ConBERT: A Concatenation of Bidirectional Transformers for Standardization of Operative Reports from Electronic Medical Records
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Data
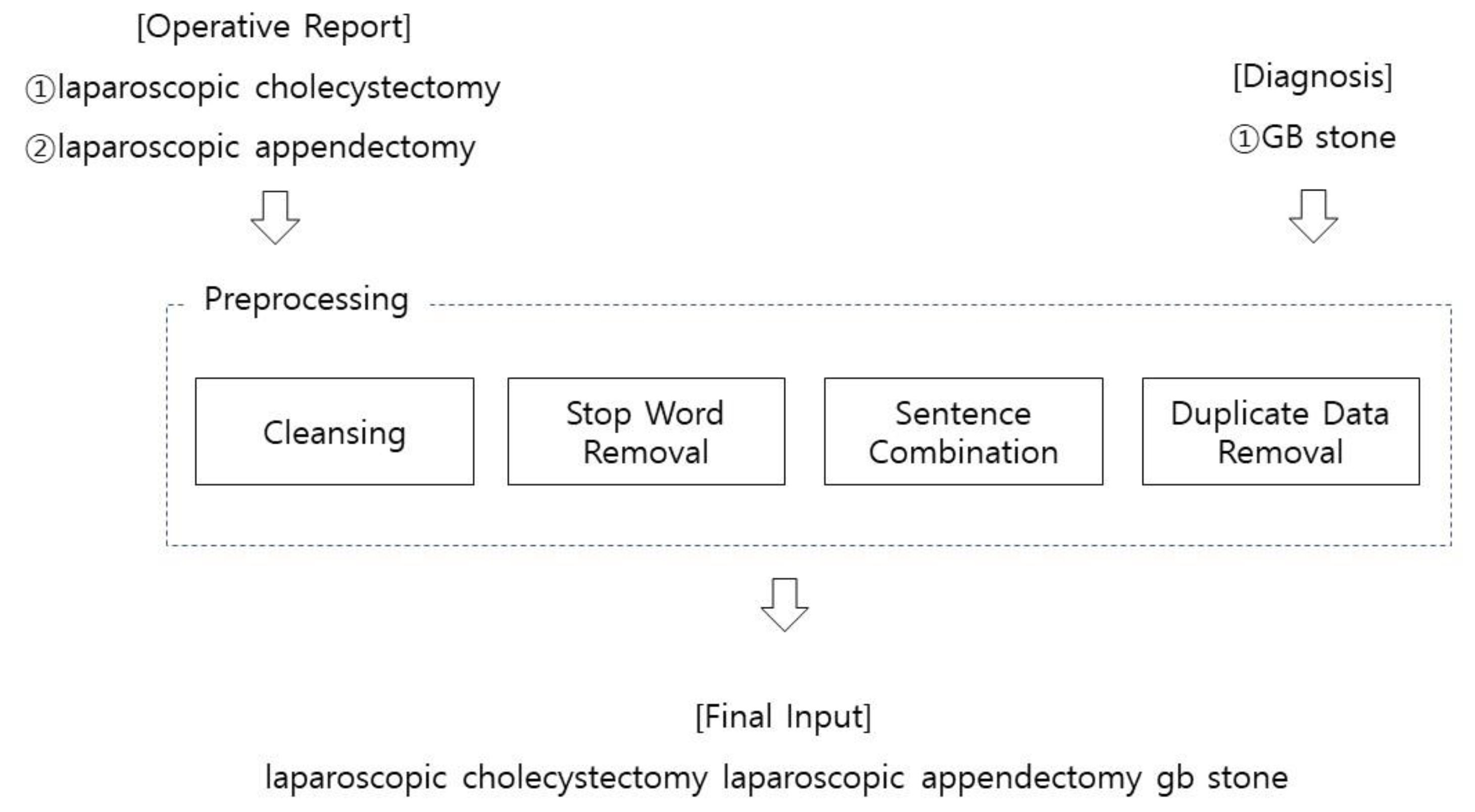
2.2. Preprocessing
2.3. Methods
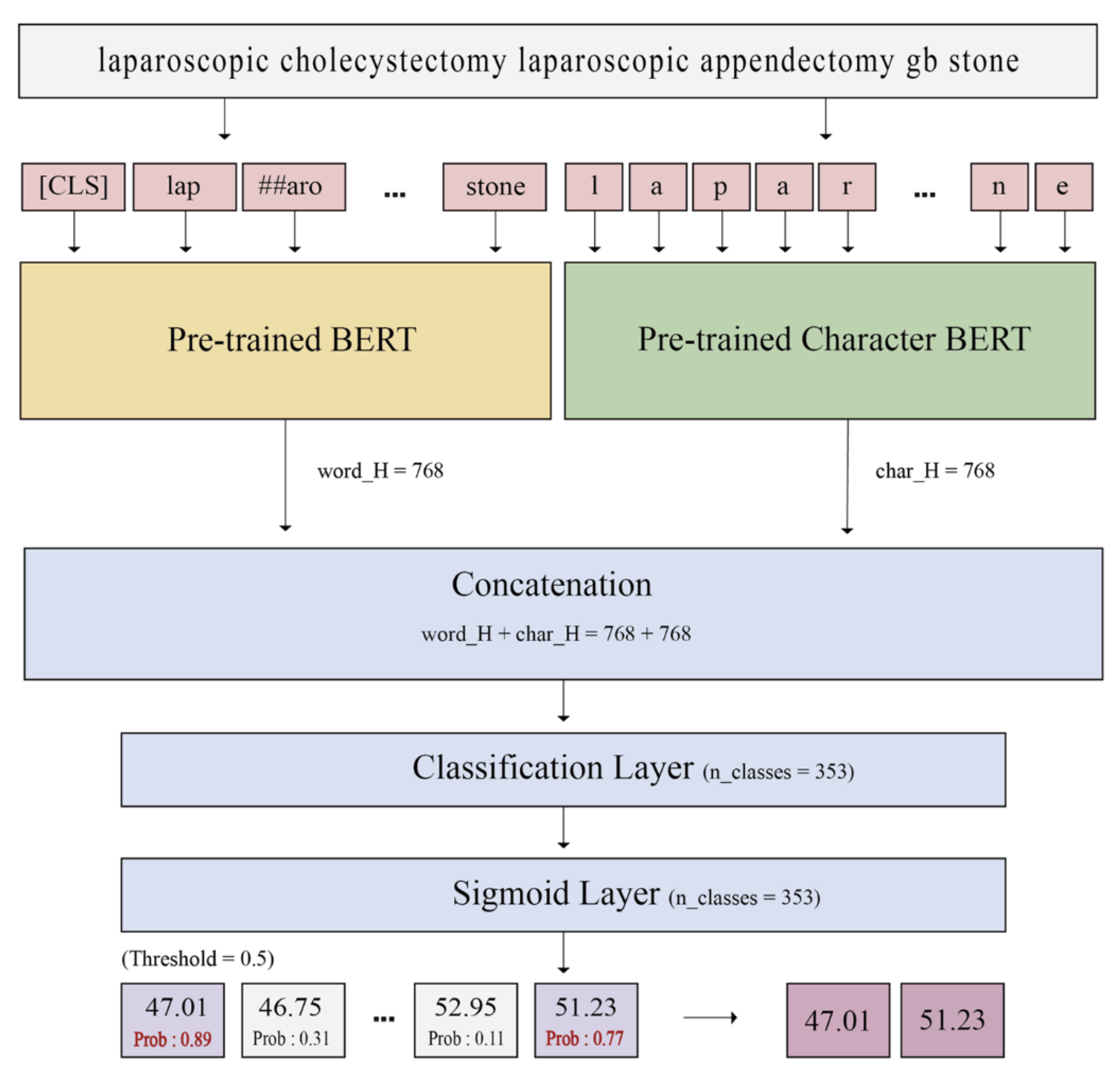
2.3.1. Embedding
2.3.2. BERT
2.3.3. Character BERT
2.3.4. Model Aggregation
2.3.5. Training Details
2.3.6. Evaluation
3. Results
3.1. Input Comparison
3.2. Comparison of Pre-Trained Models
3.3. Comparison of Aggregated Models
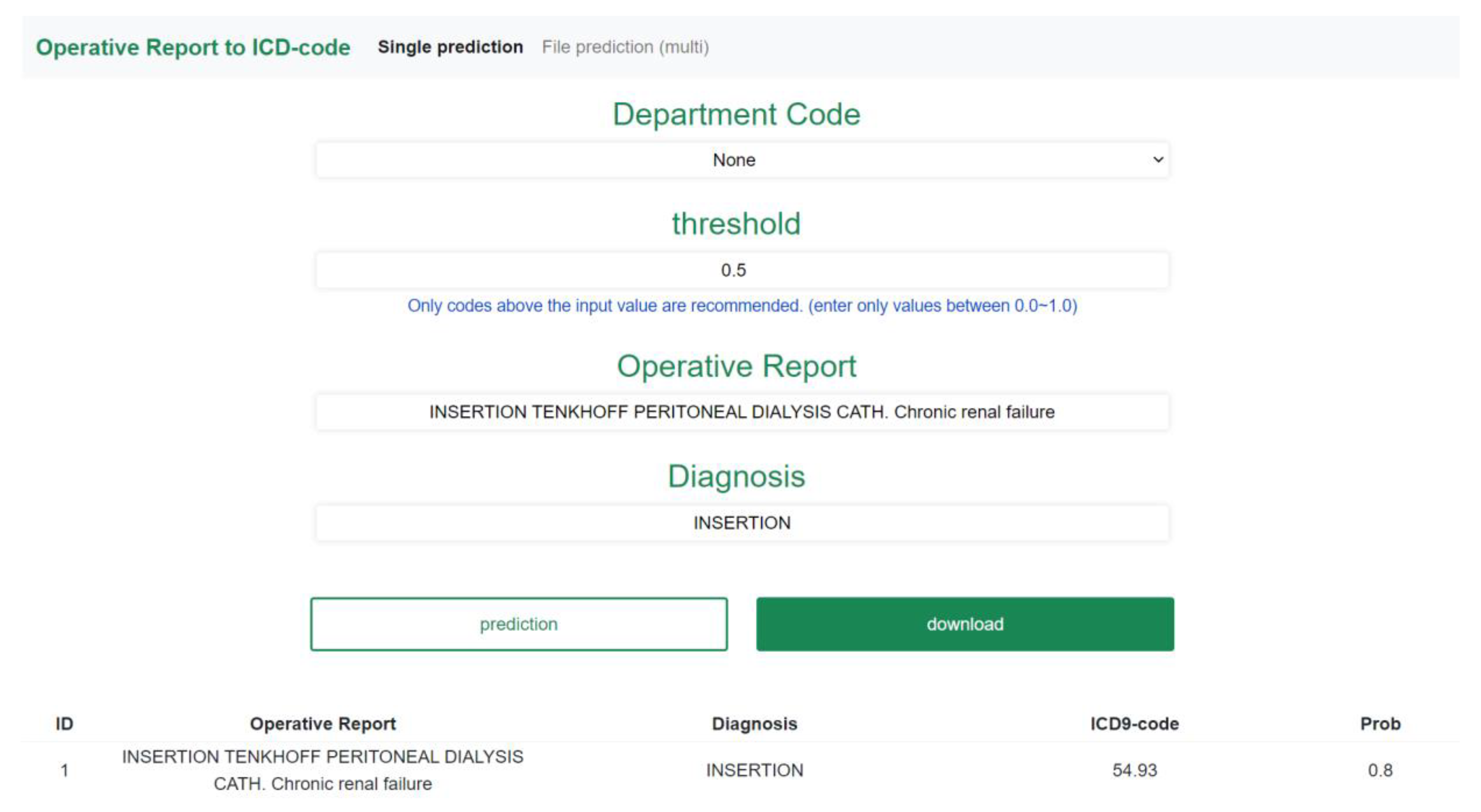
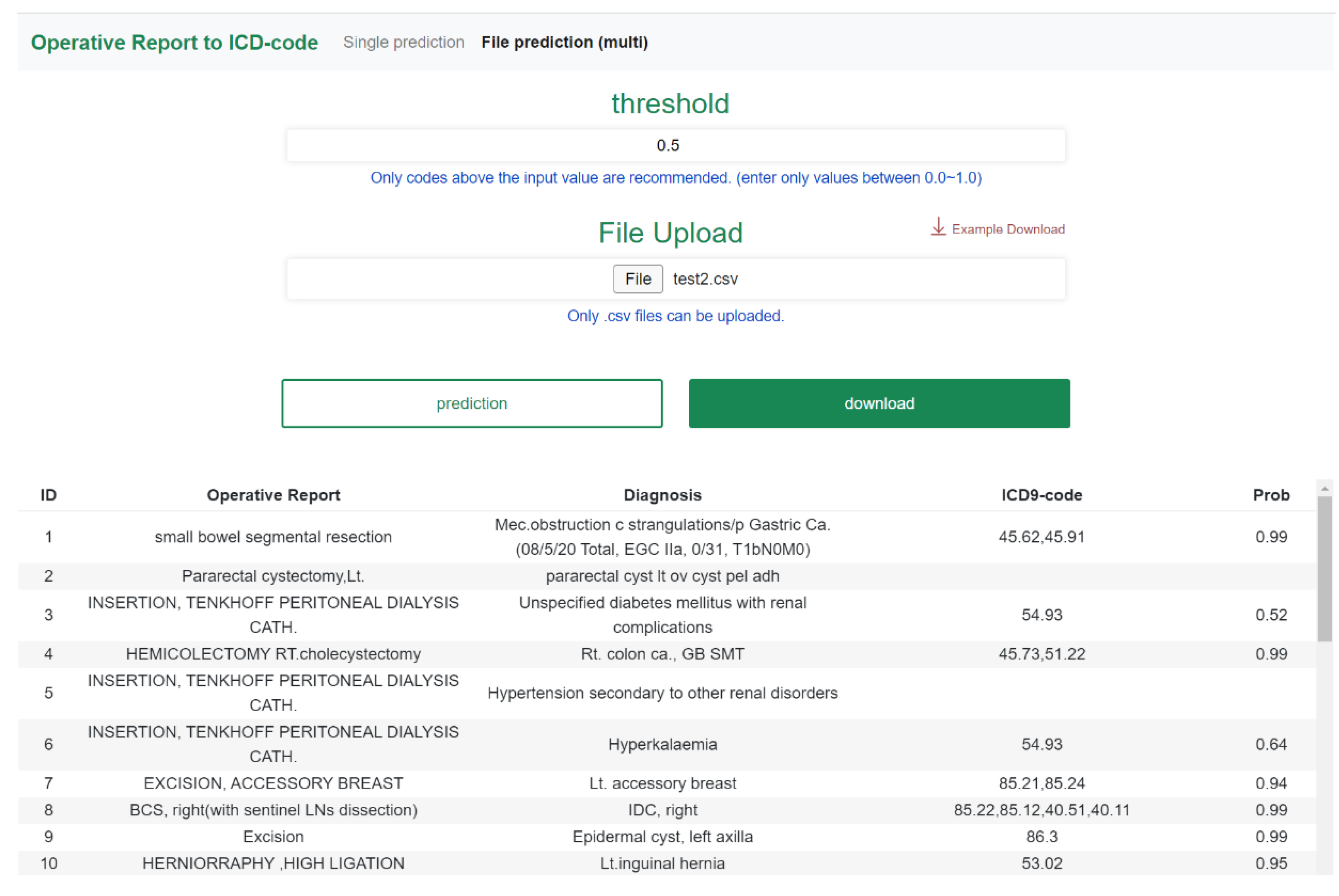
3.4. Web-Based Application
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Safiejko, K.; Tarkowski, R.; Koselak, M.; Juchimiuk, M.; Tarasik, A.; Pruc, M.; Smereka, J.; Szarpak, L. Robotic-assisted vs. standard laparoscopic surgery for rectal cancer resection: A systematic review and meta-analysis of 19,731 patients. Cancers 2021, 14, 180. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Park, J.W.; Lee, M.A.; Lim, H.K.; Kwon, Y.H.; Ryoo, S.B.; Park, K.J.; Jeong, S.Y. Two dominant patterns of low anterior resection syndrome and their effects on patients’ quality of life. Sci. Rep. 2021, 11, 3538. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.S.C.; Sousa, L.F.; Rabello, P.M.; Santiago, B.M. International Classification of Diseases—11th revision: From design to implementation. Rev. Saude Publica 2020, 54, 104. [Google Scholar] [CrossRef] [PubMed]
- Baumel, T.; Nassour-Kassis, J.; Cohen, R.; Elhadad, M.; Elhadad, N. Multi-label classification of patient notes: Case study on ICD code assignment. AAAI Workshops 2018, arXiv:1709.09587, 409–416. [Google Scholar]
- Wang, G.; Li, C.; Wang, W.; Zhang, Y.; Shen, D.; Zhang, X.; Henao, R.; Carin, L. Joint embedding of words and labels for text classification. arXiv 2018, arXiv:1805.04174v1, 2321–2331. [Google Scholar]
- Song, C.; Zhang, S.; Sadoughi, N.; Xie, P.; Xing, E. Generalized zero-shot ICD coding. arXiv 2019, arXiv:1909.13154. [Google Scholar]
- Haoran, S.; Xie, P.; Hu, Z.; Zhang, M.; Xing, E.P. Towards automated ICD coding using deep learning. arXiv 2017, arXiv:1711.04075. [Google Scholar]
- Fei, L.; Hong, Y. ICD Coding from Clinical Text Using Multi-Filter Residual Convolutional Neural Network. In Proceedings of the AAAI, New York, NY, USA, 7–12 February 2020. [Google Scholar]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. BERT: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Boukkouri, H.E.; Ferret, O.; Lavergne, T.; Noji, H.; Zweigenbaum, P.; Tsujii, J. Character BERT: Reconciling ELMo and BERT for word-level open-vocabulary representations from characters. arXiv 2020, arXiv:2010.10392. [Google Scholar]
- NLTK (Natural Language Toolkit). Available online: https://www.nltk.org/ (accessed on 8 September 2022).
- Heo, T.S.; Yongmin, Y.; Park, Y.; Jo, B.-C. Medical Code Prediction from Discharge Summary: Document to Sequence BERT Using Sequence Attention. In Proceedings of the 20th IEEE International Conference on Machine Learning and Applications (ICMLA), Pasadena, CA, USA, 13–16 December 2021; pp. 1239–1244. [Google Scholar]
- Scikit-Learn. Available online: https://scikit-learn.org/stable/ (accessed on 8 September 2022).
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient estimation of word representations in vector space. arXiv 2013, arXiv:1301.3781. [Google Scholar]
- Pennington, J.; Socher, R.; Manning, C. GLoVe: Global Vectors for Word Representation. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014. [Google Scholar]
- Bojanowski, P.; Grave, E.; Joulin, A.; Mikolov, T. Enriching Word Vectors with Subword Information. Trans. Assoc. Comput Linguist 2017, 5, 135–146. [Google Scholar] [CrossRef]
- Peters, M.E.; Neumann, M.; Iyyer, M.; Gardner, M.; Clark, C.; Lee, K.; Zettlemoyer, L. Deep contextualized word representations. arXiv 2018, arXiv:1809.09795. [Google Scholar]
- Michalopoulos, G.; Wang, Y.; Kaka, H.; Chen, H.; Wong, A. Umlsbert: Clinical domain knowledge augmentation of contextual embeddings using the unified medical language system metathesaurus. arXiv 2020, arXiv:2010.10391. [Google Scholar]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020, 36, 1234–1240. [Google Scholar] [PubMed]
- Loshchilov, I.; Hutter, F. Decoupled weight decay regularization. arXiv 2017, arXiv:1711.05101. [Google Scholar]
- He, T.; Zhang, Z.; Zhang, H.; Zhang, Z.; Xie, J.; Li, M. Bag of Tricks for Image Classification with Convolutional Neural Networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019. [Google Scholar]
- Pytorch Lightning. Available online: https://www.pytorchlightning.ai/ (accessed on 24 October 2022).
- Kim, B.H.; Ganapathi, V. Read, Attend, and Code: Pushing the Limits of Medical Codes Prediction from Clinical Notes by Machines. In Proceedings of the Machine Learning for Healthcare Conference, Online, 6–7 August 2021. [Google Scholar]
- Vu, T.; Nguyen, D.Q.; Nguyen, A. A Label Attention Model for ICD Coding from Clinical Text. In Proceedings of the IJCAI, Yokohama, Japan, 11–17 July 2020. [Google Scholar]
- Johnson, A.E.W.; Pollar, T.J.; Shen, L.; Lehmen, L.H.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Celi, L.A.; Mark, R.G. MIMIC-III, a freely accessible critical care database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef]
- Hripcsak, G.; Duke, J.D.; Shah, N.H.; Reich, C.G.; Huser, V.; Schuemie, M.J.; Suchard, M.A.; Park, R.W.; Wong, I.C.K.; Rijnbeek, P.R.; et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers. Stud. Health Technol. Inform. 2015, 216, 574–578. [Google Scholar]
- Ryu, B.; Yoo, S.; Kim, S.; Choi, J. Thirty-day hospital readmission prediction model based on common data model with weather and air quality data. Sci. Rep. 2021, 11, 23313. [Google Scholar] [CrossRef]
- Reps, J.M.; Schuemie, M.J.; Suchard, M.A.; Ryan, P.B.; Rijnbeek, P.R. Design and implementation of a standardized framework to generate and evaluate patient-level prediction models using observational healthcare data. J. Am. Med. Inform. Assoc. 2018, 25, 969–975. [Google Scholar] [CrossRef]
- Jung, H.; Yoo, S.; Kim, S.; Heo, E.; Kim, B.; Lee, H.Y.; Hwang, H. Patient-level fall risk prediction using the observational medical outcomes partnership’s common data model: Pilot feasibility study. JMIR Med. Inform. 2022, 10, e35104. [Google Scholar] [CrossRef]
- Biedermann, P.; Ong, R.; Davydov, A.; Orlova, A.; Solovyev, P.; Sun, H.; Wetherill, G.; Brand, M.; Didden, E.M. Standardizing registry data to the OMOP common data model: Experience from three pulmonary hypertension databases. BMC Med. Res. Methodol. 2021, 21, 238. [Google Scholar] [CrossRef] [PubMed]
- Lamer, A.; Abou-Arab, O.; Bourgeois, A.; Parrot, A.; Popoff, B.; Beuscart, J.B.; Tavernier, B.; Moussa, M.D. Transforming anesthesia data into the observational medical outcomes partnership common data model: Development and usability study. J. Med. Internet Res. 2021, 23, e29259. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]
- Self-Adj-Dice. Available online: https://github.com/fursovia/self-adj-dice/ (accessed on 8 September 2022).
| ID | Date | Division | Postoperative Diagnosis | Operative Report (Original) | Operative Report (ICD-9) | Code (ICD-9) |
|---|---|---|---|---|---|---|
| 1 | 6 August 2019 | Hepatobiliary | GB stone | lap. cholecystectomy | Cholecystectomy; laparoscopic | 51.23 |
| 2 | 21 May 2012 | Colorectal | rectal ca. (AV 4 cm) | ULAR | Resection; rectum, other anterior | 48.63 |
| 3 | 29 January 2020 | Colorectal | r/o appendiceal cancer | Lap. RHC | Hemicolectomy; right | 45.73 |
| Laparoscopy | 54.21 | |||||
| 4 | 22 June 2018 | Endocrine and Breast | Rt. PTC | THYROIDECTOMY, TOTAL | Thyroidectomy; complete | 06.4 |
| central LN dissection | dissection; neck, not otherwise specified, radical | 40.40 | ||||
| 5 | 19 August 2020 | Transplantation and Vascular | HBV LC with HCC | LDLT | Transplant; liver, other | 50.59 |
| Model | Corpus |
|---|---|
| MedicalBERT [10] | MIMIC-III Clinical note, PMC OA biomedical paper abstract |
| UmlsBERT [18] | Intensive Care III (MIMIC-III), MedNLi, i2b2 2006, i2b2 2010, i2b2 2012, i2b2 2014 |
| BioBERT [19] | English Wikipedia, BooksCorpus, PubMed Abstracts, PMC Full-text articles |
| MedicalCharacterBERT [10] | MIMIC-III Clinical note, PMC OA biomedical paper abstract |
| Proposed Model | AP | F1 | AUC | |||
|---|---|---|---|---|---|---|
| Micro | Macro | Micro | Macro | Micro | Macro | |
| Diagnosis | 0.4552 | 0.2347 | 0.4694 | 0.1736 | 0.9710 | 0.9282 |
| Operative report | 0.7692 | 0.4574 | 0.7292 | 0.4037 | 0.9889 | 0.9692 |
| Operative report + Diagnosis | 0.7647 | 0.4891 | 0.7509 | 0.4382 | 0.9812 | 0.9675 |
| Model | AP | F1 | AUC | ||||
|---|---|---|---|---|---|---|---|
| Micro | Macro | Micro | Macro | Micro | Macro | ||
| Single Model | * Base | 0.7854 | 0.5718 | 0.7570 | 0.3210 | 0.9860 | 0.7565 |
| Umls | 0.7872 | 0.5815 | 0.7603 | 0.3383 | 0.9863 | 0.7575 | |
| Medical | 0.7836 | 0.5786 | 0.7584 | 0.3333 | 0.9854 | 0.7564 | |
| Bio | 0.7895 | 0.5815 | 0.7592 | 0.3369 | 0.9863 | 0.7557 | |
| * MC | 0.7751 | 0.5445 | 0.7495 | 0.2965 | 0.9853 | 0.7566 | |
| Aggregated Model | Medical + * MC | 0.7898 | 0.5891 | 0.7604 | 0.3578 | 0.9867 | 0.7580 |
| Umls + * MC | 0.7937 | 0.5959 | 0.7622 | 0.3643 | 0.9876 | 0.7587 | |
| Bio + * MC | 0.7922 | 0.5905 | 0.7593 | 0.3637 | 0.9877 | 0.7583 | |
| ICD-9 Code | F1 | Number of Data in Test Set | ||
|---|---|---|---|---|
| Umls | Character | |||
| Majority labels | 51.23 | 0.9324 | 0.9245 | 628 |
| 06.4 | 0.9751 | 0.9638 | 264 | |
| 50.12 | 0.4096 | 0.3871 | 111 | |
| Minority labels | 46.43 | 0.7059 | 0.7200 | 27 |
| 38.03 | 0.7600 | 0.8077 | 23 | |
| 45.52 | 0.7857 | 0.8148 | 12 | |
| 39.25 | 0.6000 | 0.6667 | 6 | |
| Model | AP | F1 | AUC | ||||
|---|---|---|---|---|---|---|---|
| Micro | Macro | Micro | Macro | Micro | Macro | ||
| Proposed Model | Umls + * MC | 0.7672 | 0.4899 | 0.7415 | 0.3975 | 0.9842 | 0.9703 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.; Bong, J.-W.; Park, I.; Lee, H.; Choi, J.; Park, P.; Kim, Y.; Choi, H.-S.; Kang, S. ConBERT: A Concatenation of Bidirectional Transformers for Standardization of Operative Reports from Electronic Medical Records. Appl. Sci. 2022, 12, 11250. https://doi.org/10.3390/app122111250
Park S, Bong J-W, Park I, Lee H, Choi J, Park P, Kim Y, Choi H-S, Kang S. ConBERT: A Concatenation of Bidirectional Transformers for Standardization of Operative Reports from Electronic Medical Records. Applied Sciences. 2022; 12(21):11250. https://doi.org/10.3390/app122111250
Chicago/Turabian StylePark, Sangjee, Jun-Woo Bong, Inseo Park, Hwamin Lee, Jiyoun Choi, Pyoungjae Park, Yoon Kim, Hyun-Soo Choi, and Sanghee Kang. 2022. "ConBERT: A Concatenation of Bidirectional Transformers for Standardization of Operative Reports from Electronic Medical Records" Applied Sciences 12, no. 21: 11250. https://doi.org/10.3390/app122111250
APA StylePark, S., Bong, J.-W., Park, I., Lee, H., Choi, J., Park, P., Kim, Y., Choi, H.-S., & Kang, S. (2022). ConBERT: A Concatenation of Bidirectional Transformers for Standardization of Operative Reports from Electronic Medical Records. Applied Sciences, 12(21), 11250. https://doi.org/10.3390/app122111250








