Abstract
Exosomes are a subpopulation of extravascular vesicles with a diameter of 30–150 nm. They are cellular-communication mediators, often reaching very distant organism tissues. Information is transferred by exosomal cargo, composed of a wide variety of macromolecules such as nucleic acids, proteins, and lipids. Exosomes possess natural specific cell targeting properties that are desirable in designing targeted macromolecules (DNA and RNA) and drug delivery systems (doxorubicin, paclitaxel, and taxol). In this context, exosomes can be defined as bio-derived drug transporting and protecting devices for the treatment of bacterial (toxoplasmosis and salmonellosis), viral (AIDS and hepatitis B), and cancer (lung, pancreatic, colon, brain, and breast) diseases. Extensive research proves that exosomes’ natural cargo can double-act, both increasing and decreasing the disease severity. In this case, the exosomes need to be prepared, namely, their origin and their cargo need to be screened and known. Thus, appropriate methods for intact and price-effective exosome isolation are needed with further exosome properties description. Among many utilized isolation methods, the most common are ultracentrifugation, polymer-based precipitation, and affinity precipitation-isolation systems, but novel microfluidic methods compromising high efficacy and purity are being developed. In this review, we state the current knowledge and trends in exosome-based drug delivery systems.
1. Introduction
Exosomes can serve as a drug delivery platform for macromolecules (DNA, RNA, proteins, and lipids) and drugs in hastily developing pharmaceutical units. Currently, they are under extensive research to adjust their properties to overcome specific drug delivery obstacles [1,2].
For over the last two decades, exosomes have been isolated from many cell types including normal and cancer cells, and their impact on other cells has been studied. It is a well-known fact that their main properties come from the cargo that can vary between distinct cells. These cargoes are cell-derived particles, namely nucleic acids, proteins, and lipids, which can influence other cells’ metabolism and functionality [3,4]. Due to the similarity of exosomes to artificial lipid microvesicles, researchers started to incorporate drugs, macromolecules, and other substances, which, for clarity, are altogether stated as drugs in this article [5,6]. This attitude resulted in an improvement in therapy efficiency employing increased cell-drug response, a decrease in necessary drug concentration, and an unwanted systemic organism response. The possibility of additional targeted delivery is another advantage of exosomes [7]. Some researchers proposed advanced combined delivery strategies that improve therapeutics efficiency by additional cell expression pattern manipulation with macromolecules such as RNA or DNA [8,9,10].
In this review, we present current strategies for preparing exosomal drug delivery systems including all necessary steps and examples of exosomes used as drug delivery vehicles.
2. Exosomes and Their Release Routes
Exosomes are recognized as a source of various nucleic acids and proteins, acting as local and body-wide signalization and regulation systems. Exosomes (30–150 nm in diameter) are the smallest subpopulation of extravascular vesicles, which also comprise microvesicles (50 nm–1 μm) and apoptotic bodies (50 nm–5 μm) [3,4].
The source of the exosomes is in the endosomal system. In the beginning, the so-called early endosomes form as a result of inward membrane budding, and they are subjected to sorting, facing their final destination. The early endosomes can fuse with endocytic vesicles, thus leading their cargo toward recycling, degradation, or secretion. If their cargo is meant to be recycled, they form recycling endosomes [11]. Other early endosomes maturate into late endosomes. During their maturation to late endosomes, the inward budding of the vesicle membrane occurs, resulting in multivesicular bodies (MVBs) creation, containing numerous intraluminal vesicles (ILVs) [6]. At this stage, the MVB can either fuse with the lysosome—with onward degradation of ILVs—or fuse with a plasma membrane, resulting in the release of ILVs to the extracellular space (Figure 1). These released ILVs are called exosomes and contain many proteins, nucleic acids, lipids, and polysaccharides derived from the cell interior [12]. Another release mechanism is due to the outward budding of the plasma membrane, resulting in the formation of so-called shedding microvesicles or exosomes [12].
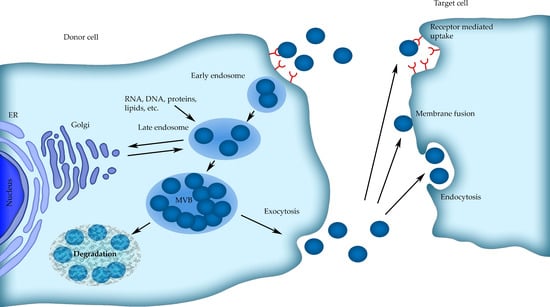
Figure 1.
Scheme of the exosomes formation, release, and internalization by cells.
3. Methods for Exosomes Characterization
Characterization of the exosomes structure in terms of drug delivery systems (DDSs) is crucial because it sets the further properties of DDS, e.g., cells/tissues affinity, stress response, absorption routes, and drug release. The International Society for Extracellular Vesicles in 2014 and 2018 (MISEV2018) presented guidelines for basic requirements that all exosomes including studies and exosomes preparation should meet [13]. Parameters such as amount, size, morphology, membrane composition, and proteins including receptors must be considered while developing exosome-based DDS. These parameters are characterized by techniques divided into optical, non-optical, and microfluidics [6].
Currently, the most widely utilized methods for exosomes characterization are optical, namely dynamic light scattering (DLS), multi-angle light scattering (MALS), and nanoparticle tracking analysis (NTA) [14,15]. These methods allow for high-resolution measurement of size, (DLS 0.5–200 nm; MALS 10–500 nm; NTA 10–1000 nm), size distribution, and concentration. The combination of DLS and MALS greatly increases the range (0.5–500 nm) and precision of measurements. Thus, it leads to the detection of three subsequent exosomes populations, described as small (Exo-S), large (Exo-L), and non-membranous exomeres [16]. In the NTA method also, the inclusion of fluorescent dyes enhances the resolution and allows the characterization of the surface immunophenotype by the use of fluorescence labeling [17,18]. Optical methods are the most popular but the downsides are low sensitivity, high reagent consumption, and the requirement for specialized equipment.
Non-optical methods include scanning electron microscopy (SEM), transmission electron microscopy (TEM), cryo-electron microscopy (Cryo-EM), atomic force microscopy (AFM), immunodetection methods (ELISA), Fourier transform infrared spectroscopy (FTIR), tunable resistive pulse sensing (TRPS), and the single-particle interferometric reflectance imaging sensor (SP-IRIS) [19,20,21]. SEM, TEM, and AFM methods are routinely utilized for direct membrane structure and morphology determination, while ELISA assays provide detection and quantification of various structural particles, mostly proteins and receptors, e.g., for exosome markers presence confirmation [20,22,23,24,25,26]. FTIR spectroscopy and attenuated total reflection-FTIR (ATR-FTIR) are proposed for exosomes quality quantification and the overall estimation of lipid and protein content [20]. With TRPS, the simultaneous measurement of size, concentration, and zeta potential is possible. This method is rapidly growing in the field of exosomes research [21]. A single-particle interferometric reflectance imaging sensor (SP-IRIS) is also used for exosomes quantification but can also be utilized for the detection of specific markers and the determination of exosomes subpopulation [26,27]. Non-optical methods also include the use of specialized equipment and the need for laborious sample preparation but provide high-quality surface, structure, and composition data.
Further development of exosomes investigation methods leads to the use of microfluidics methods in exosomes research. These methods provide high-quality, high-specificity data with subsequent low reagent consumption and high throughput [6,28,29]. Microfluidics-based methods are methods modified to suit the nanoscale for proper exosomes determination. These methods require specialized, prefabricated microchips, generally designed and prepared exclusively for a specific application in exosomes research. Microfluidics chips are generally fabricated out of a glass base and polydimethylsiloxane (PDMS) membrane and contain a lot of microchannels whose size is suited for the analyzed sample. The main difference is the inside surface of the chip, which can be functionalized in various ways, e.g., by coating, multi-layer depositing, electrodepositing, and etching [30,31,32,33]. For different microfluidic characterization methods, different types of microchips are fabricated, including immunochips, magnetic, and electrochemical chips [31,34,35]. Thus far, for exosomes research, fluorescence correlation microscopy (FCM), surface plasmon resonance (SPR), nuclear magnetic resonance (NMR and microNMR) methods, and atomic force microscopy (AFM) were used [6,29,36,37]. These methods are based on antibody-mediated detection and further quantification of exosomes and exosomal proteins either stained by fluorescence dyes (FCM), chip-membrane-bound and SPR imaged, using the NMR, or scanning mica-adhered exosomes (AFM) [6,37,38]. These methods are attracting increasing attention but still require further development and validation. Other authors report the use of liquid chromatography tandem mass spectrometry (LC-MS/MS) and flow cytometry for further proteins identification and exosomes characterization [26]. Exosomes and their proteins can also be detected with colorimetry (labeled antibodies/ELISA), direct fluorescence staining (DiO dye), changes in electrochemical properties, and optical special effects methods [29,35,38,39]. For results assessment, there is also a need for additional equipment, such as a plate-reader or fluorescence microscope [30].
Flow field fractionation (FFF) is both a characterization (in conjunction with detectors, e.g., UV) and isolation technique based on differences in flow speed of different size particles on microchips. Smaller particles will move faster, while larger ones will be more restrained and travel slower [40,41]. This method is used for assessing amount and size in a superior manner to NTA in the case of distinguishing subtle populations of exosomes [42].
4. Exosomes Isolation Methods
Currently, there are many available exosomes isolation methods (Table 1). The oldest and most commonly used exosome isolation method is ultracentrifugation stated by many as a gold standard. Other techniques are mostly based on size exclusion or antibody-mediated isolation of labeled exosomes. Every isolation method differs in terms of purity, amount, efficiency, and throughput. The most common disadvantages are inconsistency and damage or alteration in the structure of exosomes [6,43].

Table 1.
Exosome isolation methods.
Ultracentrifugation is based on the size (weight) of particles and their sedimentation under centrifugal force (100,000–110,000× g). The previous removal of cellular debris and unwanted particles can be obtained by a few steps of slower-speed centrifugation named differential centrifugation. It can be used for large pool processing. This method can also be conducted in a sucrose gradient (1.13–1.19 g/mL) or cushions for higher enrichment, yield, and purity increase [44,45].
Ultrafiltration methods include the utilization of filters with decreasing pore size; thus, larger particles will be excluded from the filtrate. It can be used in combination with centrifugation to accelerate exosomes purification by the initial removal of large particles such as cell debris. The pore clogging risk makes this method unfavorable for large pool processing [46,47].
Size-exclusion chromatography is based on the size of gel pores and the size of exosomes. Particles larger than the gel pore size will be eluted in the first place; inversely, particles smaller than the gel pore size will be restrained. Isolation is commenced in a native environment and overall very high purity and yield are obtained. In combination with ultrafiltration, this method provides lesser protein contamination with a superior exosome recovery rate. The downside is the cost of the gel [48].
Polymer-based precipitation separation methods utilize super-hydrophilic polymers to enhance the precipitation of small-size particles such as exosomes. Polyethylene glycol is commonly used in concentrations varying between 8 and 15%. With the use of this method, the solution containing exosomes is overnight-incubated with polymer and further centrifugated at about 10,000× g. Commercial kits are also available [6].
Immunological separation methods utilize labeling of the exosomes with antibodies subjected to selected protein markers located in the exosomal membrane. These labeled exosomes can be further easily handled and purified, e.g., with microchips or magnetic beads [49,50]. After magnetic separation, exosomes are purified and magnetic beads are dissolved and removed. With this simple and quick method, high-purity extracts can be obtained but exosomes lacking surface markers recognized by magnetic-labeled antibodies will not be isolated, which may lead to poor exosome pool representation [51]. This method is time-saving and straightforward with high-purity output but involves expensive reagents.
In terms of affinity-isolation methods, there are also proposed lectins and synthetic Vn (venceremin) peptides. Lectins will bind glycosylated proteins presented at the surface of exosomes, leading to exosomes precipitation. Vn peptide isolation technology is based on its high affinity to HSP-containing extracellular particles. However, this method may also lead to the contamination of extracts by cells and other particles containing mentioned markers on the membrane surface [44,52].
Methods mentioned below are conducted on a microchip and belong to microfluidic methods of exosome isolation. Principles of microfluidics were discussed in the previous part, but we need to mention that the same methods can be used for exosomes isolation (immuno- and magnetic-binding, filtering) with relatively high efficiency (about 90%) [53].
FFF is currently a very rarely used but promising method of exosomes isolation. The separation is driven by crossflow forces and is based on the particles’ molecular weight or hydrodynamic diameter. It includes both high purity, high efficiency, and short-time processing, but to date, the utilization of FFF in exosomes isolation is insignificant and awaits further development [16,54,55,56].
The acoustic fluid separation technique utilizes an acoustic field to separate particles depending on their size, density, and compressibility. Biological samples are not damaged during this procedure and the separation itself is contactless, efficient, label-free, and biocompatible. Nevertheless, further development and an increase in throughput are still postulated [57].
Deterministic lateral displacement relies on the change in particle flow path dependent on the particle size. This method is relatively simple but requires special equipment. The most common problems are clogging and poor separation but extensive efforts are taken to overcome these downsides [58]. On the other hand, small sample volumes are appropriate for relatively fast isolation, which may be handy for the isolation of biological samples [59].
The dielectrophoretic separation method employs the differences in location of different-size particles in a nonuniform electric field. Exosomes will be attracted to high electric regions while larger particles will be located in low electric regions. This method is fast and can be used in high-throughput screening but needs heating, limiting its use [60,61].
5. Exosomes Natural Cargo and Structure
Exosomes are generally recognized as cell-to-cell signaling molecules containing numerous proteins, nucleic acids, cytokines, transcription factors, and other cell-derived particles (Figure 2). Exosomes may deliver information not only to the local environment but also to greatly distanced cells and tissues. This may serve as a general signaling and communication pathway, but may also take part in oncogenes spread and further tumor progression and malignancy increase [62]. To this date, exosomes presence was confirmed in body fluids such as amniotic liquid, blood serum, breast milk, epididymal fluid, saliva, urine, effusions of ascites and pleural, bronchoalveolar lavage fluid, synovial fluid, and cell culture supernatant in vitro [63,64,65,66]. Due to these facts, exosomes are proposed as novel therapeutic and diagnostic (theranostics) devices [6]. The excreted exosomes also serve as a way of unnecessary proteins and nucleic acids removal, e.g., during cell maturation (reticulocytes) or excretory system (guts and tubules of the kidney) [67,68,69].
For proper usage of exosomes as a DDS, we first have to consider their natural cargo and the routes of their formation. Then, the characteristics of exosomes affinity and cell uptake should also be described. Exosomal cargo exhibit different purposes and functions, thanks to exosomes being basic transporters presenting outstanding properties themselves. This can be utilized in cell targeting and also as an additional enhancement of drugs. To date, ExoCarta estimates about 10,000 proteins, 3500 mRNA, and over 1100 distinct lipids found in exosomes. For the systematic review, many great databases arise such as Vesiclepedia and ExoCarta where exosome isolation procedures and sources are described along with identified cargo [70,71]. The most important proteins for DDS design are those influencing exosomes transport and cell intake.
Proteins characteristic for exosomes are membrane-bound tetraspanins (CD9, CD63, CD81, and CD82), as well as EpCAM and Rab5, used routinely for isolation. Other commonly recognized proteins are receptors (CD46 and CD55), heat shock proteins (HSP; Hsc70, Hsp70, and Hsp90), proteins taking part in exosomes formation (Alix, TSG101), and membrane proteins responsible for fusion and transport (GTPases, annexins, and flotillin; ATP7A, ATP7B, MRP2, SLC1A4, SLC16A1, and CLIC1) [6,44]. Detection of these markers with ELISA is used for confirmation of exosomes presence.
There are also reports about exosomes containing other particles such as cholesterol (B lymphocytes derived), sphingolipids, phosphoglycerides, and ceramide. The presence of the latter also distinguishes the exosomes from the lysosomes [72].
Exosomes also transport morphogens (Hedgehog, Wingless, and Wingless-like) taking part in tissue patterning development described for Drosophila melanogaster [64].
Nucleic acids are another major group of particles found in exosomes, namely DNA and RNA, which onward can take the functional role in cells. About ~1300 mRNA, 121 miRNA, and >100 small RNA were found in exosomes derived from mouse and human mast cells, while no DNA and rRNA were detected [73]. Other miRNAs found in exosomes were lin-4 and let-7, as well as miR-181a and miR-155 in breast milk [73,74].
Some of the exosomal miRNAs can be used as biomarkers in diagnostics (Table 2).

Table 2.
Examples of miRNA-level changes proposed for diagnosing diseases.
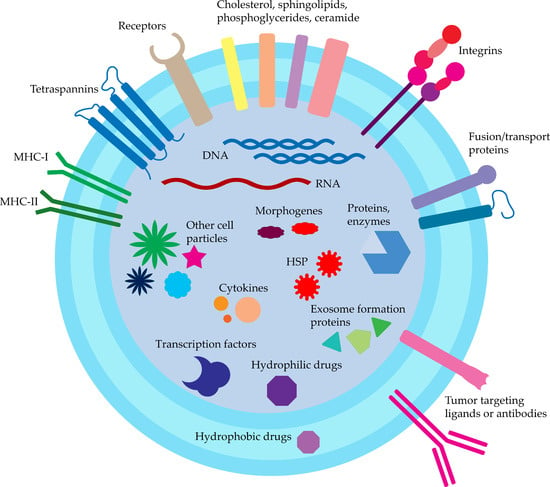
Figure 2.
Exosomes structure and cargo.
Stimulation of human cells by mouse exosomes leads to the synthesis of mouse proteins in human cells. In addition, the mRNA profile of exosomes and recipient cells was different, suggesting that mRNA is subjected to active sorting into MVB. This can be crucial in terms of therapeutic mRNA delivery by exosomes [74].
To summarize, exosomal cargo can further improve its therapeutic properties when properly designed. This can be used for the fabrication of double-action exosomes, containing both therapeutic substances with the additional effect enhancement by incorporating proper nucleic acids, lipids, and proteins into exosomes. To this date, only a few concepts of this simultaneous strategy were proposed [8,84].
6. Exosomes Innate Functions
Exosomes function will vary depending on their origin, cargo, and recipient cells. These innate properties can serve for further development and upgrading of exosome-based DDS. Exosomes’ most fundamental and widely recognized function is to mediate cell–cell signalization, thus regulating cell physiological functions and responses. This mechanism relies on the exchange of molecules described in the previous chapter.
Additionally, to date, many great articles arise with exosome function/effect description [65,85]. Thus, in this article, we only mention properties influencing DDS. Exosomes derived from bone marrow, macrophages, and tumors were described as influencing the inflammatory response, either prolonging allograft rejection time or activating inflammatory cells to the target tumor [86,87]. Exosomes also take part in the immune response and antigen presentation. Due to this fact, they can stimulate the immune response by MHC class I or II antigens presentation, which is not desirable in the case of DDS or allogenic therapeutics and can decrease their body circulation time. The current solution to this problem is exosomes isolated from mesenchymal or stromal stem cells [88].
As mentioned, the natural exosomal cargo consists of various nucleic acids, proteins, and lipids, which can directly influence cell behavior and expression pattern. Similarly, artificial siRNA, mRNA, and miRNA delivered by exosomes will regulate cell expression as reported [89]. In a similar manner, proteins can also act as procancerogenic agents, e.g., exosomes containing high levels of TGF-β and PGE2 will promote tumor growth [90]. On the other hand, dendritic-cell-derived exosomes containing MHC class I molecules and tumor antigens are able to activate T-cell anticancer response [87].
Exosomes are superior to artificial lipovesicles in terms of biocompatibility and internalization. They can also serve to reach hard-to-access and brain tissues due to the possibility of targeted delivery and blood–brain-barrier crossing, respectively [28].
7. Targeted Delivery
Another factor influencing exosomes targeted delivery is the cell origin and membrane-incorporated targeting moieties [91,92]. Isolated exosomes are proposed for targeted therapy and drug delivery to the same type of cells the exosomes were derived from. It is due to the fact that different membrane moieties are differently expressed by various cells. Surface moieties can be either expressed via genetic engineering of the endosomal sorting mechanism or fixed at the surface of released exosomes with click chemistry (Table 3) [85,92]. A similar approach is described for other DDS devices such as fibrin [93].

Table 3.
Examples of moieties for exosome targeting.
Another problem in targeted delivery is the insufficient half-life time of exosomes administered in vivo. Studies have shown the rapid clearance of exosomes by macrophages limiting the half-life of unmodified exosomes to 2–30 min [103]. To overcome this problem, additional proteins (such as CD47, PD-L1, CD31, and CD24) prolonging circulation time are added to the surface, or exosomes are coated with PEG to avoid recognition of the immune system [92,103]. Macrophage, monocyte, and MSC-derived exosomes were found to be especially immune to phagocytosis and fast body clearance [88,104].
There are also reports that certain particles, such as sphingolipids, can affect exosomes cell uptake [105].
7.1. Cell Origin
Exosomes properties depend on source cells as they are closely related to overall cell processes necessary for metabolism. Crude exosomes can act as a double-agents for cancer development according to their source. Exosomes derived from normal cells will act as anticancer/antimetastatic in opposition to the reports that cancer exosomes will further increase proliferation, metastasis, and angiogenesis of cancer cells, promoting both in vitro and in vivo growth [89,106,107]. Careful selection of the exosomes source is crucial for proper exosomal-based DDS development. Due to their bilateral mode of action, exosomes can both increase or restrain the proper DDS mode of action.
7.2. Cancer Cells
Targeted delivery of cancer exosomes is mediated by expressed surface proteins, e.g., tetraspanins, interacting with different cell types in a different manner [108]. Tetraspanins also seem responsible for further malignancy spread; thus, therapeutic usage of this type of exosomes is not recommended [109]. It is worth mentioning that cancerous exosomes were used for stimulating the immune response to prevent the remaining cancer cells to grow after resection. The immune response was mediated by tumor-specific antigens presented at the surface of exosomes administered to patients after surgery [110,111]. Additionally, exosomes released from a dorsally implanted cell-graft composed of HT29 and HCT116 cell lines preferably locate at the stomach and intestine. This may be due to the origin of the cells composing the implant, which were derived from colorectal and colon tumors [112]. This effect is also supported by higher autologous exosomes uptake by the colorectal cell line than allogenic cell-derived exosomes both in vitro and in vivo. Latter studies also revealed an increased accumulation of autologous exosomes in the tumor site [113]. This effect can be overcome by expressing on the vesicles surface cancer-targeting particles such as integrins. Designed in this manner, exosomes will specifically target cells other than autologous cells, also increasing their uptake ratio [114].
7.3. Normal Cells
Exosomes isolated from immune cells are recognized as safe, serving as delivery vehicles, and were proposed for use as vehicles for drugs, vaccines, and immunotherapy. The most common types of cells are dendritic cells, macrophages, monocytes, B cells, and T cells. These cells’ native function regulates the immunologic response and, thus, exosomes derived from them will also target cells recognized as hostile, e.g., cancer cells. Dendritic-cells-derived exosomes contain numerous membrane moieties that can induce cancer cells’ death themself. Macrophage culture purity needs to be maintained at a high profile due to the potential influence of pathogens on exosomes properties and mode of action [115]. These exosomes also convert M2 macrophages to M1 type, increasing phagocytosis of tumor cells. Additionally, macrophage-derived exosomes stimulate the inflammatory response by the release of Th-1 type cytokines, which can be utilized in cancer vaccines, where exosomes may play the role of an adjuvant [116]. Human dermal fibroblasts are also used as an exosomes source [117].
7.4. Stem Cells
MSC-derived exosomes seem to be the most suitable immunologically for DDS design. These cells were isolated from various kinds of tissues such as adipose, liver, amniotic fluid, placenta, umbilical cord, and menstrual blood stem cells [117,118]. They do not express MHC antigens at the surface, thus evading phagocytosis and fast clearance from organisms [119]. Another advantage of MSCs is the production of exosomes in greater amounts than other cells. Currently, MSCs and dendritic cells are favored for use in clinical studies [120].
7.5. Plants, Fruits, and Milk
Due to the high cost and problems with the isolation of large amounts of exosomes from the mentioned cells, plants and fruits are currently recognized as promising exosomes sources [121]. In a similar manner, bovine milk is also an easily accessible, cost-effective source, containing significant amounts of microvesicles [122]. They are generally stable in acidic environments; thus, they are great candidates for oral drugs development [118]. There was no record of modifying plants or bovines in order to obtain modified exosomes.
8. Loading Drugs to Exosomes
While designing DDS, one of the most important features is the method of drug loading. It should be chosen carefully based on the drug and the exosomes properties. The biogenesis of exosomes makes them promising drug carriers due to their biocompatibility, stability, and preferred tumor targeting [123]. They are built similarly to double-membraned liposomes and the same methods can be used for drug loading.
Exosomes can be loaded with numerous drugs, macromolecules, and other substances either by passive or active methods (Figure 3). Passive methods are simple and based on drug incubation with exosomes or with cells with further isolation of released exosomes containing drugs (Figure 3I,II). This method depends on the difference in the drug concentration gradient; thus, during the incubation with exosomes, the substance itself will migrate to the vesicle. This process can be intensified by applying an additional force, such as shaking or stirring [9,124]. In a similar matter, the incubation of cells with a drug will result in the release of exosomes loaded with the drug. The downsides of this method are low loading efficiency and low-output drug concentration (about 1%) [124]. Incubation of drugs with exosomes or other DDS vehicles is a very common and simple method, especially popular with hydrophobic drugs [122,125].
Higher loading efficiency can be achieved with active loading methods, including sonication (~28%), electroporation (~5%), extrusion, freeze–thaw, pH gradient (1.7%), and conjugation methods [124]. Both sonication and electroporation will result in a temporary disruption of the exosome membrane, mediated by sonic waves or electric pulses, respectively (Figure 3III,IV). Afterward, exosomes will accumulate the drug from the solution through small membrane pores induced by mentioned agents. These methods do not seem to alter exosome membrane properties but cause an increase in exosomes size [124,126]. The extrusion method is also based on an exosome membrane disruption by a syringe-based lipid extruder (Figure 3V) [127]. In one experiment, the freeze–thaw loading method was used with unknown efficacy but observable therapeutic effects (Figure 3VI) [128]. In addition, a method based on a pH-driven drug loading was presented but needs extended research due to the very low drug loading efficiency [129]. Another approach is to conjugate drugs to exosomes via antibodies or click-chemistry. The antibodies will bind non-covalently to the exosomes surface via their affinity to structural elements of the exosomes (Figure 3VII). The chemical synthesis will result in covalently bound drugs to exosomes via, e.g., PEG or membrane proteins, and the further release dependent on exosomes degradation. These methods can also serve to target exosomes to desired cells by the introduction of specific receptors to the membrane [7].
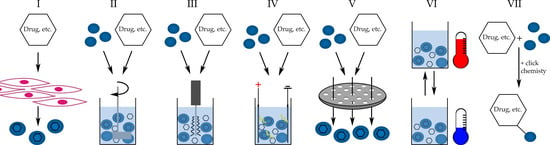
Figure 3.
Methods of loading drugs into exosomes. (I)—drug loaded exosomes released from drug exposed cells, (II)—drug incubation with exosomes with additional stirring, (III)—sonication, (IV)—electroporation, (V)—extrusion, (VI)—freeze-thaw cycles, (VII)—chemical conjugation of drug to exosome.
Mentioned methods allow for the incorporation of drugs into the inside of exosomes or the phospholipid bilayer depending on their hydro- and lipophilic properties. Hydrophilic substances will preferably locate in the lumen of exosomes, whilst substances soluble in lipids will accumulate at the membrane of exosomes (Figure 2). Some studies report that hydrophilic substances are easily loaded into exosomes, while lipophilic substances are loaded less efficiently. Generally, the substances are released from exosomes in about 50% during 24 h in vitro [122].
An extensive evaluation is needed for the assessment of the influence of mentioned loading methods on the structure and properties of exosomes. To date, aggregation and fusion were confirmed for electroporation [130].
Thus far, exosomes were used to deliver a variety of substances including chemotherapeutics (paclitaxel, doxorubicin, and taxol), RNA, and DNA (Table 4). Exosome-based designs of vaccines against bacterial (toxoplasmosis and salmonellosis), viral (AIDS and hepatitis B), cancer (lung, pancreatic, colon, brain, and breast), and other diseases were also proposed [131]. Exosomes were also used as a device for gene editing tools delivery such as Cas9 [132]. Exosomes are either collected from the disease culture medium (bacterial vaccines) or modified with genetic engineering methods. The mode of action of these vaccines is mainly by the presentation of immunogenic surface particles. There are some reports on completed and running clinical trials including exosome-mediated drug delivery with promising outcomes [120,133].
Route of Administration
According to current studies, exosomes are mostly administrated intravenously and locate preferably in the liver. Only a few studies used another administration method, namely nasal, oral, and intraperitoneal. In these cases, exosomes are preferably located in the lungs and brain, stomach, and liver, respectively. There is a clear connection between the administration route and exosomes accumulation [88,134,135]. This is also confirmed in currently completed clinical studies, as reported at clinicaltrails.gov (accessed on 15 November 2022). For instance, few studies described the use of exosomal inhalations in the treatment of pulmonary diseases such as SARS-CoV-2-related ones [136]. These studies reported a very good tolerance of exosomes. Other clinically tested applications were: ointment, oral applications, intra-disc injection, and intravenous injection.

Table 4.
Examples of drugs, macromolecules, and potential targeted therapy strategies with exosomes.
Table 4.
Examples of drugs, macromolecules, and potential targeted therapy strategies with exosomes.
| Drug/Macromolecule | Loading Efficiency and Method | Origin | Targeted Cells/Tissue | Effect | Ref. |
|---|---|---|---|---|---|
| Paclitaxel | 1.4%; RT incubation; 5.3%; electroporation; 28.3%; mild sonication | RAW 264.7 cell line | Madin–Darby canine kidney MDCKWT and MDCKMDR1 cells, Murine Lewis lung carcinoma cell subline (3LL-M27) | Over ×50 cytotoxicity increase for multiple-drug-resistant cell line. | [124] |
| Doxorubicin | 7.4%; sonication and subsequent extrusion | 4T1 cell line | MCF-7 cell line | Near-infrared laser-triggered doxorubicin release from exosomes modified with Fe3O4. | [137] |
| Doxorubicin | 6.5%; sonication | murine bone marrow of male KM mice | Zebrafish, C6-Luc glioma-bearing mice | Rapid blood–brain-barrier crossing and brain accumulation. Targeting of infiltrating brain tumor cells. | [138] |
| hsa-miR148a-3p | ND; chemical transfection (Lipofectamine 2000) | Bovine milk | HepG2, Caco-2 cell lines | Cost-effective source of exosomes. Time-dependent cell incorporation. | [139] |
| Paclitaxel | 8%; RT incubation | Bovine milk | Lung tumor xenograft in nude mice | Oral delivery. Tumor growth inhibition. Lower systemic and immunogenic toxicities compared to intravenous administration. | [140] |
| Taxol | 14%; cells incubation with drug | MSC from umbilical cord | A549, SK-OV-3, MDA-hyb1 cell lines; MDA-hyb1 breast tumors in NODscid mice | Reduced cancer growth and metastasis similar to ×1000 higher concentration of free taxol administration than exosomes containing taxol. | [141] |
| Erastin | 3.2 mg erastin/mg protein; sonication | HFL-1 cell line | MDA-MB-231 cell line | Folate-labeled exosomes for targeted delivery, promotion of ferroptosis, decreased proliferation, and migration of cancer cells. | [101] |
| CRISPR/Cas9 plasmid DNA | chemical transfection of exosomes from MSC (Exo-Fect™ Exosome Transfection Kit); chemical transfection (Lipofectamine 2000) of cells (HEK293T) and isolation of exosomes from conditioned medium | MSC; HEK293T cell line | KPC689 cell line | Successfully disrupted KrasG12D oncogenic allele in pancreatic cancer cells. Inhibition of proliferation and tumor growth | [142] |
| siRNA targeting KRAS in PEI matrix; Plasmid DNA coding p53 | >90%; incubation with PEI matrix; <5%; electroporation; 35% chemical transfection with Exo-Fect™ (about 35%) | Bovine colostrum | H1299, A549, H522, Panc-1, MiaPaCa-2 cell lines; in vivo A549 xenograft models | Inhibition of tumor growth and KRAS expression. Induced expression of p53 in p53-null H1299 cells. | [143] |
| Doxorubicin; cholesterol-modified miRNA159 | 74.5–160.6 ng/μg exosomes; incubation in triethylamine solution;1.2% of miRNA, 5.3% of cholesterol-modified miRNA | THP-1 cell line | MDA-MB-231 cell line | Targeting properties of exosomes, synergistic therapeutic effects on cancer cells, inhibition of growth and motility. Silencing of TCF-7 gene. | [8] |
| 5-fluorouracil; miR-21 inhibitor oligonucleotide | 3.1%; 0.5%; electroporation | 293T cell line | HCT-116SFR cell line; in vivo BALB/c nude mice | Successful co-delivery, down-regulation of miR-21 in cells, induction of cycle arrest, reduction of proliferation, drug resistance renversement, 5-FU cytotoxicity increase, reduction in tumor growth in vivo | [10] |
| miR-31-5p | N/A; electroporation | Bovine milk | HUVEC cell line; in vivo BALB/c mice | Improved cells function in vitro, enhancement of angiogenesis, and wound healing in diabetic mice in vivo | [144] |
| CD47 and SIRPα antibodies | Conjugated to exosome surface by through pH-sensitive linker | RAW264.7 cell line | RAW264.7 cell line, in vivo BALB/C mice | Targeting to CD47 expressing cells, improvement of macrophages phagocytosis, exosomal reprogramming of macrophages towards anti-cancer activity | [145] |
| galectin-9 siRNA, oxaliplatin | 13.17% N/A electroporation of galectin 9, 13.17% maleimide-thiol conjugates | BM-MSC | PANC-02 cell line, in vivo C57BL/6 mice, SD rats | Significant anticancer activity, Improvement of macrophage tumor suppressive activity, increase in recruitment of cytotoxic T lymphocytes, Treg downregulation | [146] |
| Berberine | 17.13%, sonication | Primary macrophages | C57BL/6J mice | Induction of macrophages to anti-inflammatory and anti-apoptotic M2 phenotype, improvement in mice movement after spinal injury | [147] |
| Erastin, Rose Bengal, CD47 surface labeled exosomes | 60%, 84% encapsulation rates, sonication CD 47 plasmid transfected donor cells | HEK293T cell line | RAW264.7 Hepa1-6 cell lines, in vivo C57BL/6 | Deterred exosome phagocytosis, in vivo and in vitro ferroptosis induction after laser irradiation, decrease in liver and kidney cytotoxicity of exosomes | [148] |
| miR-138-5p | Lentivirus transfected donor cells | Adipose-derived stem cells | T24, 5637 cell line, in vivo BALB/C nude mice | Bladder cancer cells proliferation, migration, and invasion decrease, suppression of tumor growth in vivo | [149] |
PEI—polyethyleneimine.
9. Conclusions and Further Directions
Due to extensive research, exosomes are becoming promising novel drug delivery devices. Many great advancements are achieved in the field of novel exosomes isolation techniques, thus achieving rapid, efficient, and precise isolation from a small-volume sample. Clinically, this is very promising for the future designing of exosome-based assays, biomarkers screening, and also disease diagnosis and differentiation, delivering results in a short matter of time. Nevertheless, these methods still require further validation for specificity, batch-to-batch variations, and selectivity toward specific exosomes types. Another problem is cell source and cost of purification, as gradual progress is continuously performed in this manner. Currently, milk and fruits are the most promising candidates for large-scale, commercial isolation of exosomes.
A vast variety of characterization methods is currently available, which is essential for the proper discrimination of exosomes in terms of drug delivery. This includes exosomes markers confirmation, cargo screening, and surface receptors recognition. These properties are especially important while comparing exosomes isolated from different donor cells. Especially, MSC-derived exosomes are the most promising devices for drug delivery.
The utilization of both therapeutics and exosomes benefits allows for a drug concentration decrease with the adjacent increase in efficiency and reduction in systemic side effects. Nevertheless, there is still space for upgrading drug loading efficiency to exosomes.
More attention should be given to the innate exosomes properties and their utilization in combined therapy development and improvement. Further efforts are key to better exosomes properties recognition, thus allowing a better understanding of exosomes-mediated targeted delivery. Despite the recognized natural exosome targeting properties, they are not relevant clinically, mostly due to low precision of delivery. There are significant efforts in the modification of exosome donor cells and exosomes itself in order to improve targeted delivery, but this requires further research. In addition, despite the significant number of clinical studies in the field of exosome-based drug delivery systems, there should be effort made to access the long-term toxicity, immunogenicity, and safety of exosomes. In addition, a sort of “gold standard” for therapeutic exosomes in terms of isolation, description, and route of administration should be stated as a reference for further research. The optimal route of administration is still not recognized, as a majority of studies apply exosomes intravenously. Summarizing, exosomes have great potential as theranostics devices, serving both as disease discovery and further therapy.
Author Contributions
Conceptualization: J.R.; Investigation: J.R.; writing—original draft preparation: J.R., A.G.-P., S.G. and I.B.; writing—review and editing: J.R. and I.B.; critical review: I.B.; visualization: J.R.; supervision: I.B.; funding acquisition: J.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Medical University of Silesia, grant number PCN-2-091/N/1/I.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ferreira, D.; Moreira, J.N.; Rodrigues, L.R. New advances in exosome-based targeted drug delivery systems. Crit. Rev. Oncol. Hematol. 2022, 172, 103628. [Google Scholar] [CrossRef] [PubMed]
- Amiri, A.; Bagherifar, R.; Ansari Dezfouli, E.; Kiaie, S.H.; Jafari, R.; Ramezani, R. Exosomes as bio-inspired nanocarriers for RNA delivery: Preparation and applications. J. Transl. Med. 2022, 20, 125. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Bi, J.; Huang, J.; Tang, Y.; Du, S.; Li, P. Exosome: A review of its classification, isolation techniques, storage, diagnostic and targeted therapy applications. Int. J. Nanomed. 2020, 15, 6917–6934. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef]
- Bunggulawa, E.J.; Wang, W.; Yin, T.; Wang, N.; Durkan, C.; Wang, Y.; Wang, G. Recent advancements in the use of exosomes as drug delivery systems. J. Nanobiotechnol. 2018, 16, 81. [Google Scholar] [CrossRef]
- Patil, S.M.; Sawant, S.S.; Kunda, N.K. Exosomes as drug delivery systems: A brief overview and progress update. Eur. J. Pharm. Biopharm. 2020, 154, 259–269. [Google Scholar] [CrossRef]
- Liang, Y.; Duan, L.; Lu, J.; Xia, J. Engineering exosomes for targeted drug delivery. Theranostics 2021, 11, 3183–3195. [Google Scholar] [CrossRef]
- Gong, C.; Tian, J.; Wang, Z.; Gao, Y.; Wu, X.; Ding, X.; Qiang, L.; Li, G.; Han, Z.; Yuan, Y.; et al. Functional exosome-mediated co-delivery of doxorubicin and hydrophobically modified microRNA 159 for triple-negative breast cancer therapy. J. Nanobiotechnol. 2019, 17, 93. [Google Scholar] [CrossRef]
- Qi, H.; Liu, C.; Long, L.; Ren, Y.; Zhang, S.; Chang, X.; Qian, X.; Jia, H.; Zhao, J.; Sun, J.; et al. Blood exosomes endowed with magnetic and targeting properties for cancer therapy. ACS Nano 2016, 10, 3323–3333. [Google Scholar] [CrossRef]
- Liang, G.; Zhu, Y.; Ali, D.J.; Tian, T.; Xu, H.; Si, K.; Sun, B.; Chen, B.; Xiao, Z. Engineered exosomes for targeted co-delivery of mir-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J. Nanobiotechnol. 2020, 18, 10. [Google Scholar] [CrossRef]
- Morelli, A.E.; Larregina, A.T.; Shufesky, W.J.; Sullivan, M.L.G.; Stolz, D.B.; Papworth, G.D.; Zahorchak, A.F.; Logar, A.J.; Wang, Z.; Watkins, S.C.; et al. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood 2004, 104, 3257–3266. [Google Scholar] [CrossRef] [PubMed]
- Abels, E.R.; Breakefield, X.O. Introduction to extracellular vesicles: Biogenesis, RNA cargo selection, content, release, and uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, V.; Ludwig, A.-K.; Hornung, S.; Rotan, O.; Horn, P.A.; Epple, M.; Giebel, B. Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids Surf. B Biointerfaces 2011, 87, 146–150. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, Y.; Chang, X.; Ba, X.; Hu, N.; Liu, Q.; Fang, L.; Wang, Z. Melanoma-derived exosomes endow fibroblasts with an invasive potential via mir-21 target signaling pathway. Cancer Manag. Res. 2020, 12, 12965–12974. [Google Scholar] [CrossRef]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L.; et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef]
- Shimomura, T.; Seino, R.; Umezaki, K.; Shimoda, A.; Ezoe, T.; Ishiyama, M.; Akiyoshi, K. New lipophilic fluorescent dyes for exosome labeling: Monitoring of cellular uptake of exosomes. bioRxiv 2020, 4–6. [Google Scholar] [CrossRef]
- Thane, K.E.; Davis, A.M.; Hoffman, A.M. Improved methods for fluorescent labeling and detection of single extracellular vesicles using nanoparticle tracking analysis. Sci. Rep. 2019, 9, 12295. [Google Scholar] [CrossRef]
- Noble, J.M.; Roberts, L.M.; Vidavsky, N.; Chiou, A.E.; Fischbach, C.; Paszek, M.J.; Estroff, L.A.; Kourkoutis, L.F. Direct comparison of optical and electron microscopy methods for structural characterization of extracellular vesicles. J. Struct. Biol. 2020, 210, 107474. [Google Scholar] [CrossRef]
- Romanò, S.; Di Giacinto, F.; Primiano, A.; Mazzini, A.; Panzetta, C.; Papi, M.; Di Gaspare, A.; Ortolani, M.; Gervasoni, J.; De Spirito, M.; et al. Fourier transform infrared spectroscopy as a useful tool for the automated classification of cancer cell-derived exosomes obtained under different culture conditions. Anal. Chim. Acta 2020, 1140, 219–227. [Google Scholar] [CrossRef]
- Ramos-Garcia, V.; Ten-Doménech, I.; Moreno-Giménez, A.; Gormaz, M.; Parra-Llorca, A.; Shephard, A.P.; Sepúlveda, P.; Pérez-Guaita, D.; Vento, M.; Lendl, B.; et al. ATR-FTIR spectroscopy for the routine quality control of exosome isolations. Chemom. Intell. Lab. Syst. 2021, 217, 104401. [Google Scholar] [CrossRef]
- Wu, Y.; Deng, W.; Klinke II, D.J. Exosomes: Improved methods to characterize their morphology, RNA content, and surface protein biomarkers. Analyst 2015, 140, 6631–6642. [Google Scholar] [CrossRef] [PubMed]
- Cizmar, P.; Yuana, Y. Detection and characterization of extracellular vesicles by transmission and cryo-transmission electron microscopy. In Extracellular Vesicles. Methods in Molecular Biology; Kuo, W., Jia, S., Eds.; Humana Press: New York, NY, USA, 2017; Volume 1660. [Google Scholar] [CrossRef]
- Khodashenas, S.; Khalili, S.; Forouzandeh Moghadam, M. A cell ELISA based method for exosome detection in diagnostic and therapeutic applications. Biotechnol. Lett. 2019, 41, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Davie, E.; Kulman, J. An overview of the structure and function of thrombin. Semin. Thromb. Hemost. 2006, 32, 003–015. [Google Scholar] [CrossRef] [PubMed]
- Corso, G.; Heusermann, W.; Trojer, D.; Görgens, A.; Steib, E.; Voshol, J.; Graff, A.; Genoud, C.; Lee, Y.; Hean, J.; et al. Systematic characterization of extracellular vesicle sorting domains and quantification at the single molecule—Single vesicle level by fluorescence correlation spectroscopy and single particle imaging. J. Extracell. Vesicles 2019, 8, 1663043. [Google Scholar] [CrossRef]
- Jung, H.H.; Kim, J.-Y.; Lim, J.E.; Im, Y.-H. Cytokine profiling in serum-derived exosomes isolated by different methods. Sci. Rep. 2020, 10, 14069. [Google Scholar] [CrossRef]
- Mittal, R.; Bencie, N.; Langlie, J.; Mittal, J.; Eshraghi, A.A. Exosomes as drug delivery vehicles and biomarkers for neurological and auditory systems. J. Cell. Physiol. 2021, 236, 8035–8049. [Google Scholar] [CrossRef]
- Shao, H.; Im, H.; Castro, C.M.; Breakefield, X.; Weissleder, R.; Lee, H. New technologies for analysis of extracellular vesicles. Chem. Rev. 2018, 118, 1917–1950. [Google Scholar] [CrossRef]
- Kanwar, S.S.; Dunlay, C.J.; Simeone, D.M.; Nagrath, S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip 2014, 14, 1891–1900. [Google Scholar] [CrossRef]
- Zhang, P.; He, M.; Zeng, Y. Ultrasensitive microfluidic analysis of circulating exosomes using a nanostructured graphene oxide/polydopamine coating. Lab Chip 2016, 16, 3033–3042. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, H.; Fine, D.; Schmulen, J.; Hu, Y.; Godin, B.; Zhang, J.X.J.; Liu, X. Ciliated micropillars for the microfluidic-based isolation of nanoscale lipid vesicles. Lab Chip 2013, 13, 2879. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Yu, Z.; Chen, D.; Wang, Z.; Miao, J.; Li, Q.; Zhang, D.; Song, J.; Cui, D. Progress in microfluidics-based exosome separation and detection technologies for diagnostic applications. Small 2020, 16, 1903916. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Tian, H.; Li, X.; Jin, D.; Li, X.; Kong, J.; Yang, C.; Yang, X.; Lu, Y.; Luo, Y.; et al. Clinical application of a microfluidic chip for immunocapture and quantification of circulating exosomes to assist breast cancer diagnosis and molecular classification. PLoS ONE 2017, 12, e0175050. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; Park, J.; Pathania, D.; Castro, C.M.; Weissleder, R.; Lee, H. Integrated magneto–electrochemical sensor for exosome analysis. ACS Nano 2016, 10, 1802–1809. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Chung, J.; Balaj, L.; Charest, A.; Bigner, D.D.; Carter, B.S.; Hochberg, F.H.; Breakefield, X.O.; Weissleder, R.; Lee, H. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat. Med. 2012, 18, 1835–1840. [Google Scholar] [CrossRef]
- Ashcroft, B.A.; de Sonneville, J.; Yuana, Y.; Osanto, S.; Bertina, R.; Kuil, M.E.; Oosterkamp, T.H. Determination of the size distribution of blood microparticles directly in plasma using atomic force microscopy and microfluidics. Biomed. Microdevices 2012, 14, 641–649. [Google Scholar] [CrossRef]
- Im, H.; Shao, H.; Park, Y.I.; Peterson, V.M.; Castro, C.M.; Weissleder, R.; Lee, H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014, 32, 490–495. [Google Scholar] [CrossRef]
- Yoshioka, Y.; Kosaka, N.; Konishi, Y.; Ohta, H.; Okamoto, H.; Sonoda, H.; Nonaka, R.; Yamamoto, H.; Ishii, H.; Mori, M.; et al. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using exoscreen. Nat. Commun. 2014, 5, 3591. [Google Scholar] [CrossRef]
- Petersen, K.E.; Manangon, E.; Hood, J.L.; Wickline, S.A.; Fernandez, D.P.; Johnson, W.P.; Gale, B.K. A review of exosome separation techniques and characterization of b16-f10 mouse melanoma exosomes with AF4-UV-MALS-DLS-TEM. Anal. Bioanal. Chem. 2014, 406, 7855–7866. [Google Scholar] [CrossRef]
- Kim, Y.B.; Yang, J.S.; Lee, G.B.; Moon, M.H. Evaluation of exosome separation from human serum by FRIT-inlet asymmetrical flow field-flow fractionation and multiangle light scattering. Anal. Chim. Acta 2020, 1124, 137–145. [Google Scholar] [CrossRef]
- Zhou, M.; Weber, S.R.; Zhao, Y.; Chen, H.; Sundstrom, J.M. Methods for exosome isolation and characterization. In Exosomes; Elsevier: Amsterdam, The Netherlands, 2020; pp. 23–38. [Google Scholar]
- Gardiner, C.; Vizio, D.D.; Sahoo, S.; Théry, C.; Witwer, K.W.; Wauben, M.; Hill, A.F. Techniques used for the isolation and characterization of extracellular vesicles: Results of a worldwide survey. J. Extracell. Vesicles 2016, 5, 32945. [Google Scholar] [CrossRef] [PubMed]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta—Gen. Subj. 2012, 1820, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Jung, R.E.; Schmoekel, H.G.; Zwahlen, R.; Kokovic, V.; Hammerle, C.H.F.; Weber, F.E. Platelet-rich plasma and fibrin as delivery systems for recombinant human bone morphogenetic protein-2. Clin. Oral Implant. Res. 2005, 16, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Cai, J.; Zitkovsky, H.S.; Chen, B.; Guo, L. Comparison of yield, purity, and functional properties of large-volume exosome isolation using ultrafiltration and polymer-based precipitation. Plast. Reconstr. Surg. 2022, 149, 638–649. [Google Scholar] [CrossRef]
- Barreiro, K.; Huber, T.B.; Holthofer, H. Isolating urinary extracellular vesicles as biomarkers for diabetic disease. In Diabetic Nephropathy; Humana: New York, NY, USA, 2020; pp. 175–188. [Google Scholar]
- La Shu, S.; Yang, Y.; Allen, C.L.; Hurley, E.; Tung, K.H.; Minderman, H.; Wu, Y.; Ernstoff, M.S. Purity and yield of melanoma exosomes are dependent on isolation method. J. Extracell. Vesicles 2020, 9, 1692401. [Google Scholar] [CrossRef]
- Doldán, X.; Fagúndez, P.; Cayota, A.; Laíz, J.; Tosar, J.P. Electrochemical sandwich immunosensor for determination of exosomes based on surface marker-mediated signal amplification. Anal. Chem. 2016, 88, 10466–10473. [Google Scholar] [CrossRef]
- Bohmer, N.; Demarmels, N.; Tsolaki, E.; Gerken, L.; Keevend, K.; Bertazzo, S.; Lattuada, M.; Herrmann, I.K. Removal of cells from body fluids by magnetic separation in batch and continuous mode: Influence of bead size, concentration, and contact time. ACS Appl. Mater. Interfaces 2017, 9, 29571–29579. [Google Scholar] [CrossRef]
- Mousavi, S.M.; Amin Mahdian, S.M.; Ebrahimi, M.S.; Taghizadieh, M.; Vosough, M.; Nahand, J.S.; Hosseindoost, S.; Vousooghi, N.; Javar, H.A.; Larijani, B.; et al. Microfluidics for detection of exosomes and microRNAs in cancer: State of the art. Mol. Ther.—Nucleic Acids 2022, 28, 758–791. [Google Scholar] [CrossRef]
- Ghosh, A.; Davey, M.; Chute, I.C.; Griffiths, S.G.; Lewis, S.; Chacko, S.; Barnett, D.; Crapoulet, N.; Fournier, S.; Joy, A.; et al. Rapid isolation of extracellular vesicles from cell culture and biological fluids using a synthetic peptide with specific affinity for heat shock proteins. PLoS ONE 2014, 9, e110443. [Google Scholar] [CrossRef]
- Wang, J.; Li, W.; Zhang, L.; Ban, L.; Chen, P.; Du, W.; Feng, X.; Liu, B.-F. Chemically edited exosomes with dual ligand purified by microfluidic device for active targeted drug delivery to tumor cells. ACS Appl. Mater. Interfaces 2017, 9, 27441–27452. [Google Scholar] [CrossRef]
- Oh, S.; Kang, D.; Ahn, S.-M.; Simpson, R.J.; Lee, B.-H.; Moon, M.H. Miniaturized asymmetrical flow field-flow fractionation: Application to biological vesicles. J. Sep. Sci. 2007, 30, 1082–1087. [Google Scholar] [CrossRef]
- Kang, D.; Oh, S.; Ahn, S.-M.; Lee, B.-H.; Moon, M.H. Proteomic analysis of exosomes from human neural stem cells by flow field-flow fractionation and nanoflow liquid chromatography−tandem mass spectrometry. J. Proteome Res. 2008, 7, 3475–3480. [Google Scholar] [CrossRef] [PubMed]
- Paulaitis, M.; Guzman, N.; Agarwal, K.; Saji, M. Abstract P3-09-04: Exosome-specific microRNA signatures in combination with characteristic surface markers on the circulating exosomes themselves provide new insights into the EMT. In Poster Session Abstracts; American Association for Cancer Research: Philadelphia, PA, USA, 2010; p. P3-09-04. [Google Scholar]
- Gao, Y.; Wu, M.; Lin, Y.; Xu, J. Acoustic microfluidic separation techniques and bioapplications: A review. Micromachines 2020, 11, 921. [Google Scholar] [CrossRef] [PubMed]
- Salafi, T.; Zhang, Y.; Zhang, Y. A review on deterministic lateral displacement for particle separation and detection. Nano-Micro Lett. 2019, 11, 77. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.T.; Wunsch, B.H.; Dogra, N.; Ahsen, M.E.; Lee, K.; Yadav, K.K.; Weil, R.; Pereira, M.A.; Patel, J.V.; Duch, E.A.; et al. Integrated nanoscale deterministic lateral displacement arrays for separation of extracellular vesicles from clinically-relevant volumes of biological samples. Lab Chip 2018, 18, 3913–3925. [Google Scholar] [CrossRef]
- Ayala-Mar, S.; Perez-Gonzalez, V.H.; Mata-Gómez, M.A.; Gallo-Villanueva, R.C.; González-Valdez, J. Electrokinetically driven exosome separation and concentration using dielectrophoretic-enhanced PDMS-based microfluidics. Anal. Chem. 2019, 91, 14975–14982. [Google Scholar] [CrossRef]
- Chen, H.; Yamakawa, T.; Inaba, M.; Nakano, M.; Suehiro, J. Characterization of extra-cellular vesicle dielectrophoresis and estimation of its electric properties. Sensors 2022, 22, 3279. [Google Scholar] [CrossRef]
- Al-Nedawi, K.; Meehan, B.; Micallef, J.; Lhotak, V.; May, L.; Guha, A.; Rak, J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008, 10, 619–624. [Google Scholar] [CrossRef]
- van Niel, G.; Porto-Carreiro, I.; Simoes, S.; Raposo, G. Exosomes: A common pathway for a specialized function. J. Biochem. 2006, 140, 13–21. [Google Scholar] [CrossRef]
- Lakkaraju, A.; Rodriguez-Boulan, E. Itinerant exosomes: Emerging roles in cell and tissue polarity. Trends Cell Biol. 2008, 18, 199–209. [Google Scholar] [CrossRef]
- Schorey, J.S.; Bhatnagar, S. Exosome function: From tumor immunology to pathogen biology. Traffic 2008, 9, 871–881. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Rasool, H.I.; Palanisamy, V.; Mathisen, C.; Schmidt, M.; Wong, D.T.; Gimzewski, J.K. Structural-mechanical characterization of nanoparticle exosomes in human saliva, using correlative afm, fesem, and force spectroscopy. ACS Nano 2010, 4, 1921–1926. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.T.; Teng, K.; Wu, C.; Adam, M.; Johnstone, R.M. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol. 1985, 101, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, R.M. Exosomes biological significance: A concise review. Blood Cells Mol. Dis. 2006, 36, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Pisitkun, T.; Shen, R.-F.; Knepper, M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA 2004, 101, 13368–13373. [Google Scholar] [CrossRef]
- Exocarta. Available online: http://www.exocarta.org/ (accessed on 8 September 2022).
- Vesiclepedia. Available online: http://microvesicles.org/ (accessed on 8 September 2022).
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Kosaka, N.; Izumi, H.; Sekine, K.; Ochiya, T. MicroRNA as a new immune-regulatory agent in breast milk. Silence 2010, 1, 7. [Google Scholar] [CrossRef]
- Tanaka, Y.; Kamohara, H.; Kinoshita, K.; Kurashige, J.; Ishimoto, T.; Iwatsuki, M.; Watanabe, M.; Baba, H. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer 2013, 119, 1159–1167. [Google Scholar] [CrossRef]
- Wang, H.; Hou, L.; Li, A.; Duan, Y.; Gao, H.; Song, X. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. Biomed Res. Int. 2014, 2014, 864894. [Google Scholar] [CrossRef]
- Machida, T.; Tomofuji, T.; Maruyama, T.; Yoneda, T.; Ekuni, D.; Azuma, T.; Miyai, H.; Mizuno, H.; Kato, H.; Tsutsumi, K.; et al. MiR-1246 and miR-4644 in salivary exosome as potential biomarkers for pancreatobiliary tract cancer. Oncol. Rep. 2016, 36, 2375–2381. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Aswath, K.; Schroeder, S.G.; Lippolis, J.D.; Reinhardt, T.A.; Sonstegard, T.S. MicroRNA expression profiles of bovine milk exosomes in response to staphylococcus aureus infection. BMC Genom. 2015, 16, 806. [Google Scholar] [CrossRef]
- Huang, X.; Yuan, T.; Liang, M.; Du, M.; Xia, S.; Dittmar, R.; Wang, D.; See, W.; Costello, B.A.; Quevedo, F.; et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur. Urol. 2015, 67, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Perez-Hernandez, J.; Martinez-Arroyo, O.; Ortega, A.; Galera, M.; Solis-Salguero, M.A.; Chaves, F.J.; Redon, J.; Forner, M.J.; Cortes, R. Urinary exosomal miR-146a as a marker of albuminuria, activity changes and disease flares in lupus nephritis. J. Nephrol. 2021, 34, 1157–1167. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Mao, J.; Wang, B.; Wang, L.; Wen, H.; Xu, L.; Fu, J.; Yang, H. Exosomal miR-1910-3p promotes proliferation, metastasis, and autophagy of breast cancer cells by targeting MTMR3 and activating the NF-κB signaling pathway. Cancer Lett. 2020, 489, 87–99. [Google Scholar] [CrossRef]
- Yang, H.; Fu, H.; Wang, B.; Zhang, X.; Mao, J.; Li, X.; Wang, M.; Sun, Z.; Qian, H.; Xu, W. Exosomal miR-423-5p targets SUFU to promote cancer growth and metastasis and serves as a novel marker for gastric cancer. Mol. Carcinog. 2018, 57, 1223–1236. [Google Scholar] [CrossRef]
- Sun, S.; Chen, H.; Xu, C.; Zhang, Y.; Zhang, Q.; Chen, L.; Ding, Q.; Deng, Z. Exosomal miR-106b serves as a novel marker for lung cancer and promotes cancer metastasis via targeting PTEN. Life Sci. 2020, 244, 117297. [Google Scholar] [CrossRef]
- Zhan, Q.; Yi, K.; Cui, X.; Li, X.; Yang, S.; Wang, Q.; Fang, C.; Tan, Y.; Li, L.; Xu, C.; et al. Blood exosomes-based targeted delivery of cPLA2 siRNA and metformin to modulate glioblastoma energy metabolism for tailoring personalized therapy. Neuro Oncol. 2022, 24, 1871–1883. [Google Scholar] [CrossRef]
- He, J.; Ren, W.; Wang, W.; Han, W.; Jiang, L.; Zhang, D.; Guo, M. Exosomal targeting and its potential clinical application. Drug Deliv. Transl. Res. 2021, 12, 2385–2402. [Google Scholar] [CrossRef]
- Pêche, H.; Heslan, M.; Usal, C.; Amigorena, S.; Cuturi, M.C. Presentation of donor major histocompatibility complex antigens by bone marrow dendritic cell-derived exosomes modulates allograft rejection. Transplantation 2003, 76, 1503–1510. [Google Scholar] [CrossRef]
- Hao, S.; Bai, O.; Yuan, J.; Qureshi, M.; Xiang, J. Dendritic cell-derived exosomes stimulate stronger CD8+ CTL responses and antitumor immunity than tumor cell-derived exosomes. Cell. Mol. Immunol. 2006, 3, 205–211. [Google Scholar] [PubMed]
- Yi, Y.W.; Lee, J.H.; Kim, S.-Y.; Pack, C.-G.; Ha, D.H.; Park, S.R.; Youn, J.; Cho, B.S. Advances in analysis of biodistribution of exosomes by molecular imaging. Int. J. Mol. Sci. 2020, 21, 665. [Google Scholar] [CrossRef]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Croce, C.M. A new role for microRNAs, as ligands of Toll-like receptors. RNA Biol. 2013, 10, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.; Poliakov, A.; Liu, C.; Liu, Y.; Deng, Z.; Wang, J.; Cheng, Z.; Shah, S.V.; Wang, G.-J.; Zhang, L.; et al. Induction of myeloid-derived suppressor cells by tumor exosomes. Int. J. Cancer 2009, 124, 2621–2633. [Google Scholar] [CrossRef] [PubMed]
- Wiklander, O.P.B.; Nordin, J.Z.; O’Loughlin, A.; Gustafsson, Y.; Corso, G.; Mäger, I.; Vader, P.; Lee, Y.; Sork, H.; Seow, Y.; et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J. Extracell. Vesicles 2015, 4, 26316. [Google Scholar] [CrossRef] [PubMed]
- Luan, X.; Sansanaphongpricha, K.; Myers, I.; Chen, H.; Yuan, H.; Sun, D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol. Sin. 2017, 38, 754–763. [Google Scholar] [CrossRef] [PubMed]
- Rech, J.; Rogóż, W.; Borecka, A.; Turek, A. Application of fibrin in tissue engineering. Achievements and perspectives. Postep. Hig. Med. Dosw. 2021, 75, 749–761. [Google Scholar] [CrossRef]
- Cui, G.; Guo, H.; Li, H.; Zhai, Y.; Gong, Z.; Wu, J.; Liu, J.; Dong, Y.; Hou, S.; Liu, J. RVG-modified exosomes derived from mesenchymal stem cells rescue memory deficits by regulating inflammatory responses in a mouse model of Alzheimer’s disease. Immun. Ageing 2019, 16, 10. [Google Scholar] [CrossRef]
- Escrevente, C.; Keller, S.; Altevogt, P.; Costa, J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer 2011, 11, 108. [Google Scholar] [CrossRef]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef]
- Zhao, L.; Gu, C.; Gan, Y.; Shao, L.; Chen, H.; Zhu, H. Exosome-mediated siRNA delivery to suppress postoperative breast cancer metastasis. J. Control. Release 2020, 318, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Laulagnier, K.; Javalet, C.; Hemming, F.J.; Chivet, M.; Lachenal, G.; Blot, B.; Chatellard, C.; Sadoul, R. Amyloid precursor protein products concentrate in a subset of exosomes specifically endocytosed by neurons. Cell. Mol. Life Sci. 2018, 75, 757–773. [Google Scholar] [CrossRef] [PubMed]
- Toda, Y.; Takata, K.; Nakagawa, Y.; Kawakami, H.; Fujioka, S.; Kobayashi, K.; Hattori, Y.; Kitamura, Y.; Akaji, K.; Ashihara, E. Effective internalization of U251-MG-secreted exosomes into cancer cells and characterization of their lipid components. Biochem. Biophys. Res. Commun. 2015, 456, 768–773. [Google Scholar] [CrossRef] [PubMed]
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; De Milito, A.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A.; et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J. Biol. Chem. 2009, 284, 34211–34222. [Google Scholar] [CrossRef]
- Yu, M.; Gai, C.; Li, Z.; Ding, D.; Zheng, J.; Zhang, W.; Lv, S.; Li, W. Targeted exosome-encapsulated erastin induced ferroptosis in triple negative breast cancer cells. Cancer Sci. 2019, 110, 3173–3182. [Google Scholar] [CrossRef]
- Adamus, T.; Hung, C.-Y.; Yu, C.; Kang, E.; Hammad, M.; Flores, L.; Nechaev, S.; Zhang, Q.; Gonzaga, J.M.; Muthaiyah, K.; et al. Glioma-targeted delivery of exosome-encapsulated antisense oligonucleotides using neural stem cells. Mol. Ther.—Nucleic Acids 2022, 27, 611–620. [Google Scholar] [CrossRef]
- Parada, N.; Romero-Trujillo, A.; Georges, N.; Alcayaga-Miranda, F. Camouflage strategies for therapeutic exosomes evasion from phagocytosis. J. Adv. Res. 2021, 31, 61–74. [Google Scholar] [CrossRef]
- Haney, M.J.; Klyachko, N.L.; Zhao, Y.; Gupta, R.; Plotnikova, E.G.; He, Z.; Patel, T.; Piroyan, A.; Sokolsky, M.; Kabanov, A.V.; et al. Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J. Control. Release 2015, 207, 18–30. [Google Scholar] [CrossRef]
- Charoenviriyakul, C.; Takahashi, Y.; Morishita, M.; Matsumoto, A.; Nishikawa, M.; Takakura, Y. Cell type-specific and common characteristics of exosomes derived from mouse cell lines: Yield, physicochemical properties, and pharmacokinetics. Eur. J. Pharm. Sci. 2017, 96, 316–322. [Google Scholar] [CrossRef]
- Keller, S.; König, A.-K.; Marmé, F.; Runz, S.; Wolterink, S.; Koensgen, D.; Mustea, A.; Sehouli, J.; Altevogt, P. Systemic presence and tumor-growth promoting effect of ovarian carcinoma released exosomes. Cancer Lett. 2009, 278, 73–81. [Google Scholar] [CrossRef]
- Umezu, T.; Tadokoro, H.; Azuma, K.; Yoshizawa, S.; Ohyashiki, K.; Ohyashiki, J.H. Exosomal miR-135b shed from hypoxic multiple myeloma cells enhances angiogenesis by targeting factor-inhibiting HIF-1. Blood 2014, 124, 3748–3757. [Google Scholar] [CrossRef]
- Rana, S.; Yue, S.; Stadel, D.; Zöller, M. Toward tailored exosomes: The exosomal tetraspanin web contributes to target cell selection. Int. J. Biochem. Cell Biol. 2012, 44, 1574–1584. [Google Scholar] [CrossRef] [PubMed]
- Zomer, A.; Maynard, C.; Verweij, F.J.; Kamermans, A.; Schäfer, R.; Beerling, E.; Schiffelers, R.M.; de Wit, E.; Berenguer, J.; Ellenbroek, S.I.J.; et al. In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell 2015, 161, 1046–1057. [Google Scholar] [CrossRef]
- Wolfers, J.; Lozier, A.; Raposo, G.; Regnault, A.; Théry, C.; Masurier, C.; Flament, C.; Pouzieux, S.; Faure, F.; Tursz, T.; et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat. Med. 2001, 7, 297–303. [Google Scholar] [CrossRef] [PubMed]
- ClinicalTrials. Available online: https://clinicaltrials.gov/ct2/show/NCT01550523 (accessed on 15 November 2022).
- Hikita, T.; Miyata, M.; Watanabe, R.; Oneyama, C. Sensitive and rapid quantification of exosomes by fusing luciferase to exosome marker proteins. Sci. Rep. 2018, 8, 14035. [Google Scholar] [CrossRef] [PubMed]
- Emam, S.E.; Abu Lila, A.S.; Elsadek, N.E.; Ando, H.; Shimizu, T.; Okuhira, K.; Ishima, Y.; Mahdy, M.A.; Ghazy, F.S.; Ishida, T. Cancer cell-type tropism is one of crucial determinants for the efficient systemic delivery of cancer cell-derived exosomes to tumor tissues. Eur. J. Pharm. Biopharm. 2019, 145, 27–34. [Google Scholar] [CrossRef]
- Nie, H.; Xie, X.; Zhang, D.; Zhou, Y.; Li, B.; Li, F.; Li, F.; Cheng, Y.; Mei, H.; Meng, H.; et al. Use of lung-specific exosomes for miRNA-126 delivery in non-small cell lung cancer. Nanoscale 2020, 12, 877–887. [Google Scholar] [CrossRef]
- Arteaga-Blanco, L.A.; Bou-Habib, D.C. The role of extracellular vesicles from human macrophages on host-pathogen interaction. Int. J. Mol. Sci. 2021, 22, 10262. [Google Scholar] [CrossRef]
- Xia, Y.; Rao, L.; Yao, H.; Wang, Z.; Ning, P.; Chen, X. Engineering macrophages for cancer immunotherapy and drug delivery. Adv. Mater. 2020, 32, 2002054. [Google Scholar] [CrossRef]
- Rosenberger, L.; Ezquer, M.; Lillo-Vera, F.; Pedraza, P.L.; Ortúzar, M.I.; González, P.L.; Figueroa-Valdés, A.I.; Cuenca, J.; Ezquer, F.; Khoury, M.; et al. Stem cell exosomes inhibit angiogenesis and tumor growth of oral squamous cell carcinoma. Sci. Rep. 2019, 9, 663. [Google Scholar] [CrossRef]
- Meng, W.; He, C.; Hao, Y.; Wang, L.; Li, L.; Zhu, G. Prospects and challenges of extracellular vesicle-based drug delivery system: Considering cell source. Drug Deliv. 2020, 27, 585–598. [Google Scholar] [CrossRef] [PubMed]
- Moon, B.; Chang, S. Exosome as a delivery vehicle for cancer therapy. Cells 2022, 11, 316. [Google Scholar] [CrossRef] [PubMed]
- Rezaie, J.; Feghhi, M.; Etemadi, T. A review on exosomes application in clinical trials: Perspective, questions, and challenges. Cell Commun. Signal. 2022, 20, 145. [Google Scholar] [CrossRef] [PubMed]
- Roshancheshm, S.; Asadi, A.; Khoshnazar, S.M.; Abdolmaleki, A.; Khudhur, O.O.; Sh, W.S. Application of natural and modified exosomes as a drug delivery system. Nanomed. J. 2022, 9, 192–204. [Google Scholar] [CrossRef]
- Mehryab, F.; Rabbani, S.; Shahhosseini, S.; Shekari, F.; Fatahi, Y.; Baharvand, H.; Haeri, A. Exosomes as a next-generation drug delivery system: An update on drug loading approaches, characterization, and clinical application challenges. Acta Biomater. 2020, 113, 42–62. [Google Scholar] [CrossRef]
- Kibria, G.; Ramos, E.K.; Wan, Y.; Gius, D.R.; Liu, H. Exosomes as a drug delivery system in cancer therapy: Potential and challenges. Mol. Pharm. 2018, 15, 3625–3633. [Google Scholar] [CrossRef]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Mahajan, V.; Deygen, I.; Klyachko, N.L.; Inskoe, E.; Piroyan, A.; Sokolsky, M.; Okolie, O.; et al. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 655–664. [Google Scholar] [CrossRef]
- Rech, J.; Wilińska, J.; Borecka, A.; Turek, A. Application of fibrin in drug technology: Achievements and perspectives. Postep. Hig. Med. Dosw. 2020, 74, 322–330. [Google Scholar] [CrossRef]
- Jiang, X.-C.; Gao, J.-Q. Exosomes as novel bio-carriers for gene and drug delivery. Int. J. Pharm. 2017, 521, 167–175. [Google Scholar] [CrossRef]
- Kalimuthu, S.; Gangadaran, P.; Rajendran, R.L.; Zhu, L.; Oh, J.M.; Lee, H.W.; Gopal, A.; Baek, S.H.; Jeong, S.Y.; Lee, S.-W.; et al. A new approach for loading anticancer drugs into mesenchymal stem cell-derived exosome mimetics for cancer therapy. Front. Pharmacol. 2018, 9, 1116. [Google Scholar] [CrossRef]
- Kalani, A.; Chaturvedi, P.; Kamat, P.K.; Maldonado, C.; Bauer, P.; Joshua, I.G.; Tyagi, S.C.; Tyagi, N. Curcumin-loaded embryonic stem cell exosomes restored neurovascular unit following ischemia-reperfusion injury. Int. J. Biochem. Cell Biol. 2016, 79, 360–369. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, S.; Wang, Z. High yield, scalable and remotely drug-loaded neutrophil-derived extracellular vesicles (EVS) for anti-inflammation therapy. Biomaterials 2017, 135, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Stoicheva, N.G.; Hui, S.W. Electrofusion of cell-size liposomes. Biochim. Biophys. Acta—Biomembr. 1994, 1195, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Santos, P.; Almeida, F. Exosome-based vaccines: History, current state, and clinical trials. Front. Immunol. 2021, 12, 711565. [Google Scholar] [CrossRef]
- Osteikoetxea, X.; Silva, A.; Lázaro-Ibáñez, E.; Salmond, N.; Shatnyeva, O.; Stein, J.; Schick, J.; Wren, S.; Lindgren, J.; Firth, M.; et al. Engineered cas9 extracellular vesicles as a novel gene editing tool. J. Extracell. Vesicles 2022, 11, e12225. [Google Scholar] [CrossRef]
- ClinicalTrials. Available online: https://clinicaltrials.gov/ct2/results?cond=&term=exosome&cntry=&state=&city=&dist= (accessed on 15 November 2022).
- Zhuang, X.; Xiang, X.; Grizzle, W.; Sun, D.; Zhang, S.; Axtell, R.C.; Ju, S.; Mu, J.; Zhang, L.; Steinman, L.; et al. Treatment of brain inflammatory diseases by delivering exosome encapsulated anti-inflammatory drugs from the nasal region to the brain. Mol. Ther. 2011, 19, 1769–1779. [Google Scholar] [CrossRef]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 47. [Google Scholar] [CrossRef]
- ClinicalTrials. Available online: https://clinicaltrials.gov/ct2/show/NCT04491240 (accessed on 15 November 2022).
- Yuan, A.; Ruan, L.; Jia, R.; Wang, X.; Wu, L.; Cao, J.; Qi, X.; Wei, Y.; Shen, S. Tumor exosome-mimicking iron oxide nanoparticles for near infrared-responsive drug delivery. ACS Appl. Nano Mater. 2022, 5, 996–1002. [Google Scholar] [CrossRef]
- Wang, J.; Tang, W.; Yang, M.; Yin, Y.; Li, H.; Hu, F.; Tang, L.; Ma, X.; Zhang, Y.; Wang, Y. Inflammatory tumor microenvironment responsive neutrophil exosomes-based drug delivery system for targeted glioma therapy. Biomaterials 2021, 273, 120784. [Google Scholar] [CrossRef]
- del Pozo-Acebo, L.; López de las Hazas, M.-C.; Tomé-Carneiro, J.; Gil-Cabrerizo, P.; San-Cristobal, R.; Busto, R.; García-Ruiz, A.; Dávalos, A. Bovine milk-derived exosomes as a drug delivery vehicle for mirna-based therapy. Int. J. Mol. Sci. 2021, 22, 1105. [Google Scholar] [CrossRef]
- Agrawal, A.K.; Aqil, F.; Jeyabalan, J.; Spencer, W.A.; Beck, J.; Gachuki, B.W.; Alhakeem, S.S.; Oben, K.; Munagala, R.; Bondada, S.; et al. Milk-derived exosomes for oral delivery of paclitaxel. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 1627–1636. [Google Scholar] [CrossRef] [PubMed]
- Melzer, C.; Rehn, V.; Yang, Y.; Bähre, H.; von der Ohe, J.; Hass, R. Taxol-loaded msc-derived exosomes provide a therapeutic vehicle to target metastatic breast cancer and other carcinoma cells. Cancers 2019, 11, 798. [Google Scholar] [CrossRef] [PubMed]
- McAndrews, K.M.; Xiao, F.; Chronopoulos, A.; LeBleu, V.S.; Kugeratski, F.G.; Kalluri, R. Exosome-mediated delivery of CRISPR/cas9 for targeting of oncogenic KRASG12D in pancreatic cancer. Life Sci. Alliance 2021, 4, e202000875. [Google Scholar] [CrossRef] [PubMed]
- Munagala, R.; Aqil, F.; Jeyabalan, J.; Kandimalla, R.; Wallen, M.; Tyagi, N.; Wilcher, S.; Yan, J.; Schultz, D.J.; Spencer, W.; et al. Exosome-mediated delivery of RNA and DNA for gene therapy. Cancer Lett. 2021, 505, 58–72. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Chen, J.; Wang, C.; Yuan, M.; Kang, Y.; Wu, Z.; Li, W.; Zhang, G.; Machens, H.-G.; Rinkevich, Y.; et al. Milk exosomes-mediated miR-31-5p delivery accelerates diabetic wound healing through promoting angiogenesis. Drug Deliv. 2022, 29, 214–228. [Google Scholar] [CrossRef] [PubMed]
- Nie, W.; Wu, G.; Zhang, J.; Huang, L.; Ding, J.; Jiang, A.; Zhang, Y.; Liu, Y.; Li, J.; Pu, K.; et al. Responsive exosome nano-bioconjugates for synergistic cancer therapy. Angew. Chem. Int. Ed. 2020, 59, 2018–2022. [Google Scholar] [CrossRef]
- Zhou, W.; Zhou, Y.; Chen, X.; Ning, T.; Chen, H.; Guo, Q.; Zhang, Y.; Liu, P.; Zhang, Y.; Li, C.; et al. Pancreatic cancer-targeting exosomes for enhancing immunotherapy and reprogramming tumor microenvironment. Biomaterials 2021, 268, 120546. [Google Scholar] [CrossRef]
- Gao, Z.-S.; Zhang, C.-J.; Xia, N.; Tian, H.; Li, D.-Y.; Lin, J.-Q.; Mei, X.-F.; Wu, C. Berberine-loaded M2 macrophage-derived exosomes for spinal cord injury therapy. Acta Biomater. 2021, 126, 211–223. [Google Scholar] [CrossRef]
- Du, J.; Wan, Z.; Wang, C.; Lu, F.; Wei, M.; Wang, D.; Hao, Q. Designer exosomes for targeted and efficient ferroptosis induction in cancer via chemo-photodynamic therapy. Theranostics 2021, 11, 8185–8196. [Google Scholar] [CrossRef]
- Liu, T.; Li, T.; Zheng, Y.; Xu, X.; Sun, R.; Zhan, S.; Guo, X.; Zhao, Z.; Zhu, W.; Feng, B.; et al. Evaluating adipose-derived stem cell exosomes as miRNA drug delivery systems for the treatment of bladder cancer. Cancer Med. 2022, 11, 3687–3699. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).