Abstract
In this article, a four parameter lifetime model called the Topp–Leone modified Weibull distribution is proposed. The suggested distribution can be considered as an alternative to Kumaraswamy Weibull, generalized modified Weibull, extend odd Weibull Lomax, Weibull-Lomax, Marshall-Olkin alpha power extended Weibull and exponentiated generalized alpha power exponential distributions, etc. The suggested model includes the Topp-Leone Weibull, Topp-Leone Linear failure rate, Topp-Leone exponential and Topp-Leone Rayleigh distributions as a special case. Several characteristics of the new suggested model including quantile function, moments, moment generating function, central moments, mean, variance, coefficient of skewness, coefficient of kurtosis, incomplete moments, the mean residual life and the mean inactive time are derived. The probability density function of the Topp–Leone modified Weibull distribution can be right skewed and uni-modal shaped but, the hazard rate function may be decreasing, increasing, J-shaped, U-shaped and bathtub on its parameters. Three different methods of estimation as; maximum likelihood, maximum product spacing and Bayesian methods are used to estimate the model parameters. For illustrative reasons, applications of the Topp–Leone modified Weibull model to four real data sets related to medical and engineering sciences are provided and contrasted with the fit reached by several other well-known distributions.
1. Introduction and Motivation
Over the last four decades or more, a major amount of statistical research has developed a number of innovative statistical distributions. This criterion was inspired by theoretical concepts, real-world applications, or both. The most recent advancement is to build a larger range of statistical distributions by adding one (or more) shape parameters to a baseline distribution, which tends to improve the diversity of the statistical models developed. Particularly appropriate to data sets for researching reliability experiments, medical, clinical trials, finance, infant mortality rate, and engineering applications, among other things. The Weibull distribution is a well-liked distribution that may be used to predict how long a product will last, such as a ball bearing, a car part, or electrical insulation see [,,,,,,,,,]. It is also frequently employed in biological and medical applications. This distribution, however, lacks a bathtub or upside-down shaped hrf, making it ineffective for simulating the lifetime of some systems. To overcome this shortcoming, refs. [,,] studied the modified Weibull (MW) distribution that can be used to describe various reliability models. It has three parameters, and are two scale parameters and is the shape parameter. The distribution function (cdf), the probability density function (pdf), survival function (sf) and hazard rate function (hrf) of the MW distribution due to [] are provided via
and
Recently, many authors have proposed many generalizations of the MW distribution to improve its flexibility. For example, generalized MW by [], beta MW by [], exponentiated MW by [], new MW by [], transmuted MW by [], McDonald MW by [], Kumaraswamy MW [], gamma MW by [], Burr XII MW by [], log-logistic MW by [], half logistic MW by [], beta exponentiated MW by [] and sine MW by [].
Topp–Leone () distribution was proposed by []. Recently, statisticians have been showing interest on Topp–Leone () distribution as a generator. This distribution has one parameter and its domain is restricted to . Furthermore, this distribution is not particularly adaptable. It is an extension of the triangular distribution and can be used as an alternative in circumstances where a bounded distribution, such as the uniform, triangular, trapezoid, or beta, is acceptable. Ref. [] introduced -generated (-G) family of distributions and the cdf of the TL-G can be defined as
the associated pdf is available below
The sf and hrf of the -G are provided via
and
respectively.
Many researchers have recently suggested numerous generalizations of the TL-G to increase its versatility. For example, the Weibull distribution, inverse Weibull distribution, Burr XII distribution, geometric distribution, the transmuted TL-G family of distributions, odd log-logistic family, Lomax distribution, TL-G family of distributions, the TL-G generalized exponential power series distribution, power series family of distributions, the type II TL-G family, the type II generalized TL-G family, the Fréchet TL-G family, among others. See [] for elaborate references.
In this article, the Topp–Leone modified Weibull (TLMW) distribution, which has four parameters are introduced by connecting in Equation (1) to the cdf in Equation (5). The TLMW distribution encompasses the behavior of and provides better fits than some well known lifetime distributions, such as Kumaraswamy Weibull, generalized modified Weibull, extend odd Weibull Lomax, Weibull-Lomax, Marshall-Olkin alpha power extended Weibull and exponentiated generalized alpha power exponential distributions, distributions. We are motivated to introduce the () distribution because it is capable of modeling decreasing, increasing, J-shaped, U-shaped and bathtub hrf which are common in reliability studies and lifetime data analysis. It might be a useful option for fitting skewed data that other common distributions may not be able to correctly fit, and it may also be utilized in a range of challenges in diverse sectors such as biological investigations, industrial dependability, the suggested family have also an important utilisation in the insurance industry see; [,,] and survival analysis.
The reminder of the paper is organized as follows: In Section 2, the Topp–Leone modified Weibull distribution is defined. Also, an useful expansion of the distribution and some sub-models are studied in the same section. Some statistical features of the distribution are discussed in Section 3. In Section 4, the maximum likelihood (ML), maximum product spacing (MPS) and Bayesian estimators of the unknown parameters are obtained. Section 5 concerned with simulation results. Four illustrative applications based on real data sets related to medical and engineering sciences are investigated in Section 6. Finally, some conclusions are provided in Section 7.
2. Topp–Leone Modified Weibull Distribution
In this section, we introduce the Topp–Leone modified Weibull distribution by taking in Equation (3) to the cdf of in Equation (5). Hence, the cdf and pdf of can indeed be expressed as
and
By writing where if the pdf of X can be written as (10).
The hrf is defined by
Figure 1 and Figure 2 shows the pdf and hrf for different values of and and at . The plots shows the pdf can be decreasing, uni-modal and right skewness but the hrf can be decreasing, increasing, J-shaped, U-shaped and bathtub.
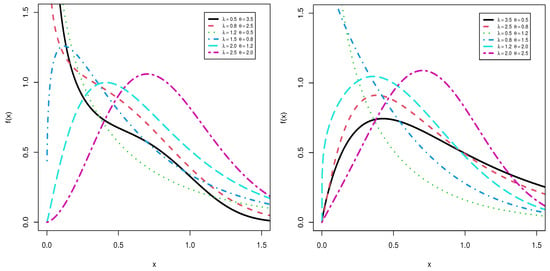
Figure 1.
Plots of the pdf for TLMW model at .
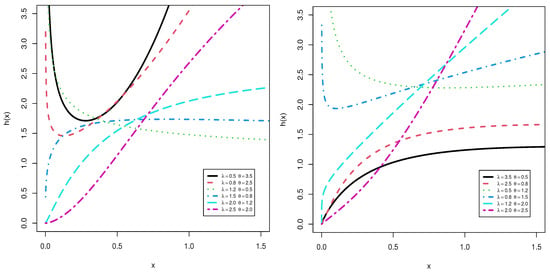
Figure 2.
Plots of the hrf for TLMW model at .
A Useful Expansion for the Density Function
In this part, the pdf representation of the distribution are discussed. The mathematical relation and the series representation are given below. If b is a positive real non-negative integer and then
The distribution is a very flexible model which contains the well-known models listed below as special models:
- If , the distribution reduces to the Weibull (TLW) distribution.
- If , then Equation (9) the distribution reduces to the Linear failure rate (TLLFR) model.
- If , the TLMW distribution reduces to the exponential (TLE) distribution.
- If and , then Equation (9) the TLMW distribution reduces to the Rayleigh (TLR) distribution.
3. Some Statistical Features
In this part, some of the statistical features of the distribution such as; quantile function (QF), moments(Ms), M generating function (MGF), central Ms, mean, variance, coefficient of skewness (CS), coefficient of kurtosis (CK), incomplete Ms (INMs), the mean residual life (MRL) and the mean inactive time are computed.
3.1. Quantile Function
The qth QF of the is the real solution of the following equation
In particular, setting in Equation (16), the median of X can be calculated. Also, using the QF given by Equation (16), the Bowley’s skewness (see []) and the Moors’ kurtosis (see []) can be determined as
and
where is the QF provided in Equation (16). Table 1, provide the median, skewness and kurtosis, of the distribution for various values of , , and . It is clear from Table 1 that for fixed , and with increasing , median and kurtosis are decreasing but the skewness is increasing.

Table 1.
Quantiles for distribution.
3.2. Moments and Moment Generating Function
The M of distribution can be computed using the following expression
Further, the central Ms and cumulants of the distribution can be determined from and , respectively, where , , , and etc. Additionally, and can be obtained according to the following relation and , respectively.
The MGF of model is provided as below
Table 2 and Table 3 show the numerical values of , , , , variance (V), skewness (CS) and kurtosis (CK) of the distribution.

Table 2.
Moments for distribution at .

Table 3.
Moments for distribution at .
3.3. Incomplete Moments
In terms of lifespan models, It is interesting to know the lower and upper INMs of X defined by and respectively, for any real The lower INMs of distribution is
Likewise, the upper INMs of distribution is
where and is the lower and upper incomplete gamma functions.
The MRL has many applications (see [,]). The MRL function is defined as
where is the first INM of X and by putting in Equation (20), then
Furthermore, the mean inactive time of X is determined as
4. Three Different Estimation Methods
In this part, ML approach, MPS approach, and Bayesian approach techniques are provided for distribution parameters.
4.1. Maximum Likelihood Method
In this subsection, parameters estimates are derived using ML method. The log likelihood function for distribution with the vector of parameters is
The corresponding scoring function is supplied via
The components of the score vector are described as
and
The estimates (MLEs) of , symbolize can indeed be produced by optimizing the log-likelihood function with regard to the involved parameters. In this approach, R-software packages such as nlm() and maxLik() might be useful (see [,]). Alternatively, the parameters can be found by concurrently solving four non-linear equations using an iterative process such as the Quasi Newton-Raphson method. Because the MLEs of the unknown parameters cannot be constructed in closed form, determining the exact distributions of the MLEs is difficult. As a result, specific confidence ranges can’t founded for the parameters. The large sample approximation must be used. It is known that the asymptotic distribution of the MLE is , see [], where , the inverse of the observed information matrix of the unknown parameters is,
and whose elements are given in Appendix A.
4.2. Maximum Product of Spacing Method
In this subsection, the MPS will be used, a substitute for the MLE method that is used to estimate the parameters of continuous uni-variate models. This method was developed by [,,,,,,]. The uniform spacing of the TLMW distribution can be characterised as follows, assuming a random sample of size n:
where denotes to the uniform spacing, , and .
The MPS estimators (MPSE) of the TLMW distribution parameters may be constructed by maximizing Equation (27) with regards to and .
and
4.3. Bayesian Estimation
The Bayesian technique is extremely helpful in the analysis of reliability since it allows for the incorporation of previous knowledge into the research process. This is important because the restricted availability of data is one of the major issues with reliability analysis. By apply the insightful before to the , and parameters, which are prior gamma distributions. , and have independent joint prior density functions that look like this:
By determining suitable and superior values for the independent joint prior’s hyper-parameters using the estimate and variance-covariance matrix of the MLE technique. By equating the gamma priors’ mean and variance as the following equations, the estimated hyper-parameters can be determined as follows:
where, L is the number of iteration and , , and .
The likelihood function and joint prior function combine to form the joint posterior density function of . The joint posterior of the TLMW distribution is thus written as
utilising a function for squared error loss (SEL), which is the most typical function for symmetric loss. The Bayesian estimators of based on the SEL function.
The integrals provided by Equation (36) cannot be directly obtained, it should be emphasised. By employing the Markov chain Monte Carlo (MCMC) technique to estimate the value of integrals. Important sub-classes of MCMC approaches include Gibbs sampling and, within Gibbs samplers, a more generic Metropolis-Hesting. The Metropolis-Hasting (M-H) algorithm and Gibbs sampling are the two most often used MCMC applications. The MH technique, like acceptance-rejection sampling, presupposes that a candidate value can be generated from a proposal distribution for each iteration of the algorithm. The M-H algorithm claims that a candidate value from a normal proposal distribution can be generated for each iteration of the process, much like acceptance-rejection sampling claims. To obtain random samples of conditional posterior densities from the TLMW distribution, the M-H will be used inside Gibbs sampling. The conditional posterior distributions are as follows:
and
For further information, read []. The credible intervals (CIs) for Bayesian estimates are also known as the highest posterior density (HPD) intervals. Researchers utilized a well recognized approach to generate HPD ranges for unknown distribution characteristics. To generate estimates, samples are drawn using the proposed MH algorithm in this method; for information on the HPD algorithm, see [].
5. Simulation
In this section, the purpose is to analyze the performance of the various estimation methods presented in sections above. Given a proposed model, a simulation study is used to analyse the behaviour of the offered methodologies and assess the statistical performances of the estimators. Calculations have been performed using the R-statistical programming language. Calculating MLEs and HPD intervals in R-language is done through utilizing the “maxLike” and “HDInterval” packages. Based on the following data produced by the TLMW distribution, where X is distributed as the TLMW distribution for various actual values of the parameters, Monte Carlo experiments were conducted:
- In Table 4, the actual values of the parameters are selected as first combination: , , and is 0.6 and 3.
- In Table 5, the actual values of the parameters are selected as second combination: , , and is 0.6 and 3.
- In Table 6, the actual values of the parameters are selected as third combination: , , and is 0.6 and 3.

Table 4.
ML, MPS, Bayes estimation methods: RB, MSE, LACI and LCCI at first combination.

Table 5.
ML, MPS, Bayes estimation methods: RB, MSE, LACI and LCCI at second combination.

Table 6.
ML, MPS, Bayes estimation methods: RB, MSE, LACI and LCCI at third combination.
The sample sizes used in this simulation were 30, 80, and 150. The optimum method is one that minimises the relative bias (RB), mean squared error (MSEr), and length of confidence interval (LCI) of the estimator. In contrast to the LCI of MLE and MPS, which is the length of asymptotic confidence intervals (LACI), and Bayes calculates the length of confidence credible intervals (LCCI). Table 4, Table 5 and Table 6 provide the results of the simulation, and the following conclusion remakes are noted as a result.
- For techniques of estimation methods, the RBias, MSE, and the LCI related to the parameter estimations drop as n rises.
- It is noticed that the measures of the Bayesian estimation method are better than MLE method, as it is recognized.
- It is noticed that MPSE measures are, on average, more accurate than MLE and Bayesian estimate measures.
- Table 4, shows that the actual value of parameter increases then the measures of , , decreases.
- Table 6, show that the actual value of parameter increases then the measures of parameters increases.
A heatmap is a graphical representation of data that uses a system of color-coding to represent different values. Figure 3 and Figure 4 show heatmaps summarizing the results of the RB and MSE of simulation results. The dark red shading indicates RB and MSE with large value. The light yellow shading indicates RB and MSE with small value of parameters for TLMW under different estimation methods.
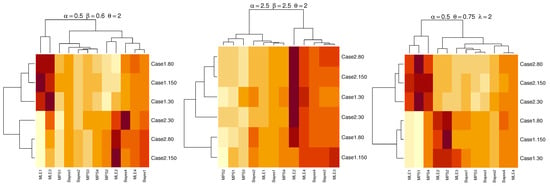
Figure 3.
Heatmaps of RB for parameters under different estimation methods.
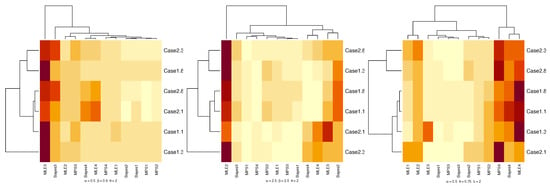
Figure 4.
Heatmaps of MSEr for parameters under different estimation methods.
6. Applications
In this section, applications of the distribution using four real data sets as COVID-19 data, survival rates sunaina Pigs, Waiting times of service of 100 bank customers, and Tensile strength for single carbon fibres (in GPa) in order to illustrate the potentiality of the model are provided. The proposed distribution will be compared with other related lifetime models as Kumaraswamy Weibull (KW) by [], generalized modified Weibull (GMW) by [], extend odd Weibull Lomax (EOWL) by [], Weibull-Lomax (WL) by [], Marshall-Olkin alpha power extended Weibull (MOAPEW) by [] and exponentiated generalized alpha power exponential (EGAPE) by [].
When comparing distributions, we consider the following discriminating criteria: Akaike information criterion (AKIC), consistent AKIC (CAKIC), Bayesian information criterion (BIC) and Hannan–Quinn information criterion (HQIC), the Kolmogrov-Smirnov distance (KSD) with Pvalue (PVKS).
where p is a number of parameters, and n is a sample size.
6.1. COVID-19 Data
The data set given as follows: 1, 1, 2, 4, 5, 1, 1, 3, 6, 6, 4, 1, 5, 6, 6, 8, 5, 7, 7, 9, 9, 15, 17, 11, 13, 5, 14, 5, 13, 9, 19, 15, 11, 14, 12, 11, 7, 13, 10, 20, 22, 21, 12, 14, 9, 14, 7, 16, 17, 13, 21, 11, 11, 8, 11, 12, 15, 21, 20, 18, 15, 14, 21, 16, 11, 28, 29, 19, 14, 19, 29, 34, 34, 46, 46, 47, 36.00, 38, 40, 32, 39, 34, 35, 36, 35, 45, 62. The most recent data, used by [], revealed the daily fatality confirmed cases attributable to COVID-19. The data consists of 89 observed values, with 18.72 reported deaths on average every day.
By using equations Equations (22)–(25) to obtain the MLEs of the model parameters with standard error (SE) and the results are reported in Table 7. The estimated density with histogram of waiting times data for distribution and other distributions are displayed in Figure 5. The estimated cumulative and the fitted distribution and other distributions for the waiting times data set are displayed in Figure 6. The estimated PP and QQ plot for distribution are displayed for waiting times data in Figure 7.

Table 7.
MLE for parameters of and comparative models for COVID-19 data.
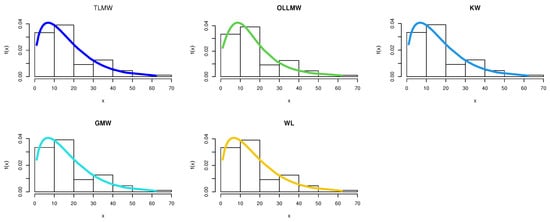
Figure 5.
Pdf for different models for COVID-19 data.
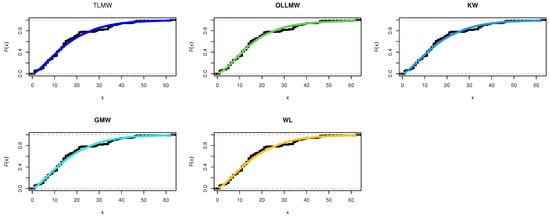
Figure 6.
Cdf for different models for COVID-19 data.
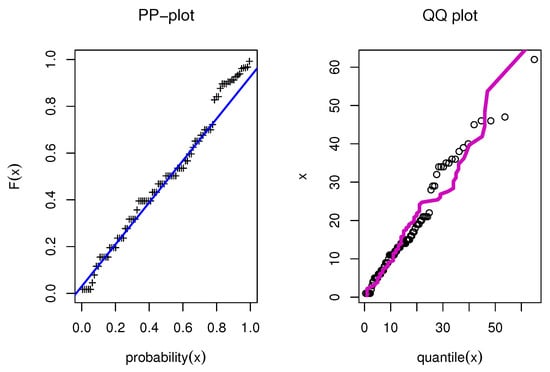
Figure 7.
PP and QQ for model for COVID-19 data.
Now, applying the formal goodness-of-fit tests in order to verify which distribution fits better to these data set. The measures AKIC, BIC, CAKIC, HQIC and the KSD with PVKS (See Table 8) are considered.

Table 8.
Discrimination criteria and KS test of the TLMW model parameters for COVID-19 data.
The CIs for the parameters , , and are presented for MLE and Bayesian estimation methods in Table 9. Figure 8 displayed contour plot of parameters for model to check uniqueness and max-value of likelihood for COVID-19 data. While, Figure 9 confirmed convergence of MCMC results, which show the MCMC samples as a pairs plot, with the trace plot, and marginal frequency of proposed distribution as normal distribution for each parameter on the diagonal for parameters of model for COVID-19 data. Figure 10 discussed Auto-correlation Function (ACF) test. Auto-correlation for MCMC is the correlation between a iteration series with a lagged version of itself. The ACF starts at a lag of 0, which is the correlation of the iteration series with itself and therefore results in a correlation of 1.

Table 9.
MLE and Bayesian estimation methods of the model parameters for COVID-19 data.
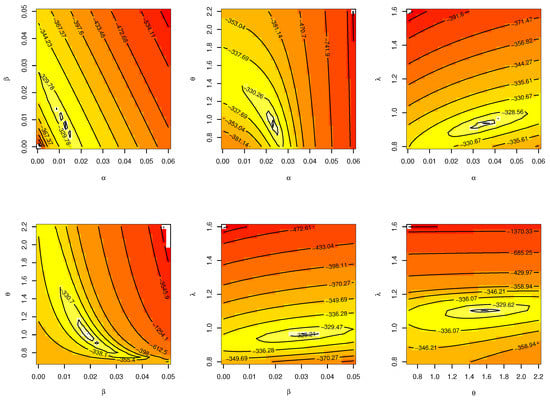
Figure 8.
Contour plot for parameters of model for COVID-19 data.
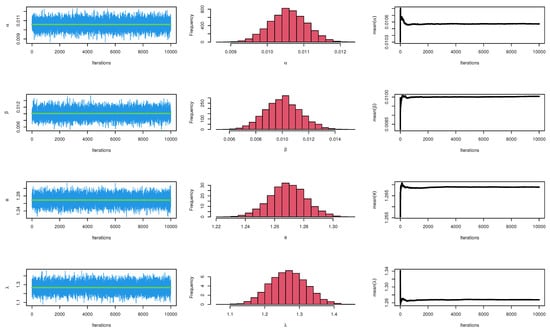
Figure 9.
MCMC plots for parameters of model for COVID-19 data.
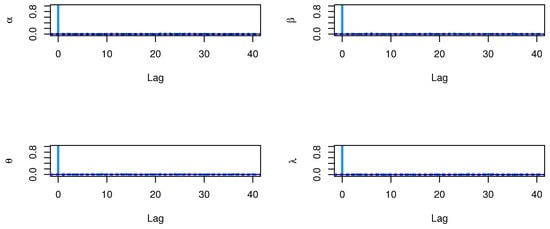
Figure 10.
ACF test plots for parameters of model for COVID-19 data.
6.2. Guinea Pigs Data
The set of data [] shows survival rates for Guinea pigs infected with virulent tubercle bacilli. One of the reasons guinea pigs were chosen for this investigation is that they are thought to be highly susceptible to human tuberculosis. The data set given as follows: 0.10, 0.33, 0.440, 0.56, 0.59, 0.72, 0.74, 0.77, 0.920, 0.93, 0.96, 1.00, 1.00, 1.02, 1.05, 1.07, 1.07, 1.08, 1.08, 1.08, 1.09, 1.12, 1.13, 1.15, 1.160, 1.200, 1.21, 1.22, 1.22, 1.24, 1.30, 1.34, 1.360, 1.390, 1.44, 1.46, 1.53, 1.59, 1.60, 1.63, 1.63, 1.680, 1.71, 1.72, 1.76, 1.83, 1.950, 1.96, 1.97, 2.02, 2.13, 2.15, 2.16, 2.22, 2.3, 2.31, 2.40, 2.45, 2.51, 2.53, 2.54, 2.54, 2.780, 2.93, 3.27, 3.42, 3.47, 3.61, 4.02, 4.32, 4.580, 5.55.
By using equations Equations (22)–(25) to obtain the MLEs of the model parameters and the results are reported in Table 10. The estimated density with histogram of waiting times data for distribution and other distributions are displayed in Figure 11. The estimated cumulative and the fitted distribution and other distributions for the Guinea Pigs data set are displayed in Figure 12. The estimated PP and QQ plot for distribution are displayed for Guinea Pigs data in Figure 13.

Table 10.
MLE for parameters of TLMW and comparative models for Guinea Pigs data.
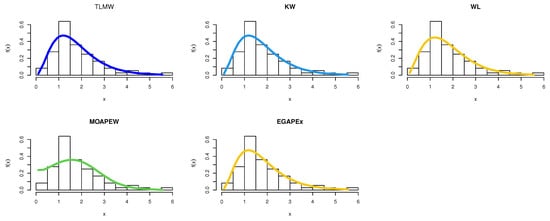
Figure 11.
Pdf for different models for Guinea Pigs data.
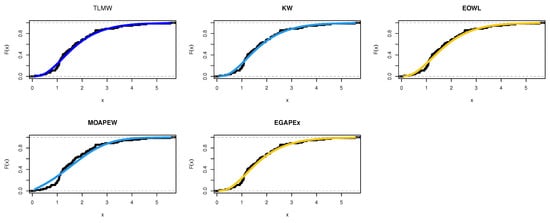
Figure 12.
Cdf for different models for Guinea Pigs data.
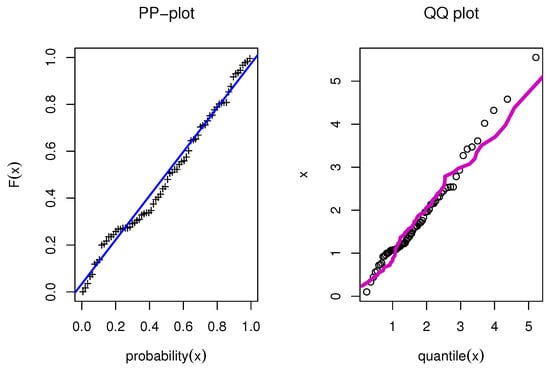
Figure 13.
PP and QQ for model for Guinea Pigs data.
Now, applying the formal goodness-of-fit tests in order to verify which distribution fits better to these data set. The measures AKIC, BIC, CAKIC, HQIC and the KSD with PVKS (See Table 11) are considered.

Table 11.
Discrimination criteria and KS test of the TLMW model parameters for Guinea Pigs data.
The CIs for the parameters , , and are presented for MLE and Bayesian estimation methods in Table 12. Figure 14 displayed contour plot of parameters for model to check uniqueness and max-value of likelihood for Guinea Pigs data. While, Figure 15 confirmed convergence of MCMC results, which show the MCMC samples as a pairs plot, with the trace plot, and marginal frequency of proposed distribution as normal distribution for each parameter on the diagonal for parameters of model for Guinea Pigs data. Figure 16 discussed ACF test. Auto-correlation for MCMC is the correlation between a iteration series with a lagged version of itself. The ACF starts at a lag of 0, which is the correlation of the iteration series with itself and therefore results in a correlation of 1.

Table 12.
MLE and Bayesian estimation methods of the model parameters for Guinea Pigs data.
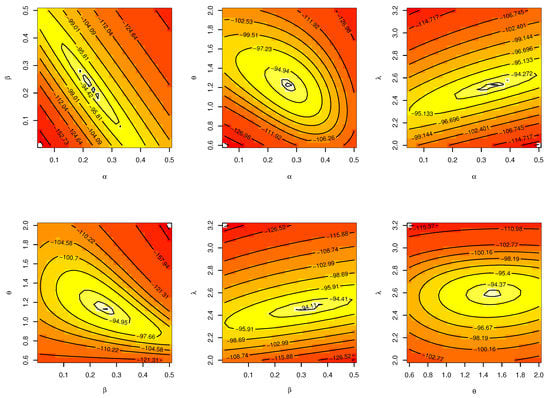
Figure 14.
Contour plot for parameters of model for Guinea Pigs data.
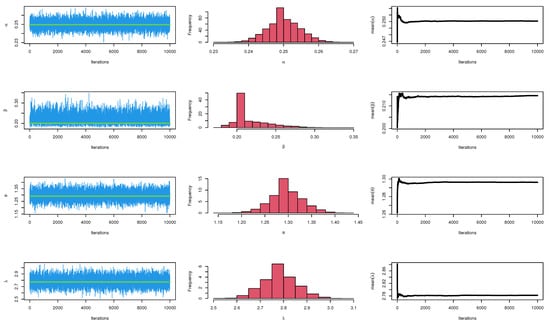
Figure 15.
MCMC plots for parameters of model for Guinea Pigs data.
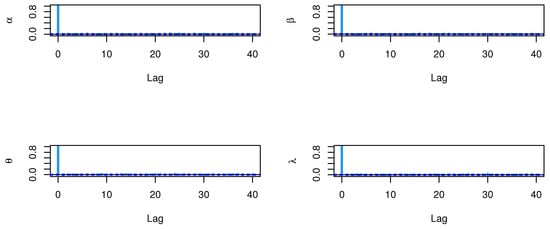
Figure 16.
ACF test plots for parameters of model for Guinea Pigs data.
6.3. Waiting Times of Service of 100 Bank Customers
The data set given in Table 13 represents the waiting times (in minutes) before service of 100 bank customers (see []).

Table 13.
Waiting times (in minutes) before service of 100 bank customers.
Summary statistics of the Waiting times data set are presented in Table 14.

Table 14.
Summary statistics of the Waiting times data set.
Table 14, show that the distribution of this data set is positively skewed. By using equations Equations (22)–(25) to obtain the MLEs of the model parameters and the results are reported in Table 15.

Table 15.
MLE for parameters of and comparative models for waiting times.
The estimated density with histogram of waiting times data for distribution and other distributions are displayed in Figure 17. The estimated cumulative and the fitted distribution and other distributions for the waiting times data set are displayed in Figure 18. The estimated PP and QQ plot for distribution are displayed for waiting times data in Figure 19.
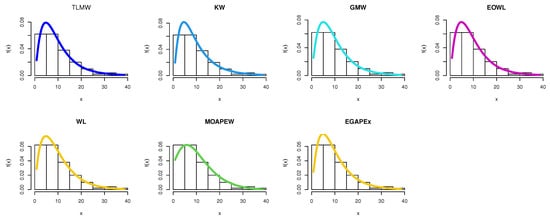
Figure 17.
Pdf for different models for waiting times.
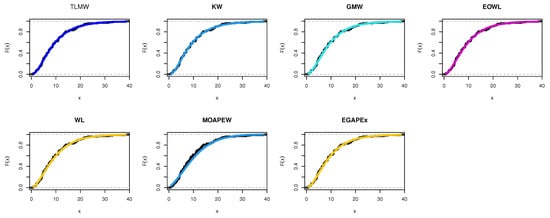
Figure 18.
Cdf for different models for waiting times.
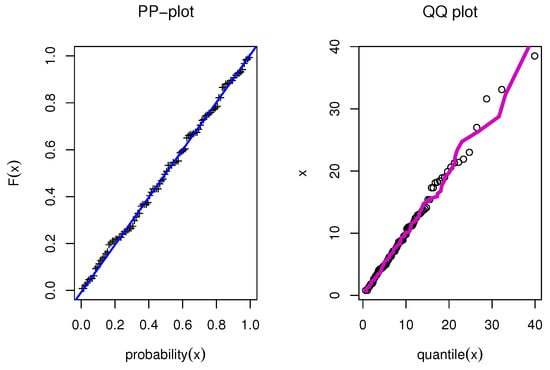
Figure 19.
PP and QQ for model for waiting times.
Now, applying the formal goodness-of-fit tests in order to verify which distribution fits better to these data set. The measures AKIC, BIC, CAKIC, HQIC and the KSD with PVKS (See Table 16) are considered.

Table 16.
Discrimination criteria and KS test of the model parameters for waiting times.
The CIs for the parameters , , and are presented for MLE and Bayesian estimation methods in Table 17. Figure 20 displayed contour plot of parameters for model to check uniqueness and max-value of likelihood for waiting times. While, Figure 21 confirmed convergence of MCMC results, which show the MCMC samples as a pairs plot, with the trace plot, and marginal frequency of proposed distribution as normal distribution for each parameter on the diagonal for parameters of model for waiting times. Figure 22 discussed ACF test. Auto-correlation for MCMC is the correlation between a iteration series with a lagged version of itself. The ACF starts at a lag of 0, which is the correlation of the iteration series with itself and therefore results in a correlation of 1.

Table 17.
MLE and Bayesian estimation methods of the model parameters for waiting times.
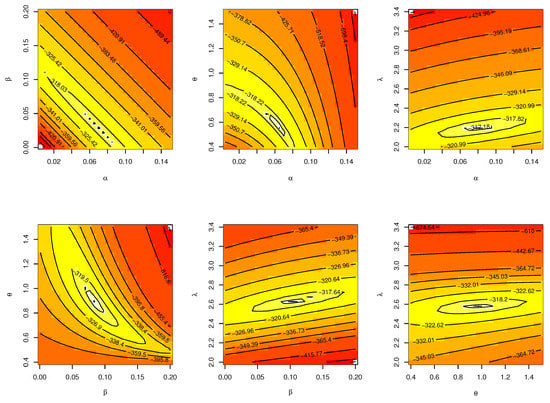
Figure 20.
Contour plot for parameters of model for waiting times.
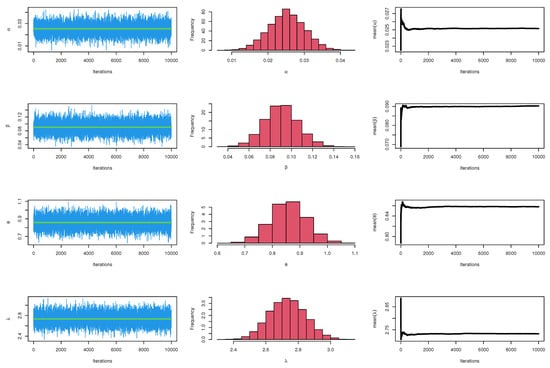
Figure 21.
MCMC plots for parameters of model for waiting times.
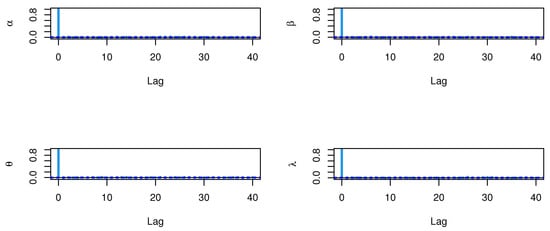
Figure 22.
ACF test plots for parameters of model for waiting times.
6.4. Tensile Strength for Single Carbon Fibres (in GPa)
The data set given in Table 18 contains the tensile strength for single carbon fibres (in GPa). The data set finds its source in [].

Table 18.
Tensile strength (with unit in GPa) for single carbon fibers.
We use equations Equations (22)–(25) to obtain the MLEs of the model parameters and the results are reported in Table 19.

Table 19.
MLE for parameters of and comparative models for single carbon fibers data.
The estimated density with histogram of single carbon fibers data for distribution and other distributions are displayed in Figure 23. The estimated cumulative and the fitted distribution and other distributions for the second set are displayed in Figure 24. The estimated PP and QQ plot for distribution are displayed for single carbon fibers data in Figure 25.
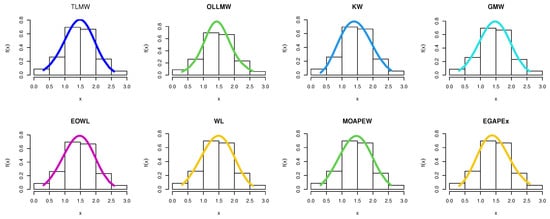
Figure 23.
Pdf for different models for single carbon fibers data.
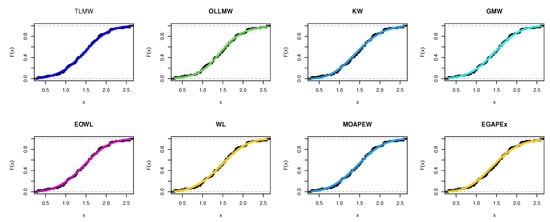
Figure 24.
Cdf for different models for single carbon fibers data.
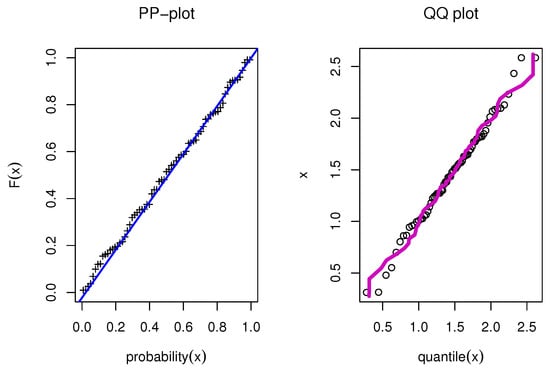
Figure 25.
PP and QQ for model for single carbon fibers data.
Now, applying the formal goodness-of-fit tests in order to verify which distribution fits better to single carbon fibers data set. The measures AKIC, BIC, CAKIC, HQIC and the KSD with PVKS (See Table 20) are considered.

Table 20.
Discrimination criteria and KS test of the model parameters for single carbon fibers.
The CIs for the parameters , , and are presented for MLE and Bayesian estimation methods in Table 21. Figure 26 displayed contour plot of parameters for model to check uniqueness and max-value of likelihood for single carbon fibers data. While, Figure 27 confirmed convergence of MCMC results, which show the MCMC samples as a pairs plot, with the trace plot, and marginal frequency of proposed distribution as normal distribution for each parameter on the diagonal for parameters of model for single carbon fibers. Figure 28 discussed ACF test. Auto-correlation for MCMC is the correlation between a iteration series with a lagged version of itself. The correlation between the iteration series and itself, which is where the ACF starts with a lag of 0, yields a correlation of 1.

Table 21.
LE and Bayesian estimation methods of the model parameters for single carbon fibers.
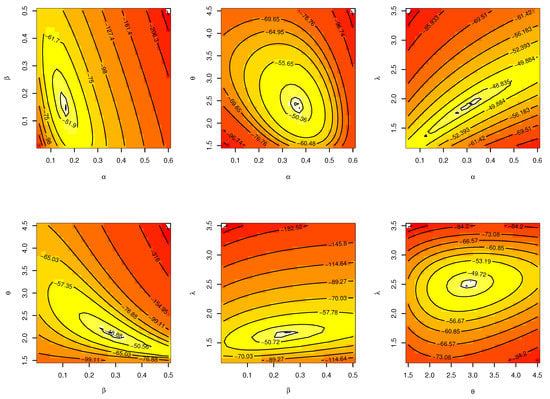
Figure 26.
Contour plot for parameters of model for single carbon fibers.
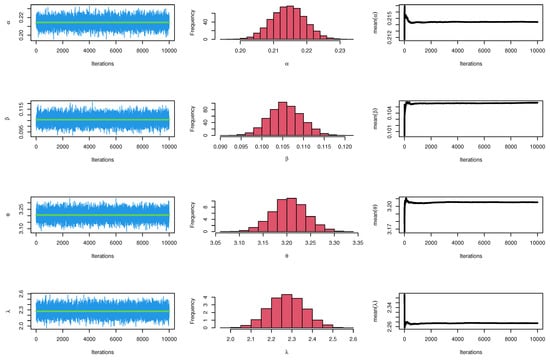
Figure 27.
MCMC plots for parameters of model for single carbon fibers.
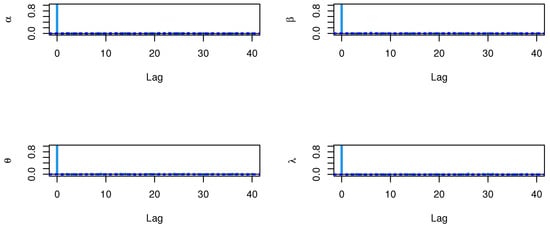
Figure 28.
ACF test plots for parameters of model for single carbon fibers.
7. Conclusions
In this paper, a four-parameter family of distributions called the Topp–Leone modified Weibull distribution () is introduced. Some of its statistical and reliability features are derived. The density function can decreasing, uni-modal and right skewed but, its hazard rate function can be decreasing, increasing, J-shaped, U-shaped and bathtub. Therefore, it can be used quite effectively in analyzing lifetime data. Three different methods of estimation as; maximum likelihood, maximum product spacing and Bayesian methods are used to estimate the model parameters. Additionally, the model can be used as a substitute for many known distributions and in some cases it can provide better results (with respect to model fitting) than the models considered in the paper. The flexibility of this distribution is evaluated by applying it to four real data sets and comparing it to various other distributions. The findings of the tables and figures show that the suggested model regularly outperforms other competitor models for these four data sets as COVID-19 data, survival rates Guinea Pigs, Waiting times of service of 100 bank customers, and Tensile strength for single carbon fibres (in GPa). Finally, the distribution will attract a broader range of applications, including medical, engineering, and social sciences, among others.
Author Contributions
These authors contributed equally to this work. All authors have read and agreed to the published version of the manuscript.
Funding
The authors extend their appreciation to the Deanship of Scientific Research at Imam Mohammad Ibn Saud Islamic University for funding this work through Research Group no. RG-21-09-15.
Data Availability Statement
Datasets can be found under the Section 6.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
The second order partial derivatives of the log likelihood function of distribution with respect to are given by:
where
References
- Piña-Monarrez, M.R. Weibull stress distribution for static mechanical stress and its stress/strength analysis. Qual. Reliab. Eng. Int. 2018, 34, 229–244. [Google Scholar] [CrossRef]
- Almarashi, A.M.; Jamal, F.; Chesneau, C.; Elgarhy, M. The Exponentiated Truncated Inverse Weibull-Generated Family of Distributions with Applications. Symmetry 2020, 12, 650. [Google Scholar] [CrossRef]
- Abouelmagd, T.H.M.; Al-mualim, S.; Elgarhy, M.; Elgarhy, M.; Afify, A.Z.; Ahmad, M. Properties of the four-parameter Weibull distribution and its Applications. Pak. J. Stat. 2017, 33, 449–466. [Google Scholar]
- Alahmadi, A.A.; Alqawba, M.; Almutiry, W.; Shawki, A.W.; Alrajhi, S.; Al-Marzouki, S.; Elgarhy, M. A New version of Weighted Weibull distribution: Modelling to COVID-19 data. Discret. Dyn. Nat. Soc. 2022, 2022, 3994361. [Google Scholar] [CrossRef]
- Algarni, A.; Elgarhy, M.; Almarashi, A.M.; Fayomi, A.; El-Saeed, A.R. Classical and Bayesian Estimation of the Inverse Weibull Distribution: Using Progressive Type-I Censoring Scheme. Adv. Civ. Eng. 2021, 2021, 5701529. [Google Scholar] [CrossRef]
- Al-Moisheer, A.S.; Elbatal, I.; Almutiry, W.; Elgarhy, M. Odd inverse power generalized Weibull generated family of distributions: Properties and applications. Math. Probl. Eng. 2021, 2021, 5082192. [Google Scholar] [CrossRef]
- Alkarni, S.H.; Afify, A.Z.; Elbatal, I.; Elgarhy, M. The Extended Inverse Weibull Distribution: Properties and Applications. Complexity 2020, 2020, 3297693. [Google Scholar] [CrossRef]
- Alotaibi, N.; Elbatal, I.; Almetwally, E.M.; Alyami, S.A.; Al-Moisheer, A.S.; Elgarhy, M. Bivariate Step-Stress Accelerated Life Tests for the Kavya–Manoharan Exponentiated Weibull Model under Progressive Censoring with Applications. Symmetry 2022, 14, 1791. [Google Scholar] [CrossRef]
- Alotaibi, N.; Elbatal, I.; Almetwally, E.M.; Alyami, S.A.; Al-Moisheer, A.S.; Elgarhy, M. Truncated Cauchy Power Weibull-G Class of Distributions: Bayesian and Non-Bayesian Inference Modelling for COVID-19 and Carbon Fiber Data. Mathematics 2022, 10, 1565. [Google Scholar] [CrossRef]
- Hassan, A.S.; Elgarhy, M. Exponentiated Weibull-Weibull Distribution: Statistical Properties and Applications. Gazi J. Sci. 2019, 32, 616–635. [Google Scholar]
- Lai, C.D.; Xie, M.; Murty, D.N.P. A modified Weibull distribution. IEEE Trans. Reliab. 2003, 52, 33–37. [Google Scholar] [CrossRef]
- Ng, H.K. Parameter estimation for a modified Weibull distribution, for progressively type-II censored samples. IEEE Trans. Reliab. 2005, 54, 374–380. [Google Scholar] [CrossRef]
- Sarhan, A.M.; Zaindin, M. Modified Weibull distribution. Appl. Sci. 2009, 11, 123–136. [Google Scholar]
- Carrasco, J.M.F.; Ortega, E.M.M.; Cordeiro, G.M. A generalized modified Weibull distribution for lifetime modeling. Comput. Stat. Data Anal. 2008, 53, 450–462. [Google Scholar] [CrossRef]
- Silva, G.O.; Ortega, E.M.M.; Cordeiro, G.M. The beta modified Weibull distribution. Lifetime Data Anal. 2010, 16, 409–430. [Google Scholar] [CrossRef]
- Sarhan, A.M.; Apaloo, J. Exponentiated modified Weibull extension distribution. Reliab. Eng. Syst. Saf. 2013, 112, 137–144. [Google Scholar] [CrossRef]
- Almalki, S.J.; Yuan, J. A new modified Weibull distribution. Reliab. Eng. Syst. Saf. 2013, 111, 164–170. [Google Scholar] [CrossRef]
- Khan, M.S.; King, R. Transmuted modified Weibull distribution: A generalization of the modified Weibull probability distribution. Eur. J. Pure Appl. Math. 2013, 6, 66–86. [Google Scholar]
- Merovci, F.; Elbatal, I. The McDonald modified Weibull distribution: Properties and applications. arXiv 2013, arXiv:1309.2961. [Google Scholar]
- Cordeiro, G.M.; Ortega, E.M.; Silva, G.O. The Kumaraswamy modified Weibull distribution: Theory and applications. J. Stat. Comput. Simul. 2014, 84, 1387–1411. [Google Scholar] [CrossRef]
- Oluyede, B.O.; Huang, S.; Yang, T. A new class of generalized modified Weibull distributions with applications. Austrian J. Stat. 2015, 44, 45–68. [Google Scholar] [CrossRef]
- Mdlongwa, P.; Oluyede, B.O.; Amey, A.; Huang, S. The Burr XII modified Weibull distribution: Model, Properties and Applications. Electron. J. Appl. Stat. Anal. 2017, 10, 118–145. [Google Scholar]
- Oluyede, B.O.; Bindele, H.F.; Makubate, B.; Huang, S. Anew generalized log-logistic and modified Weibull distribution with applications. Int. J. Stat. Probab. 2018, 7, 72–93. [Google Scholar] [CrossRef][Green Version]
- Elbatal, I.; Zayad, M.; Rasekhi, M.; Afify, A.Z.; Iqbal, Z. A new extended Weibull model for lifetime data. J. Appl. Probab. 2019, 4, 57–73. [Google Scholar]
- Shahzad, M.N.; Ullah, E.; Hussanan, A. Beta Exponentiated Modified Weibull Distribution: Properties and Application. Symmetry 2019, 11, 781. [Google Scholar] [CrossRef]
- Elbatal, I.; Alotaibi, N.; Al-Dayel, I.; Shawki, A.W.; Elgarhy, M. Statistical Analysis of COVID-19 Data in Kingdom of Saudi Arabia Using: Sine Modified Weibull Model. JP J. Biostat. 2022, 20, 11–26. [Google Scholar] [CrossRef]
- Topp, C.W.; Leone, F.C. A family of J-shaped frequency functions. J. Am. Stat. Assoc. 1955, 50, 209–219. [Google Scholar] [CrossRef]
- Al-Shomrani, A.; Arif, O.; Hanif, S.; Shahbaz, M.Q.; Shawky, A. Topp–Leone Family of Distributions: SomeProperties and Application. Pak. J. Stat. Oper. Res. 2016, 12, 443. [Google Scholar] [CrossRef]
- Hassan, A.S.; Elgarhy, M.; Ahmad, Z. Type II generalized Topp-Leone family of distributions: Properties and applications. J. Data Sci. 2019, 17, 638–659. [Google Scholar] [CrossRef]
- Pešta, M.; Okhrin, O. Conditional least squares and copulae in claims reserving for a single line of business. Insur. Math. Econ. 2014, 56, 28–37. [Google Scholar] [CrossRef]
- Gijbels, I.; Omelka, M.; Pešta, M.; Veraverbeke, N. Score tests for covariate effects in conditional copulas. J. Multivar. Anal. 2017, 159, 111–133. [Google Scholar] [CrossRef]
- Maciak, M.; Pešta, M.; Peštová, B. Change point in dependent and non-stationary panels. Stat. Pap. 2020, 61, 1385–1407. [Google Scholar] [CrossRef]
- Kenney, J.F.; Keeping, E.S. Mathematics of Statistics, 3rd ed.; Van Nostrand: Princeton, NJ, USA, 1962; Part 1; pp. 101–102. [Google Scholar]
- Moors, J.J.A. A Quantile Alternative for Kurtosis. Statistician 1998, 37, 25–32. [Google Scholar] [CrossRef]
- Lai, C.; Xie, M. Stochastic Ageing and Dependence for Reliability; Springer: New York, NY, USA, 2006. [Google Scholar]
- Guess, F.; Proschan, F. Mean residual life: Theory and applications. Handb. Stat. 1988, 5, 215–224. [Google Scholar]
- Dennis, J.E.; Schnabel, R.B. Numerical Methods for Unconstrained Optimization and Nonlinear Equations; Prentice-Hall: Englewood Cliffs, NJ, USA, 1983. [Google Scholar]
- Henningsen, A.; Toomet, O. miscTools: Miscellanneous Small Tools and Utilities. R Package Version 0.6. 2010. Available online: http://cran.r-project.org/package=miscTools (accessed on 22 September 2022).
- Lawless, J.F. Statistical Model & Methods for Lifetime Data; Wiley: New York, NY, USA, 1982. [Google Scholar]
- Cheng, R.C.H.; Amin, N.A.K. Estimating parameters in continuous univariate distributions with a shifted origin. J. R. Stat. Soc. Ser. B (Methodol.) 1983, 45, 394–403. [Google Scholar] [CrossRef]
- Singh, R.K.; Singh, S.K.; Singh, U. Maximum product spacings method for the estimation of parameters of generalized inverted exponential distribution under Progressive Type II Censoring. J. Stat. Manag. Syst. 2016, 19, 219–245. [Google Scholar]
- Almetwally, E.M.; Almongy, H.M. Maximum product spacing and Bayesian method for parameter estimation for generalized power Weibull distribution under censoring scheme. J. Data Sci. 2019, 17, 407–444. [Google Scholar] [CrossRef]
- Basu, S.; Singh, S.K.; Singh, U. Estimation of Inverse Lindley Distribution Using Product of Spacings Function for Hybrid Censored Data. Methodol. Comput. Appl. Probab. 2019, 21, 1377–1394. [Google Scholar] [CrossRef]
- Almetwally, E.M.; Almongy, H.M.; ElSherpieny, E.A. Adaptive type-II progressive censoring schemes based on maximum product spacing with application of generalized Rayleigh distribution. J. Data Sci. 2019, 17, 802–831. [Google Scholar] [CrossRef]
- El-Sherpieny, E.S.A.; Almetwally, E.M.; Muhammed, H.Z. Progressive Type-II hybrid censored schemes based on maximum product spacing with application to Power Lomax distribution. Phys. A Stat. Mech. Its Appl. 2020, 553, 124251. [Google Scholar] [CrossRef]
- Alshenawy, R.; Al-Alwan, A.; Almetwally, E.M.; Afify, A.Z.; Almongy, H.M. Progressive Type-II Censoring Schemes of Extended Odd Weibull Exponential Distribution with Applications in Medicine and Engineering. Mathematics 2020, 8, 1679. [Google Scholar] [CrossRef]
- Chen, M.H.; Shao, Q.M. Monte Carlo estimation of Bayesian credible and HPD intervals. J. Comput. Graph. Stat. 1999, 8, 69–92. [Google Scholar]
- Cordeiro, G.M.; Ortega, E.M.; Nadarajah, S. The Kumaraswamy Weibull distribution with application to failure data. J. Frankl. Inst. 2010, 347, 1399–1429. [Google Scholar] [CrossRef]
- Alsuhabi, H.; Alkhairy, I.; Almetwally, E.M.; Almongy, H.M.; Gemeay, A.M.; Hafez, E.H.; Aldallal, R.A.; Sabry, M. A superior extension for the Lomax distribution with application to Covid-19 infections real data. Alex. Eng. J. 2022, 61, 11077–11090. [Google Scholar] [CrossRef]
- Tahir, M.H.; Cordeiro, G.M.; Mansoor, M.; Zubair, M. The Weibull-Lomax distribution: Properties and applications. Hacet. J. Math. Stat. 2015, 44, 455–474. [Google Scholar] [CrossRef]
- Almetwally, E.M. Marshall olkin alpha power extended Weibull distribution: Different methods of estimation based on type I and type II censoring. Gazi Univ. J. Sci. 2022, 35, 293–312. [Google Scholar]
- ElSherpieny, E.A.; Almetwally, E.M. The Exponentiated Generalized Alpha Power Family of Distribution: Properties and Applications. Pak. J. Stat. Oper. Res. 2022, 18, 349–367. [Google Scholar] [CrossRef]
- Sivakumar, D.C.U.; Kanaparthi, R.; Rao, G.S.; Kalyani, K. The Odd generalized exponential log-logistic distribution group acceptance sampling plan. Stat. Transit. New Ser. 2019, 20, 103–116. [Google Scholar] [CrossRef][Green Version]
- Ghitany, M.E.; Atieh, B.; Nadarajah, S. Lindley distribution and its application. Math. Comput. Simul. 2008, 78, 493–506. [Google Scholar] [CrossRef]
- Raqab, M.; Madi, T.; Debasis, K. Estimation of P (Y < X) for the 3-parameter generalized exponential distribution. Commun. Stat.-Theory Methods 2008, 37, 2854–2864. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).