Multiple chronic conditions (MCC) are defined as “conditions that last a year or more and require ongoing medical attention and/or limit activities of daily living” [
1]. Among the most common chronic conditions are hypertension, heart disease, cardiovascular disease, arthritis, diabetes, cancer, dementia, and various cognitive disorders. The number of people with MCC has increased over the years and, combined with aging, has become a major global health challenge [
2]. Because people with MCC must frequently be hospitalized, expenses related to medical care have also risen. In addition, health services worldwide have been focusing on individual chronic diseases, so there are few projects that address the prediction of factors such as mortality from MCC. This situation makes it necessary to create a tool that can predict different factors, such as mortality, from MCC. This tool would help health professionals to make decisions and subsequently treat patients suffering from these conditions.
Not surprisingly, multiple chronic diseases and cardiovascular pathologies are associated with impaired quality of life, an increased risk of disability, institutionalization, and mortality, and higher healthcare utilization and associated costs. For example, the annual risk of hospitalization increases from 4% among individuals with zero to one chronic condition to more than 60% for those with six or more chronic conditions. Furthermore, 30-day readmission rates increase progressively as the number of chronic conditions rises, and per capita, Medicare expenditures increase exponentially as a function of the number of chronic conditions [
3]. Additionally, physician care is based on treating one disease at a time and does not consider the possible effects of a combination of different pathologies.
Performing mortality prediction based on multiple chronic conditions and cardiovascular diseases can be a significant challenge for a healthcare professional. Correct prediction can identify patients with a higher risk to their health, allowing physicians to take appropriate measures to avoid or minimize this risk and, in turn, improve the quality of care and avoid potential hospital admissions [
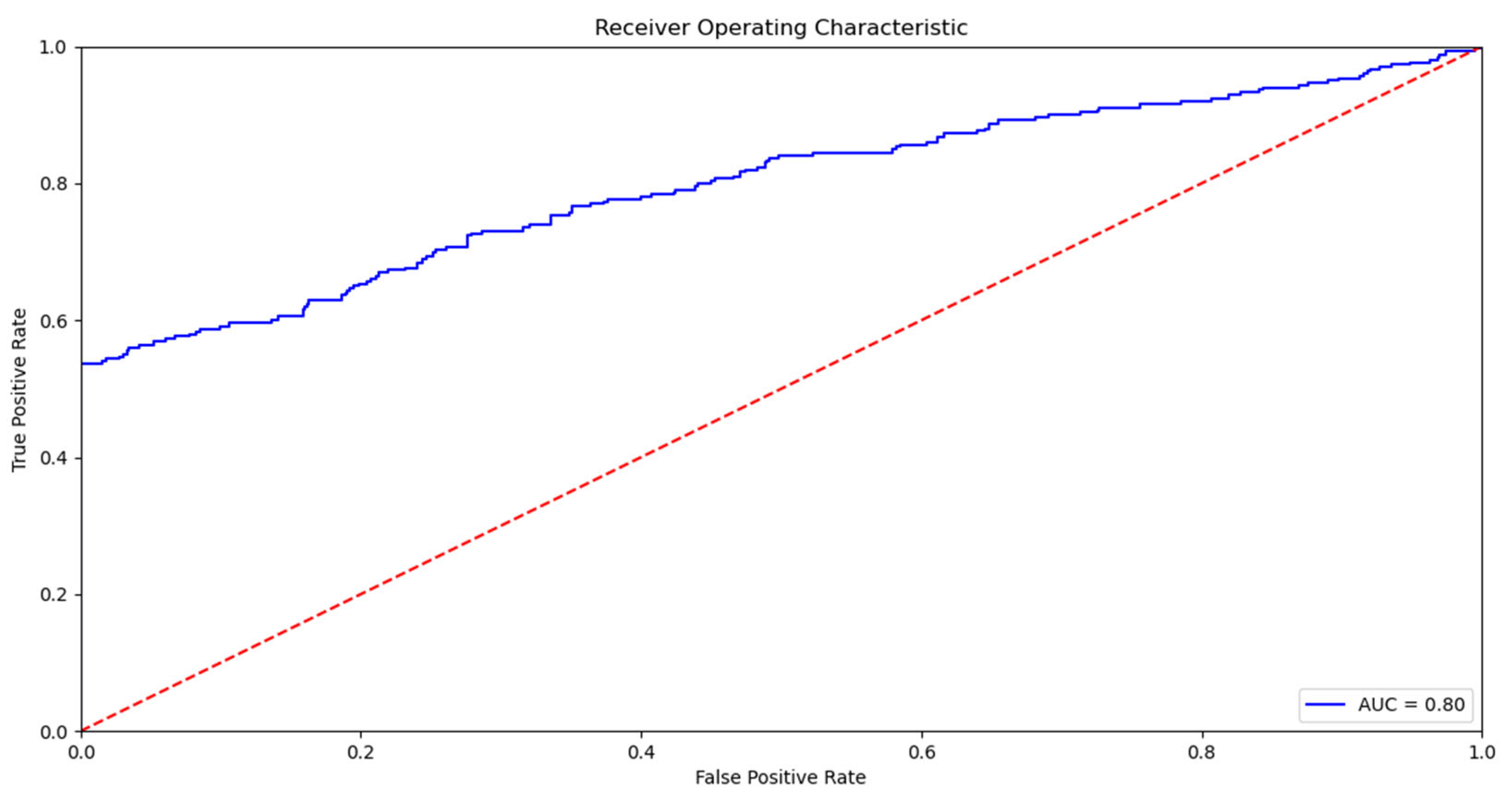
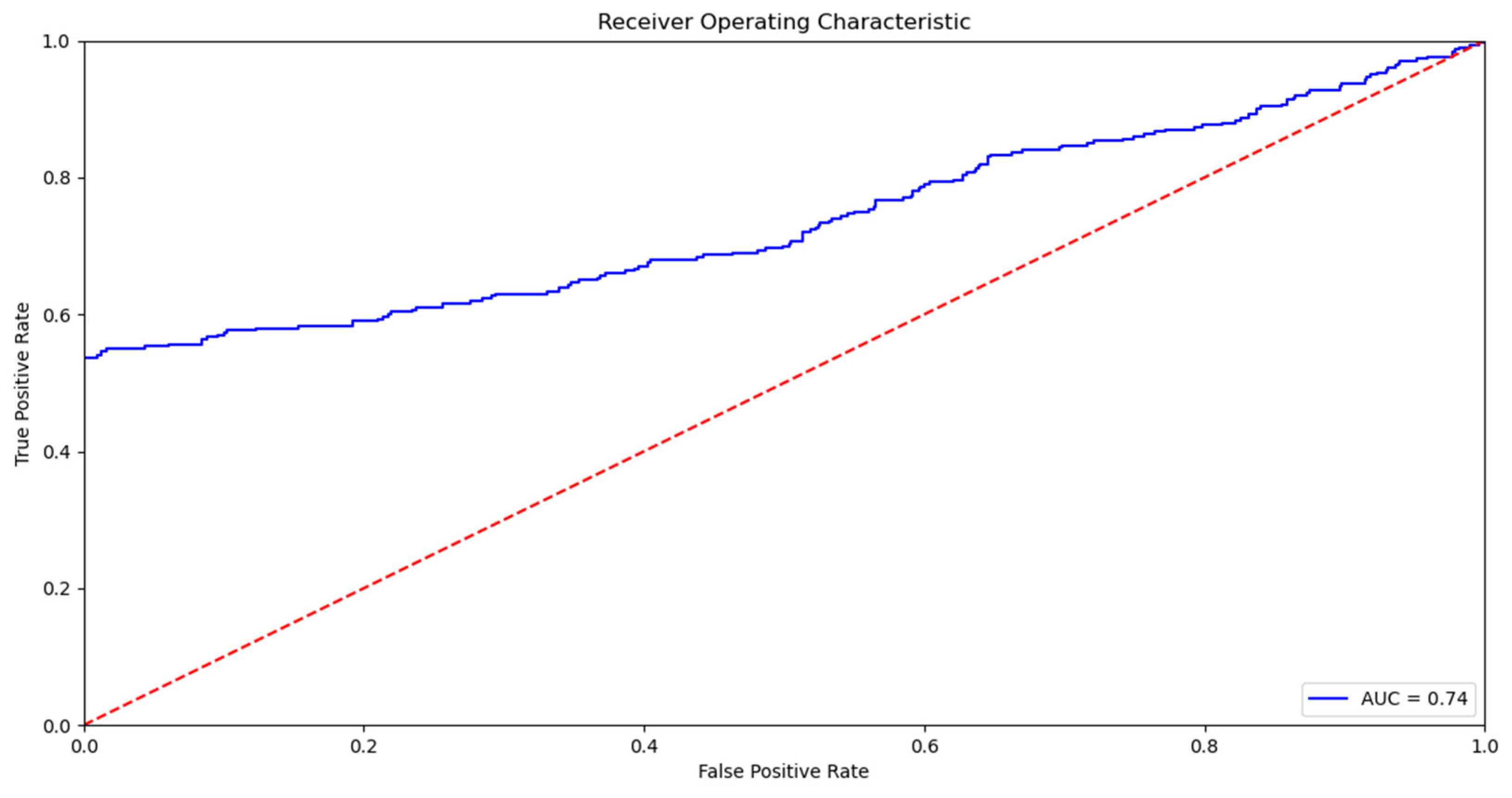
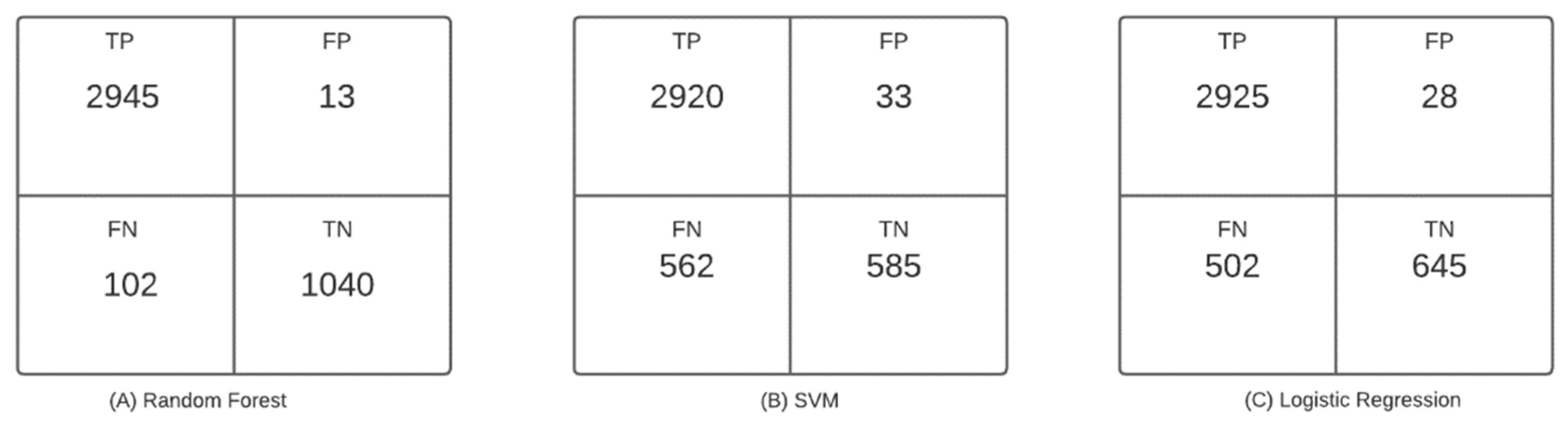
4]. The related work section shows that the use of machine learning algorithms to predict mortality and other pathologies has been increasing in recent years. This increase is because machine learning algorithms provide the tools to make an automatic and accurate mortality prediction, which is why they greatly support the decision-making process of health professionals. In addition, the literature review reveals that the most used models are those based on decision trees and support vector machines, with an increase in the use of neural networks at the present time [
5].
This study aimed to predict patients’ mortality risk from cardiovascular variables, medical history, medications, and electrocardiogram variables. Supervised learning algorithms were used to perform the predictions.
Related Work
First, there will be a discussion of the main aspects and challenges of mortality risk prediction in older adult patients with multiple chronic conditions and cardiovascular pathologies. In order to predict mortality, the effect of different variables must be taken into account. Rodgers et al. conducted a study to show the association between cardiovascular risk and the age and gender of patients. This study showed that age is an important factor in the deterioration of cardiovascular function, resulting in an increased risk of cardiovascular disease (CVD) in older adults. It also showed that the prevalence of CVD increases with age in both men and women, including the prevalence of atherosclerosis, stroke, and myocardial infarction; in fact, the incidence of CVD is 89.3% in men and 91.8% in women in adults older than 80 years. Patients’ risk may change depending on other pathologies, such as diabetes, hypertension, and obesity. A high prevalence of heart failure, atrial fibrillation, and other forms of CVD was first noted in elderly patients. The high prevalence of CVD in this population has been linked to many factors, including increased oxidative stress, inflammation, apoptosis, and general myocardial deterioration and degeneration [
6].
With the progress of sophisticated machine learning algorithms and advanced data collection and storage technologies, researchers have achieved promising results for predicting the risk of having a single chronic disease. State-of-the-art machine learning algorithms, such as deep neural networks, support vector machines, logistic regression, random forest, and decision trees, have been widely implemented and validated to predict the risk of having chronic diseases, such as hypertension, cardiovascular disease, heart disease, and diabetes mellitus. Much of the research has focused on predicting a single chronic disease, but chronic diseases, especially in old age, often co-occur with other chronic diseases, a phenomenon known as multimorbidity or MCC. The lack of a model for multiple chronic conditions can be attributed to the paucity of a dataset covering MCC. Available datasets are usually dedicated to a particular type of disease. To address this challenge, Yang et al. designed interpretable machine learning algorithms to tackle the problem of classifying whether or not a participant is at high risk for CCM by identifying associated risk factors and exploring the interactions between them. Yang et al. used the Catapult Health database, which consists of 451,425 records, among which 137,118 records, 77,149 records, and 237,159 records were collected in 2018, 2017, and 2012–2016, respectively. For this study, 301,631 patients were selected, of which 10,427 participants (3.5%) were flagged as high-risk individuals with at least one chronic condition, and 291,204 participants (96.5%) were flagged as low-risk individuals without CCM. Twenty variables were selected for analysis: diastolic blood pressure (DBP), systolic blood pressure (SBP), glucose (GLU), triglycerides (TGS), total cholesterol (TCHOL), body mass index (BMI), low-density lipoprotein (LDL) cholesterol, weight, age, alanine aminotransferase (ALT), blood pressure assessment, abdominal circumference, metabolic syndrome risk, diabetes assessment, and BMI assessment. Seven machine learning algorithms, namely, k-nearest neighbors (kNN), decision tree (DT), random forest (RF), gradient boosting tree (GBT), logistic regression (LR), support vector machine (SVM), and Naïve Bayes (NB), were used and evaluated for this study. Regarding the results, the AUC values achieved by the GBT classifier, RF classifier, SVM classifier, DT classifier, NB classifier, LR classifier, and kNN classifier were 0.850, 0.846, 0.833, 0.837, 0.821, 0.823, and 0.826, respectively [
7].
In another study by Kawano et al., 85,361 patients were selected, and three machine learning models, XGBoost, artificial neural network, and logistic regression, were built to predict mortality based on different cardiac variables and chronic conditions. Regarding the results, the AUC values were 0.811 for XGBoost, 0.774 for neural networks, and 0.772 for logistic regression models, indicating that the predictive capacity of XGBoost was the highest. This study showed that the machine-learning-based model has a higher predictive ability than the conventional logistic regression model and can be helpful in risk assessment and health counseling for health checkup participants [
8].
In order to design the algorithm, a review of previous studies was carried out to establish the main variables affecting mortality and other medical events and the algorithms and supervised learning methods used in these situations. Di Castelnuovo et al. and Mirkin et al. performed a study to identify the cardiovascular characteristics, comorbidities, and laboratory measures associated with in-hospital mortality in European countries [
9,
10]. The random forest model used by the authors found that among the most important variables for predicting mortality were impaired renal function, elevated C-reactive protein, advanced age, diabetes, hypertension, and myocardial infarction. Cox’s multivariate survival analysis confirmed these results. Another study by Kaiser et al. [
11] used the information from 529 patients from the CHS database to describe the effect of social support on survival after heart failure. The authors built a Cox survival model using the following as variables: age, gender, race (white or black), medications (antihypertensive, oral hypoglycemic, and lipid-lowering), coronary heart disease status (defined as myocardial infarction, angina, coronary artery bypass grafting, or angioplasty), systolic and diastolic blood pressure, cystatin-C, general health status (excellent, good, fair, or poor), self-reported limitations in activities of daily living (ADLs), body mass index, and physical activity. The model results showed that participants with high scores in social networks tend to have a higher body mass index (BMI), fewer depressive symptoms, and better interpersonal support scores. Another interesting result was that participants with a high social support score had a 29% lower mortality rate than those with a low score. These findings suggest that structural social support prior to diagnosis may modestly buffer mortality in patients with heart failure. In [
12,
13,
14], to analyze risk factors for hospital readmission, a study was conducted with 100 patients and Chi-square tests. The results showed that the most influential variables in hospital readmission were noncompliance with medication (25.8%), poor dietary compliance (22.6%), uncontrolled diabetes mellitus (22.6%), ischemia (19.4%), anemia (16.1%) and worsening left ventricular function (16.1%).
The most used supervised learning methods are decision-tree-based algorithms such as random forest, support vector machine, and Naive Bayes and survival algorithms such as Cox regression, as shown in the studies conducted by Guo et al., Segar et al., and Mezzatesta et al. [
14,
15,
16]. The variables commonly used to train the mentioned models are demographic variables, laboratory examination data, and clinical, cardiovascular, and electrocardiogram variables. Finally, da Silva et al. [
17] present a deep learning algorithm to predict the deterioration of patients’ health. This DeepSigns model predicts vital signs using LSTM networks and calculates the prognostic index for the early diagnosis of patients’ worsening health status. To construct the predictor, the authors used fifteen attributes, such as temperature, blood pressure, heart rate, oxygenation, hematocrit, and age, among others. In general, the main contributions of their work were (i) the development of a method to predict patient data, feeding back new data to the model to obtain greater accuracy, and (ii) the evaluation of many prognostic indices using predicted vital signs instead of specific values measured over time. The final model was able to predict vital signs with 80% accuracy with regard to the results obtained, identifying states of possible future deterioration. As can be seen, there are studies in the literature that used machine learning models to predict mortality based on sociodemographic and clinical variables. However, studies were not found using a cardiovascular study database (CHS) to make such associations and predictions using machine learning. Articles using similar methodology and variables but with other datasets show performance ranging from 66% to 81% [
18,
19,
20,
21].
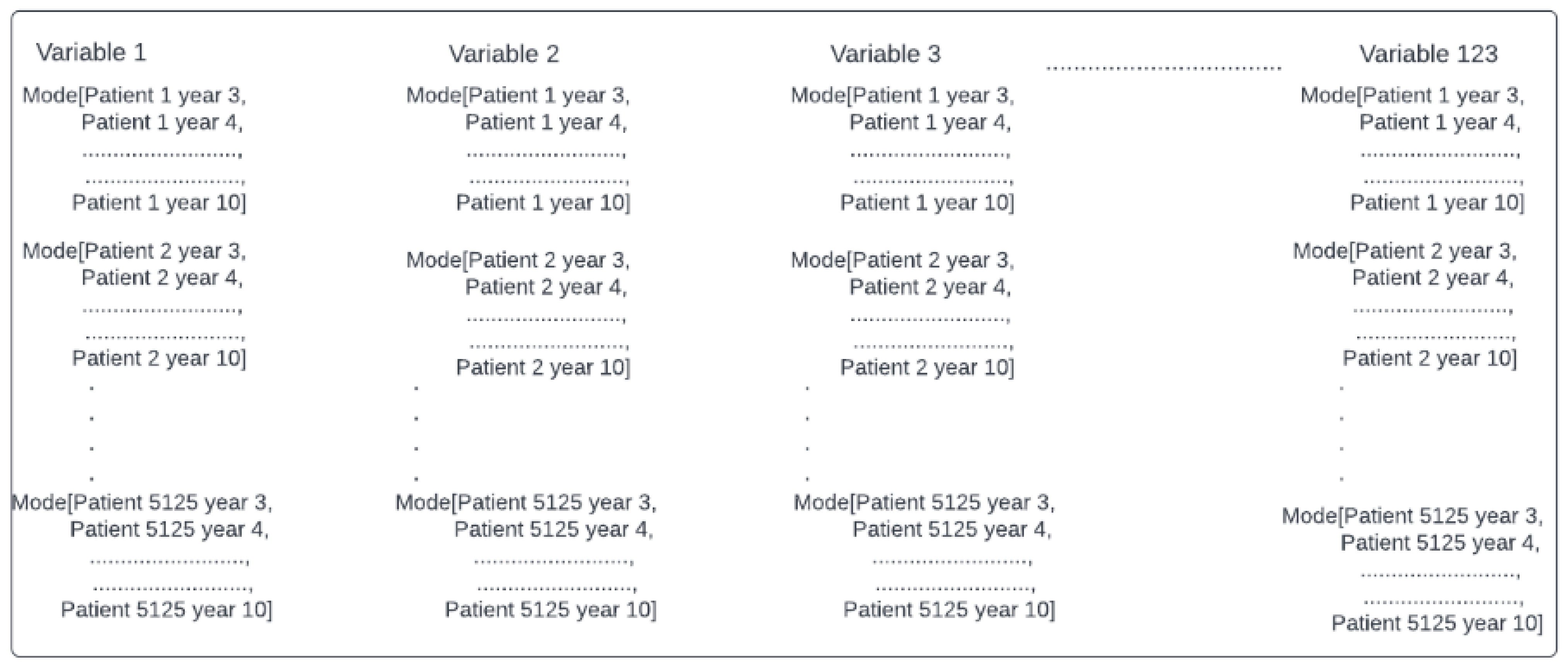
As seen in the literature review, mortality predictions using machine learning algorithms from multiple chronic conditions, including cardiovascular and other physiological variables, have been increasing. Some of the variables used for mortality predictions have already been described, and the main algorithms used have been discussed. However, one of the main prediction challenges is to acquire suitable data. The Cardiovascular Health Study was an observational study to identify risk factors for cardiovascular disease in older adults. The results of this study are available to researchers to investigate conditions that affect older adults and constitute a valuable source for secondary use. This database provides 20 years of data for 5125 patients. This article aims to study mortality risks by relating, through machine learning models, multiple chronic conditions and other clinical variables in older adults using the CHS dataset [
22].