Abstract
Protein-based studies contribute significantly to gathering functional information about biological systems; therefore, the protein–protein interaction detection task is one of the most researched topics in the biomedical literature. To this end, many state-of-the-art systems using syntactic tree kernels (TK) and deep learning have been developed. However, these models are computationally complex and have limited learning interpretability. In this paper, we introduce a linguistic-pattern-representation-based Gradient-Tree Boosting model, i.e., LpGBoost. It uses linguistic patterns to optimize and generate semantically relevant representation vectors for learning over the gradient-tree boosting. The patterns are learned via unsupervised modeling by clustering invariant semantic features. These linguistic representations are semi-interpretable with rich semantic knowledge, and owing to their shallow representation, they are also computationally less expensive. Our experiments with six protein–protein interaction (PPI) corpora demonstrate that LpGBoost outperforms the SOTA tree-kernel models, as well as the CNN-based interaction detection studies for BioInfer and AIMed corpora.
1. Introduction
In recent years, natural-language-processing techniques have been increasingly used to study bio-entity-based interactions from the literature for a wide range of tasks, such as protein–protein interaction (PPI) [1], chemical–disease relation [2], and chemical–protein relation [3]. Protein-based interactions are the pinnacle of biological mechanisms in all organisms, and therefore the protein–protein interaction is one of the most exhaustively researched topics in the biomedical field. Over the years, multiple PPI corpora have been deployed, and each one has a varied level of bio-entity information [4]; for example, BioInfer includes gene- and RNA-based protein-association information annotated as per binding, directed-edge, and negative interactions. Similarly, HPRD50 primarily comprises human protein–gene-based interactions. Therefore, the types of interacting partners and nature of interactions vary within the literature. While some instances contain one-to-one entity-pair association, e.g., “oxytocin → insulin” in “Subcutaneous injections of oxytocin increased insulin, glucagon and glucose levels significantly”, others exhibit complex pairings, with one-to-many or many-to-many associations, e.g., “UCP2 → leptin” and “UCP3 → leptin” in “UCP2, UCP3 and leptin gene expression: modulation by food restriction and leptin”.
Many of the earlier studies developed tree-kernel (TK) classifiers for PPI association-detection tasks [1,5,6]. These models have successfully obtained a state-of-the-art (SOTA) performance in PPI tasks for both cross-corpus and within-corpus settings. Recently studies based on deep-learning architectures have also generated interesting results, particularly for larger corpuses such as AIMed and BioInfer. They employ enriched embedding developed with features such as shortest dependency path (sdpCNN), continuous bag of words (CBOW) embedding (MCNN), and dependency-tree embedding (McDeepCNN) [7,8,9]. The syntactic relevant representation generated from these embeddings helps improve the performance, using convolution neural networks (CNNs). These approaches draw a contrast to the feature-engineering techniques used by TK-based PPI detection studies. In separate studies conducted by Murugesan et al. [5] and Warikoo et al. [6], enriched syntax and semantic features were used with tree kernels to achieve improved PPI detection.
In this paper, we introduce gradient-boosted learning on semantically optimized linguistic-pattern representation. The eXtreme Gradient Boosting (XGboost) is a gradient-boosting decision tree that can be used for classification or regression problems. Gradient boosting tries to correct the residuals of all the weak learners by adding a new weak learner. Eventually, multiple learners are added together for the final prediction, and the accuracy is higher than the single one. Since the effectiveness of XGBoost, it has been validated on a real-life, large-scale, and imbalanced dataset and solves many data-science problems in a fast and accurate way. Therefore, we developed our model based on XGBoost in this research. The feature-optimization and -representation studies were developed from our previous work, using unsupervised learning to extract semantically invariant linguistic features. These invariant linguistic patterns generate a pattern-based representation, which is adapted with XGBoost for gradient-boosted tree learning. The invariant lexical patterns contribute to context-based representation for each token, in contrast to long vector embeddings based on word co-distribution. We evaluated our work on five benchmark PPI corpora, i.e., HPRD50, LLL, IEPA, AIMed, and BioInfer [10], and a new PPI corpus AIdea, https://aidea-web.tw/file/d0cec130-b15d-4c4c-b7e8-bf95a0c34dd8-553658198_train_test_dataset_1___Sample_data.7z (accessed on 13 May 2022). The results from our study show improvement in interaction detection from both tree-kernel and CNN-based studies, particularly for larger corpora.
The remaining paper is organized as follows. We first present a Related Works section, followed by the Method section, which discusses the architecture of our model. Thereafter, case studies and a result analysis are given in the Results and Discussion. Finally, the study’s conclusion is given in the last section.
2. Related Works
In last the decade, many noteworthy studies have been conducted to improve relation detection in PPI datasets by experimenting with feature representation, classification strategies, and ensemble learning architectures, as shown in Table 1. A majority of these studies have been developed on supervised tree-kernel classifiers, using semantic, lexical, or dependency graph-based features. The tree-kernel-based studies exploit dependency-graph substructures (i.e., convolved sub-graphs) for classification. The tree/graph representations are often enriched with knowledge-rich feature vectors, such as lexical, syntactic, and entity co-occurrence, as implemented in studies by Airola et al. [11] and Landeghem et al. [12]. These consolidated semantic-based features perform relatively well on smaller datasets, such as HPRD50, LLL, and IEPA. However, developing selective knowledge-rich features is laborious and time-consuming. Alternatively, some studies have experimented with enriched dependency-graph structures. Erkan et al. and Satre et al. used syntactic substructures from dependency trees to develop tree-feature labels, using a dot product [13,14]. The similarity score from tree features is used with maximum-margin classifiers for binary classification. Studies by Chang et al. [15] and Warikoo et al. [6] have experimented with features such as interaction pattern trees or linguistic patterns to generate reduced dependency-tree representations for tree-kernel classifiers. Although graph/tree-kernel-based architectures can produce SOTA results, the overall model runtime is known to be computationally intensive, as discussed by Warikoo et al. [6].

Table 1.
Summary of previous PPI works.
More recently, an increasing number of PPI studies have been developed on deep-learning models, primarily using convolutional neural network (CNN) and long short-term memory (LSTM) architectures. Peng et al. [8] and Hua et al. [7] both experimented with multichannel CNN models, each using a different channel size and embedding features for a PPI detection task. While Hua et al. chose PMC, PubMed, Medline, and Wikipedia to develop the five-channel embedding layer, Peng at al. used semantic and lexical feature information, along with word embedding layers, to initialize seven-channel embedding. The use of multichannel input captures separate views of data based on each channel representation—a methodology much similar to the use of RGB channels for image processing. Yadav et al. [16] used composite embedding layers with bidirectional LSTM architecture to deliver promising results for AIMed and BioInfer corpora. Embedding for each instance was structured around the shortest-dependency path, enriched by parts-of-speech (POS) and word-position embeddings. Single-channel word-embedding deep-learning models have limited task interpretability, and they are therefore often augmented with semantic features, such as POS, position, or chunk vectors, to improve classification, using multiple data views. In addition, with the development of pre-trained language models, transformer-based BERT models have yielded promising results in the biomedical domain. Su et al. [17] and Su et al. [18] both experimented with the fine-tuning process of the BERT model. While Su et al. [17] chose LSTM on the last layer or the attention mechanism on the last layer, Su et al. [18] tried the model to summarize the outputs of the last layer, using BiLSTM and attention mechanism. Moreover, Su et al. [19] set the external knowledge base, constructing BERT with contrastive pre-training on the training. Based on the pre-trained BERT model, Warikoo et al. [20] also proposed a lexically aware transformer-based BERT model, and it explores both local and global context representations for sentence-level classification tasks. Wu et al. [21] used cross-entropy as the loss function and applied it to drop out before each fully connected layer during training.
As evident from this brief overview, representation enriched with semantic/syntactic features has been consistently used with different learning models to improve performance in bio-entity relation tasks. From the graph-based SOTA tree-kernel models to the generation of enriched embeddings for CNN, these features have been adapted with different models to incorporate syntactic and semantic information into the feature representation [5]. Semantic features have also been developed into patterns for dependency-tree pruning as part of tree-kernel studies [6,7]. However, not many studies have implemented semantic pattern representations with decision-tree classifiers [3]. In this study, we propose a linguistic-pattern-based gradient-boosted decision-tree model which employs a semantic representation for each bio-entity interaction, consolidated and optimized with linguistic patterns [6]. Traditionally, pattern/rule-based methods involve intensive feature engineering; however, our model can automatically converge shallow semantic representations into structurally invariant linguistic patterns [22]. Earlier, a study by Lung et al. [3] used features such as the number of words in a sentence, the number edges in the shortest path between interacting entities, the number of words before an entity mention, etc., as part of semantic representation with decision trees. We aimed to understand the relevance of entity-associated semantic contexts, order of context mentions, and how these features can be employed to improve the identification of bio-entity associations.
3. Materials and Methods
We developed a linguistic-pattern-based gradient-tree-boosted (LpGBoost) model for detecting bio-entity associations mentioned in the literature. Figure 1 illustrates the system architecture of the LpGBoost model. The preprocessing module reproduces the entity-interaction-based reduced representation. The pattern embedding and universal feature matrix generate scored linguistic-pattern features, corresponding to all semantic features in the pre-trained dataset. Feature optimization optimizes the candidate instances over the universal lead features, followed by PPI interaction classification over gradient-tree boosting. Each of the modules is detailed below.

Figure 1.
System architecture for LpGBoost.
3.1. Preprocessing
3.1.1. Entity-Interaction Representation
We limit the input instance size to 2n (n = sliding window size), covering prefix and suffix “n-features” around each bio-entity interaction target pair. Earlier works in PPI have also adapted reduced representation either using shortest-path dependency graphs or entity-labeled linguistic patterns [7,14]. In order to develop an even semantic representation, the original instances are first converted to Pos-Tag sequences, using Genia Tagger [23,24]. Then a sliding window is used to generate a candidate instance representation of size 4n (2n for each target-entity). The resulting reduced sequence is called the candidate sequence.
3.1.2. Semantic Feature Convolution
A sliding window is moved over candidate sequences to extract local semantic sub-sequence features. This concept of feature detection is similar to convolution in neural networks, where a window of size “n” is moved over an input tensor to maximize the optimal representation; however, here we do not use a max-pool to obtain max feature vector [25]. Multi-width variant semantic features are extracted from the candidate sequence of length “m” given by Equation (1):
where n represents sliding window size, s indicates stride size, i represents feature variant, and is the features/direction. Bidirectionally generated 2n semantic features are concatenated to generate candidate features.
3.2. Pattern Embedding and Universal Feature Matrix
3.2.1. Invariant Linguistic-Pattern Clusters
Invariant linguistic patterns (ILPs) are n-gram semantic patterns obtained from candidate sequences, which share distributional similarity based on their respective invariant scores. Each ILP represents a separate feature within ILP clusters, and all the patterns within each cluster are denoted with the same pattern representation (pattern embedding). The ILP clusters are generated from a larger pre-training corpus containing more instances with binary associations. Invariance among n-gram semantic patterns is calculated by using natural text interpretation of algebraic invariance [6]. Each n-gram semantic pattern is projected as a homogenous polynomial given in Equation (1):
where x and y represent n and n-1 length semantic components of original n-gram sequence, respectively. Coefficients evaluate joint conditional probabilities for each sequence associated with the respective components. The invariance for P(x,y) is calculated by Equation (3):
where is the invariant score of P(x,y). The distribution similarity between P(x,y) and Q(u,v) projections of lexical patterns p and q is concluded if they exhibit the following property described in Equation (4):
where Δ = 1, and W = 1; and I(P) and I(Q) represent the invariant function forms of p and q, respectively Using Equation (4), we consolidate all the n-gram semantic patterns into a reduced ILP, where each cluster holds linguistic patterns that are semantically invariant. The size of each cluster varies according to the distribution of the constituent patterns within the corpora.
3.2.2. Universal Lead Feature Matrix
ILP clusters are employed to optimize candidate features into linguistic-pattern normalized features. The pre-training corpus is used to develop universal lead features, which can be directly used to generate classification feature vectors for training/test datasets. Each pattern-normalized feature, , from 2n candidate features is scored over a softmax-weighted joint conditional probability, as given by Equations (5) and (6):
where = softmax-weighted score for frame width, k, and protein-interaction-class type, T (i.e., “1” = protein–protein interaction pair and “−1” = not a protein–protein interaction pair); = #features of frame width, k, and type T; and x = index size ∈(1, l = length of feature ). Joint conditional probability score for feature is weighted by category class softmax score for lexical patterns of frame width, k. This normalized weighing over the variable feature-size class adds differential representation among local and global features. Thus, each lead feature is scored to generate a universal lead feature matrix.
3.3. Feature Optimization
The candidate features from training/test datasets are optimized over the universal lead feature matrix to obtain the classification feature vector. The frame width, k, of each target feature, , from 2n candidate features, is optimized over , from the lead matrix. We perform a two-tiered optimization, where and are first compared with their respective invariant function scores (3), and similarity is calculated as Equation (7):
where = invariant feature score at index i, = invariant lead feature score at index i, and n = length of each n-gram linguistic pattern. In the event that the does not match any lead feature, then a second-tier comparison is scored, as given in Equation (8):
where = index-wise pos-tag feature similarity between and . Among all optimized values, the one with maximum invariance to is adapted. Corresponding lead matrix scores are substituted to develop training/test classification feature vectors, as shown in Equation (9):
where = substituted matrix score for , and #T = category-type size, i.e., −1 and 1.
3.4. Gradient-Tree Boosting
The gradient-tree-boosting method produces a prediction model based on an ensemble of weak decision-tree prediction models. Each successive model is built on greedily minimizing loss from the previous model learned over a span of several iterations. We used XGBoost implementation of gradient-tree boosting for our study. XGBoost is scalable and works well with sparse data features; it is also computationally less intensive [26]. Therefore, we adapted it to model classification with our sparse linguistic-feature representation.
4. Experimental Results and Discussion
4.1. Dataset
We evaluated our model by using five PPI benchmark corpora for PPI detection, i.e., HPDR50, LLL, IEPA, AIMed, and BioInfer, as shown in Table 2, and the data distribution is illustrated in Figure 2. AIMed has 200 PubMed abstracts identified by the Database of Interacting Proteins (DIP) to have PPI-specific content in which the interactions were annotated manually, in addition to 25 abstracts without PPI-specific content. BioInfer has the maximum number of instances among the five corpora, with 1100 sentences, as it contains annotations not only for PPI but also for other types of events. This is achieved through retaining sentences with more than one pair of interacting entities after querying the PubMed retrieval system with extracted interacting entities in pairs from the DIP and keeping a random subset of sentences annotated for entities of protein, gene, and RNA relationships, too. IEPA comprises 486 sentences with a specific pair of co-occurring chemicals from PubMed abstracts in which the interactions were annotated, and the majority of the entities was proteins. HPRD50 contained 145 sentences with annotations and lists of positive/negative PPI and was constructed by taking 50 random abstracts referenced by the Human Protein Reference Database (HPRD) [27], in which human proteins and genes were identified by the ProMiner software, while direct physical interactions, regulatory relations, and modifications were annotated by experts. The LLL corpus was originally created for the Learning Language in Logic 2005 (LLL05) challenge, a task focusing on the extraction of protein/gene interactions from biology abstracts in the Medline bibliography database. It contains three types of Bacillus subtilis gene interactions and has only 77 sentences, making it the smallest dataset among the five corpora. In addition, we also evaluated our system using AIdea, a new corpus for PPI detection tasks that was developed from a collection of PubMed abstracts.

Table 2.
PPI corpora statistics.

Figure 2.
Distribution of PPI corpora used for performance evaluation.
4.2. Experimental Setup
In this research, we followed an evaluation setup adapted from the previous PPI studies, using a 10-fold cross-validation (CV) for each PPI corpora and cross-corpora (CC) studies between two large corpora, i.e., AIMed and BioInfer [28]. For the CC setup, each data set was used for training while other was swapped for testing. Common evaluation metrics, i.e., precision (P), recall (R), and F1-score (F1) scores, were used to evaluate the performance of our model [29]. In addition, feature importance was also studied by using XGBoost library. For our experiments, we employed Genia Tagger [23,24] for POS normalization of raw instances, as described in Section 3.1.1. The sliding window size n = 3 was set to extract n-gram subsequence semantic patterns from candidate instances. Experiments were also conducted with n = 4, 5, and 6, but n = 3 was finally used owing to its better performance in previous studies [6]. In Section 3.1.2, n = 3 and s = 1 were set for semantic feature convolution. Paired bio-entity mentions were normalized as “TRIGGERPRI” [6,28]. DrugBank corpus [30] was used as a pre-training dataset to develop universal lead features. Ten-fold cross-validation was performed on the individual PPI corpora. The model uses binary logistic objective with XGBoost, https://xgboost.readthedocs.io/en/latest/python/python_api.html#module-xgboost.plotting, learning rate = 0.1 and max tree_depth = 6. Baseline experiments were developed for AIMed, BioInfer and AIdea data sets only.
4.3. Results and Discussion
In order to understand the significance of use of linguistic features with a decision-tree classifier, we studied the performance of XGBoost with other known lexical/semantic features, as shown in Table 3; we also visualized their performance in Figure 3. The comparative analysis of the HPRD50 corpus indicates that the use of linguistic features improves decision-tree-based classification by approximately 29%. Table 3 illustrates a comparative analysis of the results from our experiments. We developed a Naïve Bayes (NB) baseline and included SOTA models from PPI studies based on tree kernel and deep learning for this analysis. For an effective comparison, we limited our analysis to the studies that explore semantic features for input representation. Among the tree-kernel studies, graph-based feature kernel by Airola et al. [11] and Murugesan et al. [5] and linguistic-pattern-tree representations by Chang et al. [14] and Warikoo et al. [6] were referenced to draw a semantic-feature-based comparative analysis. The analysis shows a 3.64% improvement in the F1-score when compared with the highest tree-kernel performer, i.e., DSTK [5].

Table 3.
XGBoost-based comparative feature analysis on HPRD50 corpus.
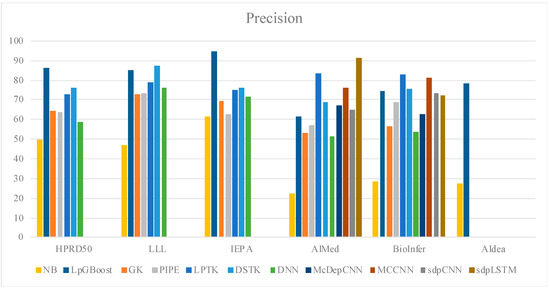
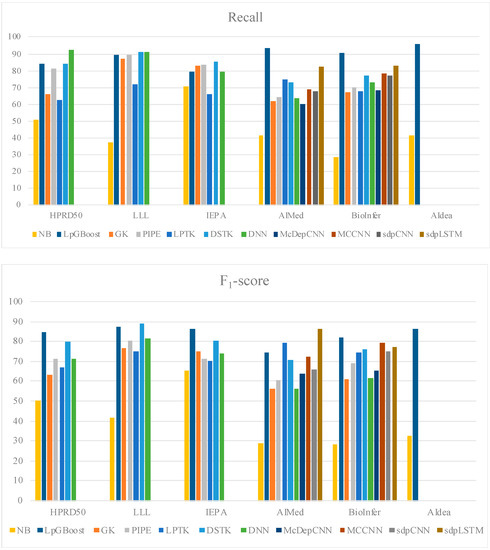
Figure 3.
Comparative analysis on precision, recall, and F1-score.
We also compared the performance of our model with semantically enriched embeddings used with deep-learning models. Both Hua et al. [7] and Peng et al. [8] used semantically enriched embeddings with CNN. Quan et al. [9] and Yadav et al. [21] studied CNN and Bi-LSTM architecture, using selective dependency graph features with embedding input. LpGBoost shows 2% and 2.4% improvement in the F1-score for AIMed and BioInfer, respectively, when compared with the MCCNN, the highest performing CNN-based model. We also developed a cross-corpus study to further examine the adaptability of our linguistic-feature representation into other cross-domain entity-association studies. As shown in Table 4 we compared the best cross-corpus reported performances from each tree-kernel and deep-learning-based study.

Table 4.
Cross-corpus evaluation of PPI datasets.
Our discussion primarily focuses on results from AIMed and BioInfer, as these have been extensively studied in several related works. The comparative analysis of the SOTA tree-kernel model (Table 5) shows a 4.55% increase in F1-average. Our approach outperforms semantically enriched CNN models; however, we do observe a 3.65% drop in F1-average when compared to sdpLSTM. We also developed a new baseline for AIdea corpus, where we achieved 86.2% on the F1-score. The performance showcases a 53.4% increase when compared to Naïve Bayes. LpGBoost exploits sparse semantic patterns (linguistic patterns) to develop decision features for gradient-boosted tree. The original instance is reduced to an entity-pair selective sematic representation; for example, “Activation of lacZ upon interaction of p85 with IR beta (delta C-43) was 4-fold less as compared to IR beta” is reduced to {[IN, NN, IN] → [NN, IN, TRIGGERPRI] → [PROTEINT, IN, TRIGGERPRI] → [PROTEINT, VBD, RB]} representation. Derived from it, the candidate features are optimized over the lead feature matrix to generate the classification vector. Table 6 shows the candidate features for the given example.

Table 5.
Ten-fold CV comparative performance evaluation on PPI corpora. A one-tailed t-test was applied to determine whether our proposed model (LpGBoost) significantly improves the performance of the F1-score of the comparisons, where * represents t-tests with alpha = 0.05.

Table 6.
Candidate feature representation developed for classification vector.
The above features combine aspects of both convolved and sequential representations, making it a robust descriptor for a tree-based decision model. The results from our XGBoost-based feature-comparison study (Table 3) also highlight that the sparse semantically relevant representation performs better with gradient-boosted trees in comparison to elaborate vectors based on term frequency and hashing. A detailed analysis of the candidate features further revealed the consistency of features f1 and f4 in improving tree-based decision-making among all the test corpora. Moreover, f1 and f4 represent sematic patterns immediately adjacent to the target-entity mention, which indicates that shorter sematic-context-based features are better descriptors in interaction detection studies over longer sequences. The results shown in Table 5 show that our model performs better than deep learning by 1.9% when studied for BioInfer, while the F1-score stagnates at 43.5% with AIMed testing. Our analysis suggests that selective labeling from interaction pattern trees used in PIPE can help improve the cross-corpus learning and general adaptability of linguistic pattern representation.
5. Conclusions
In this paper, we introduced a linguistic-pattern-representation-based boosted decision tree model to study interaction detection. The results from our extensive tests on six PPI corpora show that LpGBoost can improve prediction performance in comparison to SOTA tree-kernel models for protein interaction detection. Our model also outperformed semantically enriched CNN systems. As a part of this study, we developed linguistic-pattern-based Universal Lead Features, which were used for feature optimization in all corpora. These universal features are semi-interpretable, adaptable with other models, and can be used in cross-domain classification studies. Our study also discovered the relative significance of positional sematic features in interaction detection studies. Semantic patterns proximal to the target terms are effective in PPI detection tasks.
In the future, we would like to test the viability of our learning model on other known interaction classification tasks in the biomedical literature, such as drug–drug interaction (DDI) and chemical–protein relation (CPR) [10,21]. In addition, we also plan to conduct feature-enrichment studies by using additional information from dependency graphs to improve the cross-corpus testing and bring further interpretability to representations.
Author Contributions
Conceptualization, N.W. and Y.-C.C.; methodology, N.W. and Y.-C.C.; validation, N.W.; formal analysis, N.W.; investigation, N.W. and Y.-C.C.; resources, Y.-C.C. and S.-P.M.; writing—original draft preparation N.W. and Y.-C.C.; writing—review and editing, Y.-C.C. and S.-P.M.; visualization, N.W. and Y.-C.C.; supervision, Y.-C.C. and S.-P.M.; project administration, Y.-C.C. and S.-P.M.; funding acquisition, Y.-C.C. and S.-P.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Science and Technology Council of Taiwan, under grant NSTC 111-2221-E-038-025, and the University System of Taipei Joint Research Program under grant USTP-NTOU-TMU-109-03.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Krallinger, M.; Vazquez, M.; Leitner, F.; Salgado, D.; Chatr-aryamontri, A.; Winter, A.; Valencia, A. The Protein-Protein Interaction tasks of BioCreative III: Classification/ranking of articles and linking bio-ontology concepts to full text. BMC Bioinformatics 2011, 12 (Suppl. 8), S3. [Google Scholar] [CrossRef]
- Krallinger, M.; Rabal, O.; Akhondi, S.A.; Perez, M.P.; Santamaria, J.; Rodriguez, G.P.; Tsatsaronis, G.; Intxaurrondo, A.; Lopez, J.A.; Nandal, U.; et al. Overview of the BioCreative VI chemical-protein interaction Track. In Proceedings of the 2017 BioCreative VI Workshop, Bethesda, MD, USA, 18–20 October 2017. [Google Scholar]
- Lung, P.Y.; He, Z.; Zhao, T.; Yu, D.; Zhang, J. Extracting chemical–protein interactions from literature using sentence structure analysis and feature engineering. Database 2019, 2019, bay138. [Google Scholar] [CrossRef] [PubMed]
- Pyysalo, S.; Airola, A.; Heimonen, J.; Björne, J.; Ginter, F.; Salakoski, T. Comparative analysis of five protein-protein interaction corpora. BMC Bioinform. 2008, 9, S6. [Google Scholar] [CrossRef] [PubMed]
- Murugesan, G.; Abdulkadhar, S.; Natarajan, J. Distributed smoothed tree kernel for protein-protein interaction extraction from the biomedical literature. PLoS ONE 2017, 12, e0187379. [Google Scholar] [CrossRef] [PubMed]
- Warikoo, N.; Chang, Y.C.; Hsu, W.L. LPTK: A linguistic pattern-aware dependency tree kernel approach for the BioCreative VI CHEMPROT task. Database J. Biol. Databases Curation 2018, 2018, bay108. [Google Scholar] [CrossRef] [PubMed]
- Hua, L.; Quan, C. A shortest dependency path based convolutional neural network for protein-protein relation extraction. BioMed. Res. Int. 2016, 2016, 8479587. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Lu, Z. Deep learning for extracting protein-protein interactions from biomedical literature. In Proceedings of the 2017 Workshop on Biomedical Natural Language Processing, Vancouver, BC, Canada, 4 August 2017; pp. 29–38. [Google Scholar]
- Quan, C.; Hua, L.; Sun, X.; Bai, W. Multichannel convolutional neural network for biological relation extraction. BioMed Res. Int. 2016, 2016, 1850404. [Google Scholar] [CrossRef] [PubMed]
- Stenetorp, P.; Topi, G.; Pyysalo, S.; Ohta, T.; Kim, J.D.; Tsujii, J. BioNLP Shared Task 2011: Supporting Resources, Proceedings of BioNLP Shared Task 2011 Workshop Companion Volume for Shared Task; Association for Computational Linguistics (ACL): Stroudsburg, PA, USA, 2011. [Google Scholar]
- Airola, A.; Pyysalo, S.; Björne, J. All-paths graph kernel for protein-protein interaction extraction with evaluation of cross-corpus learning. BMC Bioinform. 2008, 9, S2. [Google Scholar] [CrossRef] [PubMed]
- Landeghem, S.V.; Saeys, Y.; Peer, Y.V.; Baets, B.D. Extracting protein-protein interactions from text using rich feature vectors and feature selection. In Proceedings of the Third International Symposium on Semantic Mining in Biomedicine (SMBM) 2008, Turku, Finland, 1–3 September 2008. [Google Scholar]
- Erkan, G.; Özgür, A.; Radev, D.R. Semi-Supervised Classification for Extracting Protein Interaction Sentences using Dependency Parsing. In Proceedings of the Conference on Empirical Methods in Natural Language Processing-Conference on Computational Natural Language Learning (EMNLP-CoNLL) 2007, Prague, Czech Republic, 28–30 June 2007; pp. 228–237. [Google Scholar]
- Satre, R.; Sagae, K.; Tsujii, J. Syntactic features for protein-protein interaction extraction. BMC Bioinform. 2007, 2016, 246. [Google Scholar] [CrossRef][Green Version]
- Chang, Y.C.; Chu, C.H.; Su, Y.C.; Chen, C.C.; Hsu, W.L. PIPE: A protein–protein interaction passage extraction module for BioCreative challenge. Database J. Biol. Databases Curation 2016, 2016, baw101. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; Kumar, A.; Ekbal, A.; Saha, S.; Bhattacharyya, P. Feature Assisted bi-directional LSTM Model for Protein-Protein Interaction Identification from Biomedical Texts. arXiv 2018, arXiv:abs/1807.02162. [Google Scholar]
- Su, P.; Vijay-Shanker, K. Investigation of BERT Model on Biomedical Relation Extraction Based on Revised Fine-tuning Mechanism. In Proceedings of the 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Seoul, Korea, 16–19 December 2020; pp. 2522–2529. [Google Scholar] [CrossRef]
- Su, P.; Vijay-Shanker, K. Investigation of improving the pre-training and fine-tuning of BERT model for biomedical relation extraction. BMC Bioinform. 2022, 23, 120. [Google Scholar] [CrossRef] [PubMed]
- Su, P.; Peng, Y.; Vijay-Shanker, K. Improving BERT Model Using Contrastive Learning for Biomedical Relation Extraction. arXiv 2021, arXiv:2104.13913. [Google Scholar]
- Warikoo, N.; Chang, Y.-C.; Hsu, W.-L. LBERT: Lexically aware Transformer-based Bidirectional Encoder Representation model for learning universal bio-entity relations. Bioinformatics 2021, 37, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; He, Y. Enriching Pre-trained Language Model with Entity Information for Relation Classification. arXiv 2019, arXiv:1905.08284. [Google Scholar]
- Dickson, L.E. Mathematical Monongraphs Algebraic Invariants, No.14.; John Wiley: New York, NY, USA, 1914. [Google Scholar]
- Tsuruka, Y.; Tateishi, Y.; Kim, J.D.; Ohta, T.; McNaught, J.; Ananiadou, S.; Tsujii, J. Developing a Robust Part-of-Speech Tagger for Biomedical Text. In Advances in Informatics. PCI 2005. Lecture Notes in Computer Science; Bozanis, P., Houstis, E.N., Eds.; Springer: Berlin/Heidelberg, Germany, 2005; Volume 3746. [Google Scholar]
- Tsuruoka, Y.; Tsujii, J. Bidirectional Inference with the Easiest-First Strategy for Tagging Sequence Data. In Proceedings of the Conference on Human Language Technology and Empirical Methods in Natural Language Processing (HLT ‘05). Association for Computational Linguistics, Stroudsburg, PA, USA, 6–8 October 2005; pp. 467–474. [Google Scholar] [CrossRef]
- Collobert, R.; Weston, J.; Bottou, L.; Karlen, M.; Kavukcuoglu, M.; Kuksa, P. Natural Language Processing (Almost) from Scratch. J. Mach. Learn. Res. 2011, 12, 2493–2537. [Google Scholar]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD ‘16), San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Fundel, K.; Küffner, R.; Zimmer, R. RelEx—Relation extraction using dependency parse trees. Bioinformatics 2007, 23, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. In Proceedings of the 25th International Conference on Neural Information Processing Systems—Volume 1 (NIPS’12); Pereira, F., Burges, C.J.C., Bottou, L., Weinberger, K.Q., Eds.; Curran Associates Inc.: Red Hook, NY, USA, 2012; Volume 1, pp. 1097–1105. [Google Scholar]
- Manning, C.D.; Schutze, H. Foundations of Statistical Natural Language Processing, 1st ed.; MIT Press: Cambridge, MA, USA, 1999. [Google Scholar]
- Segura Bedmar, I.; Martínez, P.; Herrero Zazo, M. Semeval-2013 task 9: Extraction of Drug-Drug Interactions from Biomedical Texts (Ddiextraction 2013); Association for Computational Linguistics: Stroudsburg, PA, USA, 2013. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).