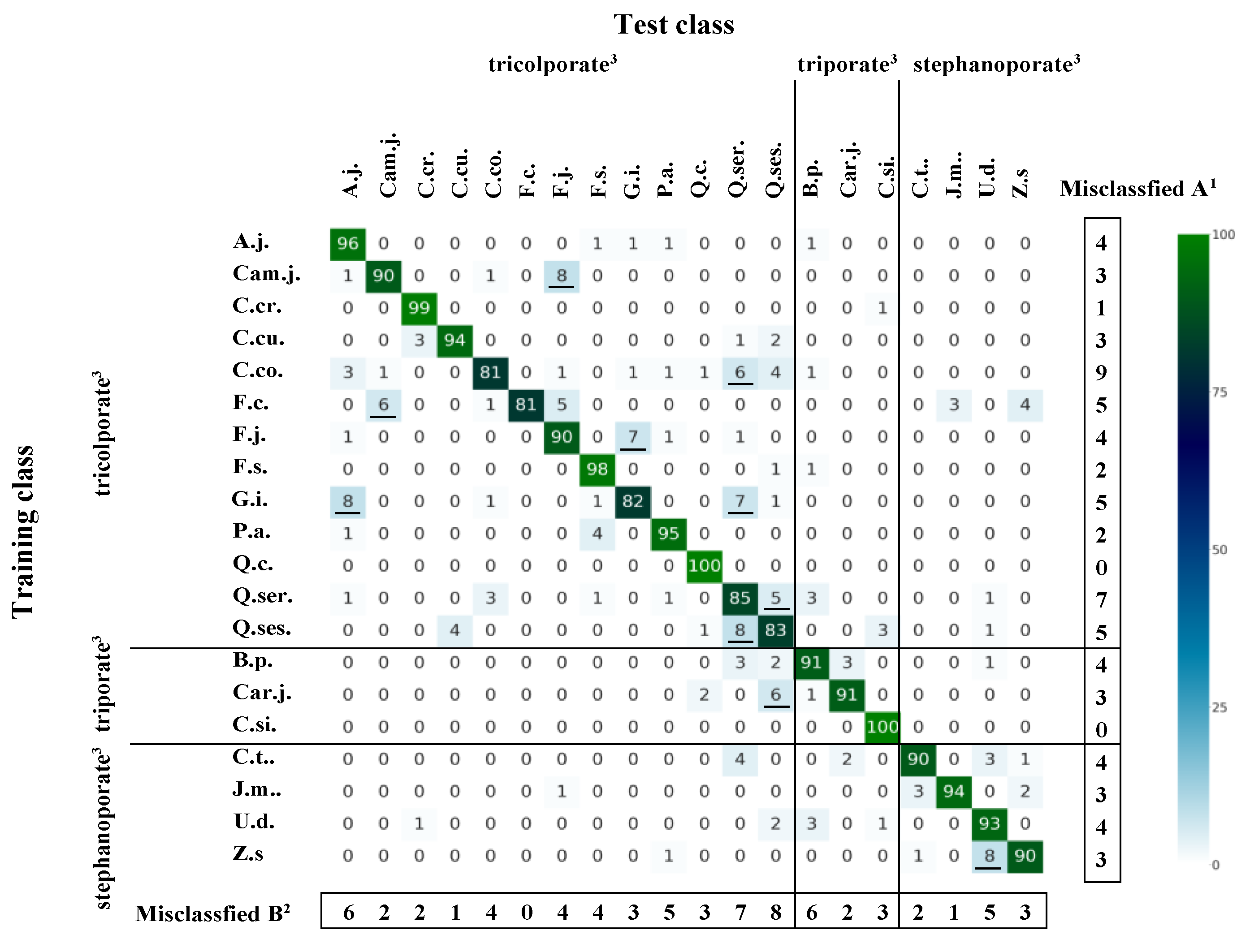
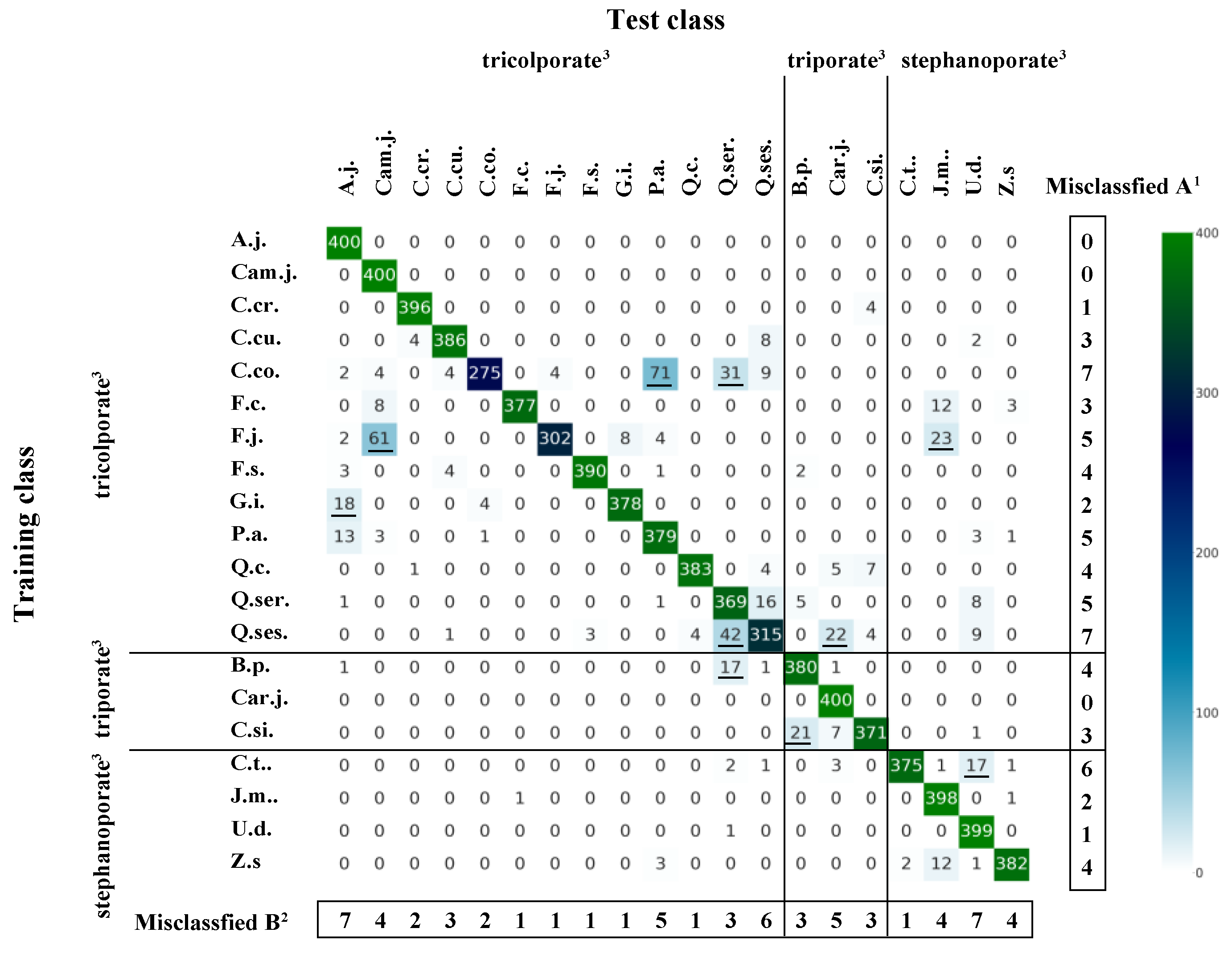
| Scientific Name | Sample Number 1 | Pollen Types 2 | Collection Date | Sampling Points | Collectors |
|---|---|---|---|---|---|
| Acer japonicum | A2909 | tricolporate | 2013/4/19 | Ashiu, Miyama, Nantan, Kyoto | Kawai |
| Camellia japonica | A1986 | tricolporate | 2002/3/31 | Mt. Tokura, Kakuda, Miyagi | Sasaki et al. |
| Castanea crenata | A1870 | tricolporate | 2002/6/19 | Kuta, Kyoto | Makino |
| Castanopsis cuspidata | A1585 | tricolporate | 1979/5/14 | Matsugasaki-kaijiri, Kyoto | Takahara |
| Cornus controversa | A1439 | tricolporate | 1978/5/22 | Imajyo, Fukui | Takahara |
| Fagus crenata | A224 | tricolporate | 1990/4/6 | Kuta, Kyoto | Takahara |
| Fagus japonica | A1589 | tricolporate | 1979/4/19 | Ohmi, Kyoto | Takahara |
| Fraxinus sieboldiana | A699 | tricolporate | 1978/4/28 | near Mizorogaike, Kyoto | Takahara |
| Gamblea innovans | A1468 | tricolporate | 1978/5/19 | Shizuichi, Kyoto | Takahara |
| Phellodendron amurense | A1444 | tricolporate | 1978/5/22 | Imajyo, Fukui | Takahara |
| Quercus crispula | A1777 | tricolporate | 2002/4/27 | Kuta, Kyoto | Makino |
| Quercus serrata | A1763 | tricolporate | 2002/4/16 | near Mizorogaike, Kyoto | Makino |
| Quercus sessilifolia | A1804 | tricolporate | 2002/5/3 | Kuta, Kyoto | Makino |
| Betula platyphylla | A1593 | triporate | 1978/4/15 | Kyoto Botanical Gardens | Takahara |
| Carpinus japonica | A1752 | triporate | 2002/4/22 | Kuta, Kyoto | Takahara |
| Celtis sinensis | A1227 | triporate | 1990/4/8 | Uji-gokasyo, Uji, Kyoto | Takahara |
| Carpinus tschonoskii | A1313 | stephanoporate | 1978/4/5 | Koishikawa Botanical Garden, Tokyo | Takahara |
| Juglans mandshurica | A1806 | stephanoporate | 2002/5/3 | Kuta, Kyoto | Makino |
| Ulmus davidiana var. japonica | A1293 | stephanoporate | 1982/3/13 | Osaka Agricultural Research Center | Takahara |
| Zelkova serrata | A1170 | stephanoporate | 1979/4/10 | Kyoto Prefectural University | Takahara |
| (1) without Data Augmentation. | ||||||||
|---|---|---|---|---|---|---|---|---|
| Focal Point | GoogLeNet | AlexNet | Average | (Max.-Min.) 2 | ||||
| 50 1 | 100 1 | 200 1 | 50 1 | 100 1 | 200 1 | |||
| A | 0.7996 | 0.8653 | 0.8891 | 0.8105 | 0.8859 | 0.8954 | 0.8576 | 0.0958 |
| B | 0.8185 | 0.8635 | 0.8774 | 0.8156 | 0.8886 | 0.9049 | 0.8614 | 0.0893 |
| C | 0.7759 | 0.8198 | 0.8338 | 0.8053 | 0.8876 | 0.8839 | 0.8344 | 0.1117 |
| D | 0.7486 | 0.8215 | 0.8252 | 0.8124 | 0.8739 | 0.9070 | 0.8314 | 0.1584 |
| E | 0.8505 | 0.8646 | 0.8681 | 0.8225 | 0.8965 | 0.8960 | 0.8664 | 0.0740 |
| Average | 0.7986 | 0.8470 | 0.8587 | 0.8133 | 0.8865 | 0.8975 | ||
| All data 3 | 0.8182 | 0.8302 | 0.8372 | 0.9009 | 0.9017 | 0.8950 | 0.8639 | 0.0836 |
| (2) with Data Augmentation | ||||||||
| Focal Point | GoogLeNet | AlexNet | Average | (Max.-Min.) 2 | ||||
| 50 1 | 100 1 | 200 1 | 50 1 | 100 1 | 200 1 | |||
| A | 0.8952 | 0.8939 | 0.9159 | 0.8274 | 0.8466 | 0.8527 | 0.8720 | 0.0885 |
| B | 0.8857 | 0.9070 | 0.9050 | 0.8586 | 0.8685 | 0.8425 | 0.8779 | 0.0645 |
| C | 0.8831 | 0.8697 | 0.9004 | 0.8453 | 0.8477 | 0.8425 | 0.8648 | 0.0579 |
| D | 0.8904 | 0.9141 | 0.9185 | 0.8437 | 0.8497 | 0.8352 | 0.8753 | 0.0833 |
| E | 0.9206 | 0.9256 | 0.9286 | 0.8430 | 0.8566 | 0.8427 | 0.8862 | 0.0859 |
| Average | 0.8950 | 0.9021 | 0.9137 | 0.8436 | 0.8538 | 0.8431 | ||
| All data 3 | 0.8850 | 0.9018 | 0.9159 | 0.7844 | 0.8035 | 0.8190 | 0.8516 | 0.1315 |
| Training Data | Test Data | No Data Augmentation | Data Augmentation | Avg Diff 2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GoogLeNet | AlexNet | GoogLeNet | AlexNet | |||||||
| MCC | Diff 1 | MCC | Diff 1 | MCC | Diff 1 | MCC | Diff 1 | |||
| A | A | 0.8891 | - | 0.8954 | - | 0.9159 | - | 0.8527 | - | - |
| B | 0.8086 | −0.0805 | 0.8396 | −0.0558 | 0.8205 | −0.0954 | 0.7739 | −0.0788 | −0.0776 | |
| C | 0.7510 | −0.1381 | 0.7842 | −0.1113 | 0.7418 | −0.1741 | 0.7118 | −0.1410 | −0.1411 | |
| D | 0.7595 | −0.1296 | 0.7771 | −0.1183 | 0.7409 | −0.1750 | 0.7023 | −0.1504 | −0.1433 | |
| E | 0.7754 | −0.1137 | 0.7560 | −0.1394 | 0.7449 | −0.1710 | 0.6745 | −0.1783 | −0.1506 | |
| Avg3 | −0.1155 | −0.1062 | −0.1539 | −0.1371 | ||||||
| B | A | 0.8536 | −0.0238 | 0.9233 | 0.0184 | 0.8796 | −0.0254 | 0.8584 | −0.0466 | −0.0193 |
| B | 0.8774 | - | 0.9049 | - | 0.9050 | - | 0.8425 | - | - | |
| C | 0.8464 | −0.0311 | 0.8725 | −0.0324 | 0.8488 | −0.0562 | 0.7996 | −0.0429 | −0.0407 | |
| D | 0.8085 | −0.0689 | 0.8432 | −0.0617 | 0.7980 | −0.1070 | 0.7934 | −0.0491 | −0.0717 | |
| E | 0.7328 | −0.1446 | 0.8048 | −0.1001 | 0.7111 | −0.1939 | 0.7515 | −0.0911 | −0.1324 | |
| Avg3 | −0.0815 | −0.0647 | −0.1191 | −0.0610 | ||||||
| C | A | 0.7893 | −0.0444 | 0.8674 | −0.0166 | 0.8307 | −0.0697 | 0.8185 | −0.0240 | −0.0387 |
| B | 0.8279 | −0.0058 | 0.8830 | −0.0010 | 0.8646 | −0.0358 | 0.8270 | −0.0155 | −0.0145 | |
| C | 0.8338 | - | 0.8839 | - | 0.9004 | - | 0.8425 | - | - | |
| D | 0.8040 | −0.0298 | 0.8725 | −0.0114 | 0.8271 | −0.0733 | 0.8197 | −0.0228 | −0.0343 | |
| E | 0.7565 | −0.0772 | 0.8561 | −0.0279 | 0.7571 | −0.1433 | 0.7861 | −0.0564 | −0.0762 | |
| Avg3 | −0.0376 | −0.0134 | −0.0841 | −0.0316 | ||||||
| D | A | 0.7621 | −0.0631 | 0.8522 | −0.0548 | 0.8258 | −0.0927 | 0.7522 | −0.0830 | −0.0734 |
| B | 0.7678 | −0.0573 | 0.8478 | −0.0592 | 0.8407 | −0.0778 | 0.7602 | −0.0750 | −0.0673 | |
| C | 0.7837 | −0.0415 | 0.8695 | −0.0375 | 0.8600 | −0.0585 | 0.7963 | −0.0389 | −0.0441 | |
| D | 0.8252 | - | 0.9070 | - | 0.9185 | - | 0.8352 | - | - | |
| E | 0.8219 | −0.0033 | 0.8977 | −0.0093 | 0.8934 | −0.0251 | 0.8190 | −0.0162 | −0.0135 | |
| Avg3 | −0.0340 | −0.0353 | −0.0538 | −0.0434 | ||||||
| E | A | 0.7488 | −0.1193 | 0.7682 | −0.1278 | 0.8055 | −0.1231 | 0.7138 | −0.1289 | −0.1248 |
| B | 0.7368 | −0.1313 | 0.7567 | −0.1394 | 0.7914 | −0.1372 | 0.6931 | −0.1496 | −0.1394 | |
| C | 0.7442 | −0.1239 | 0.7754 | −0.1206 | 0.7947 | −0.1339 | 0.7046 | −0.1381 | −0.1291 | |
| D | 0.8369 | −0.0312 | 0.8630 | −0.0330 | 0.8952 | −0.0334 | 0.8028 | −0.0399 | −0.0344 | |
| E | 0.8681 | - | 0.8960 | - | 0.9286 | - | 0.8427 | - | - | |
| Avg3 | −0.0678 | −0.0528 | −0.1017 | −0.0705 | ||||||
| Overall Avg 4 | −0.0729 | −0.0619 | −0.1001 | −0.0783 | ||||||
| Pollen Types | (1) No Data Augmentation, Focal Point D, AlexNet, 200 Epochs | (2) Data Augmentation, Focal Point E, GoogLeNet, 200 Epochs | ||||
|---|---|---|---|---|---|---|
| tricolporate 1 | triporate 1 | stephanoporate 1 | tricolporate 1 | triporate 1 | stephanoporate 1 | |
| tricolporate 1 | 8.2 | 3.3 | 2.3 | 26.2 | 16.3 | 15.3 |
| triporate1 | 1.0 | 1.3 | 0.3 | 1.5 | 9.7 | 0.3 |
| stephanoporate 1 | 0.7 | 2.0 | 4.5 | 0.6 | 1.0 | 8.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minowa, Y.; Shigematsu, K.; Takahara, H. A Deep Learning-Based Model for Tree Species Identification Using Pollen Grain Images. Appl. Sci. 2022, 12, 12626. https://doi.org/10.3390/app122412626
Minowa Y, Shigematsu K, Takahara H. A Deep Learning-Based Model for Tree Species Identification Using Pollen Grain Images. Applied Sciences. 2022; 12(24):12626. https://doi.org/10.3390/app122412626
Chicago/Turabian StyleMinowa, Yasushi, Koharu Shigematsu, and Hikaru Takahara. 2022. "A Deep Learning-Based Model for Tree Species Identification Using Pollen Grain Images" Applied Sciences 12, no. 24: 12626. https://doi.org/10.3390/app122412626
APA StyleMinowa, Y., Shigematsu, K., & Takahara, H. (2022). A Deep Learning-Based Model for Tree Species Identification Using Pollen Grain Images. Applied Sciences, 12(24), 12626. https://doi.org/10.3390/app122412626






