Abstract
Generative adversarial networks (GAN), which are fueled by deep learning, are an efficient technique for image reconstruction using under-sampled MR data. In most cases, the performance of a particular model’s reconstruction must be improved by using a substantial proportion of the training data. However, gathering tens of thousands of raw patient data for training the model in actual clinical applications is difficult because retaining k-space data is not customary in the clinical process. Therefore, it is imperative to increase the generalizability of a network that was created using a small number of samples as quickly as possible. This research explored two unique applications based on deep learning-based GAN and transfer learning. Seeing as MRI reconstruction procedures go for brain and knee imaging, the proposed method outperforms current techniques in terms of signal-to-noise ratio (PSNR) and structural similarity index (SSIM). As compared to the results of transfer learning for the brain and knee, using a smaller number of training cases produced superior results, with acceleration factor (AF) 2 (for brain PSNR (39.33); SSIM (0.97), for knee PSNR (35.48); SSIM (0.90)) and AF 4 (for brain PSNR (38.13); SSIM (0.95), for knee PSNR (33.95); SSIM (0.86)). The approach that has been described would make it easier to apply future models for MRI reconstruction without necessitating the acquisition of vast imaging datasets.
1. Introduction
Magnetic Resonance Imaging (MRI) is a non-ionizing imaging technique used in biomedical research and diagnostic medicine. A strong magnetic field and Radio Frequency (RF) pulses are the foundational elements of MRI. An image is created when antennas placed near the area of the body being examined absorb hydrogen atom radiation, which is present in abundance in all living things. Due to the greater soft-tissue contrast and non-invasive nature of MRI, it is commonly utilized to identify diseases. MRI, on the other hand, has a severe problem in that it takes a long time to acquire sufficient data in k-space. In order to address this issue, k-space imaging approaches with insufficient sampling have been proposed. Compressed sensing [1] and parallel imaging [2] are two commonly used reconstruction approaches for obtaining artifact-free images.
Numerous research organizations and well-known MRI scanner manufacturers are accelerating MRI acquisition. Hardware techniques such as several coils are utilized to sample k-space data in parallel [3]. One of the two main approaches is used in commercial MRI scanners [4] to reconstruct a picture from the coils’ under-sampled k-space data. To be more precise, aliased pictures produced by partial k-space conversion are combined into a single coherent image via the Sensitivity Encoder (SENSE) [5]. The inverse Fourier transform (IFT) is calibrated using GRAPPA [6], which uses information from signals in the complex frequency domain. These techniques are examined by [7], along with a hybrid approach that combines the advantages of SENSE with the GRAPPA method’s resilience to some flaws. Figure 1, is a summary of data from the PubMed results for “GPU reconstruction” from 2004 to 2020.
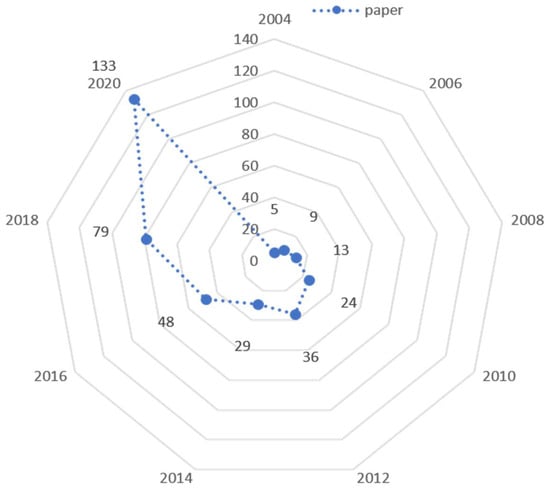
Figure 1.
Summary of data from the PubMed results for “GPU reconstruction” from 2004 to 2020. The number of papers in respective years is represented in the graph. Number of papers are 5, 9, 13, 24, 36, 29, 48, 79 and 133 in years 2004–2020.
Notable is the fact that deep learning reconstructions have notably shorter reconstruction times while maintaining higher image quality [8,9]. Using a convolutional neural network, the authors of [10] were able to determine the mapping between zero-filled (ZF) images and their corresponding fully-sampled data (CNN). Iterative processes from the ADMM algorithm were used to develop a novel deep architecture for optimizing a CS-based MRI model [11]. In [11], they recreated under-sampled 2D cardiac MR images by use of a convolutional neural network cascade. In terms of both speed and accuracy, our strategy was superior to CS approaches. De-Aliasing Generative Adversary Networks (DAGAN) was proposed by Yang et al. for fast CS-MRI reconstruction in [12]. To keep perceptual image data in the generator network, an adversarial loss was combined with a unique content loss. For MRI de-aliasing [10], created a GAN with a cyclic loss. This network’s reconstruction and refinement are carried out using cascaded residual U-Nets. In [13], the authors employed an L1/L2 norm and mixed-cost loss of Least Squares (LS) generator to train their deep residual network with skip connections as a generator for the reconstruction of high-quality MR images. A two-stage GAN technique, according to [14], can estimate missing k-space samples while also removing image artifacts. The self-attention technique was incorporated into a hierarchical deep residual convolutional neural network by [15] in order to improve the under-sampled MRI reconstruction.
Using the self-attention mechanism and the relative average discriminator (SARA-GAN), [16] constructed an artificial neural network in which half of the input data is true, and the other half is false. Research organizations and prominent MRI scanner manufacturers are working hard to speed up the acquisition of MRI scans. For example, numerous coils can be used to sample k-space data simultaneously, as demonstrated by Roemer and colleagues [17]. Under-sampled k-space data generated by the coils are used to reconstruct a picture in commercial MRI scanners [18]. Both approaches are now being applied. If the reader is interested in learning more about how the Sensitivity Encoder (SENSE) works, we suggest [19]. For example, the GeneRalized Autocalibrating Partial Parallel Acquisition (GRAPPA), developed in 2002 by Griswold et al., works on complex frequency domain signals before the IFT. For an overview of these approaches, as well as a hybrid approach that incorporates the benefits of Sense and Grappa, see [20].
The Compressed Sensing (CS) approach [21] provides efficient acquisition and reconstruction of the signal with fewer samples than the Nyquist–Shannon sampling theorem limit when a signal has a sparse representation in a specified transform domain. By selecting a tiny portion of the k-space grid, CS is employed for MRI reconstruction [22]. The IFT of the zero-filled k-space exhibits incoherent artifacts that behave like additive random noise due to the underlying premise that the under-sampling is random. CS, despite being a popular technique today, promotes smooth rebuilding, which could lead to the loss of fine, anatomically significant textural characteristics. Additionally, a sizable amount of runtime is needed. Recently, various machine learning methods for MRI acceleration were suggested. In order to reconstruct MRI from under-sampled k-space data, Ravishankar and Bressler [23] suggested a dictionary-based learning strategy that takes advantage of the sparsity of overlapping image patches highlighting local structure. Using spatio-temporal patches to reconstruct dynamic MRI [24], elaborated on this concept. Both [25,26] used compressed manifold learning based on Laplacian 75 Eigenmaps to reconstruct cardiac MRI and predict respiratory motion.
A variational network (VN) was created in 2018 by [27] to reconstruct intricate multi-channel MR data. In [28], they suggested the MoDL architecture to handle the MRI reconstruction difficulty. Meanwhile, ref. [29] created PI-CNN, which combines parallel imaging with CNN, for high-quality real-time MRI reconstruction. A method for multi-channel image reconstruction based on residual complicated convolutional neural networks was developed by [30] to expedite parallel MR imaging. Reconstructed multi-coil MR data from under-sampled data was successfully produced by [31] using a variable splitting network (VS-Net). Sensitivity encoding and generative adversarial networks (SENSE-GAN) were merged by [32] for rapid multi-channel MRI reconstruction. For the reconstruction of multi-coil MRI, ref. [33] introduced the GrappaNet architecture. The GrappaNet trained the model from beginning to end using neural networks in addition to conventional parallel imaging techniques. Dual-domain cascade U-nets were proposed by [34] for MRI reconstruction. They showed that dual-domain techniques are superior when reconstructing multi-channel data channels simultaneously. A summary of different articles regarding deep learning-based and other models for MRI reconstruction is presented in Table 1.

Table 1.
A summary of different articles regarding deep learning-based and other models for MRI reconstruction.
Training the network parameters and achieving reliable generalization results, all of the aforementioned approaches require a significant size of the dataset. On publicly accessible datasets, the majority of earlier investigations have verified their reconstruction performances. Gathering tens of thousands of multi-channel data points for model training in clinical applications is challenging, though, because retaining raw k-space data is not a common clinical flow. The generalization of learned image reconstruction networks trained on open datasets must therefore be improved. In order to address this issue, numerous transfer learning studies have been carried out lately.
To reconstruct high-quality images from under-sampled k-space data in MRI, ref. [48] created a unique deep learning approach with domain adaptability. The proposed network made use of a pre-trained network, which was then fine-tuned using a sparse set of radial MR datasets or synthetic radial MR datasets. Knoll et al. [49] investigated the effects of image content, sampling pattern, SNR, and image contrast on the generalizability of a pre-trained model in order to show the potential for transfer learning using the VN architecture. To test the ability of networks trained on normal pictures to generalize to T1- and T2-weighted brain images, ref. [50] suggested a transfer-learning approach. Meanwhile, assessed the generalization of the results of a trained U-net for the single-channel MRI reconstruction problem using MRI performed with a variety of scanners, each with a different magnetic field intensity, anatomical variations, and under-sampling masks.
This study aims to investigate the generalizability of a trained GAN model for reconstructing an MRI with insufficient samples in the following circumstances:
- ▪
- Transfer learning for a private clinical brain test dataset using the proposed GAN model.
- ▪
- Using datasets from open-source knee and private source brain tests, transfer learning of the proposed GAN model.
- ▪
- For datasets on the knee and brain with Afs of 2 and 4, transfer learning of the proposed GAN model is conducted.
2. Method and Material
The formulation of the multi-channel image reconstruction problem for parallel imaging is as follows:
is the image we intend to solve, is the under-sampling mask, is the collected k-space measurements, is the noise, is the Fourier transform and is the coil sensitivity maps.
By incorporating past knowledge, CS-MRI constricts the solution space in order to solve the inverse problem of Equation (1). Furthermore, the optimization problem can be stated as:
where the prior regularization term is denoted by , and the first term reflects data fidelity in the k-space domain, which ensures that the reconstruction results are consistent with the original under-sampled k-space data. Term is a balance parameter that establishes the trade-off between the data fidelity term and the prior knowledge. In a specific sparsity transform domain, is often an L0 or L1 norm.
Typically, an iterative strategy is necessary to tackle the above optimization issue. The regularization term), which is based on CNN, can now be used to denote, i.e.,
Utilizing the training dataset, the model’s parameters can be tuned, and the output of is with the parameters . , where H stands for the Hermitian transpose operation, and also refers to the ZF images that were reconstructed from under-sampled k-space data. Recently, MRI reconstruction has also incorporated conditional GAN.
A GAN has a discriminator D and a generator G. Both the discriminator and the generator need to be trained. The generator G can be taught, through training, to predict the distribution of the genuine data that are provided and to produce data that will deceive the discriminator D. Distinguishing between the output of the generator G and the actual data is the discriminator D’s goal. Then, after training, the generator can be used independently to generate new samples that are comparable to the original ones.
The conditional GAN loss was therefore applied to the reconstruction of MRI images, which is:
is the fully sampled ground truth, and is the equivalent reconstructed image produced by the generator. is the ZF image that serves as the generator’s input.
2.1. Datasets
The provincial institutional review committee approved the study, and all subjects provided their informed consent for inclusion prior to participating in it. The MRI scanning was authorized by the institutional review board (Miu Hospital Lahore). Private brain tumor MRI datasets were collected from 19 participants utilizing various imaging sequences. We chose 6 participants at random for network testing and 13 for tuning, which corresponded to 218 and 91 images, respectively. The “Stanford Fully Sampled 3D FSE Knees” repository provided the knee datasets used in this inquiry. The raw data were collected using an 8-coil, 3.0T full-body MR system in conjunction with a 3D TSE sequence with proton density weighting and fat saturation comparison. For network tuning, we randomly selected 18 individuals, and for testing, we randomly selected 2 subjects, which corresponded to 1800 and 200 2D images, respectively.
2.2. Model Architecture
Each of the generator networks had the same design and was based on the proposed GAN model residual of CNN. Our proposed GAN architecture, which includes a generator and discriminator, is depicted in Figure 2 in detail. Five convolutional encoding layers and five deconvolutional decoding layers made up the network, with batch normalization and leaky-ReLU activation functions following each layer. The final layer of the k-space network generator G entailed 2 output channels corresponding to real and fake components (see Figure 3).
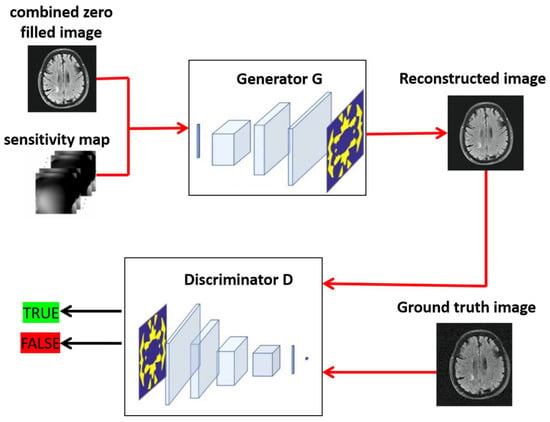
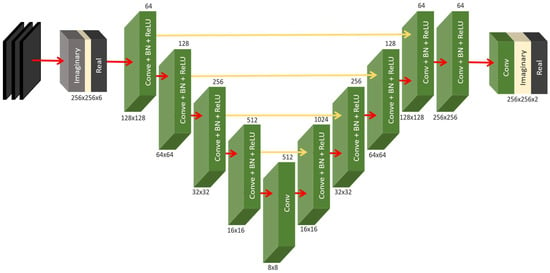
Figure 3.
Generator architecture.
The discriminator was made up of nine different blocks of convolution layers, followed by leaky-ReLU activation functions and batch normalization, and the final stage is a fully linked layer. The training was conducted using the Adam optimizer (see Figure 4). Using our proposed GAN model and transfer learning, we were able to recover the undersampled MRI data for two different circumstances, as shown in Figure 5. The dataset contains 1800 images from 18 participants and 4500 images from 45 subjects for testing. We divided the training and validation datasets during the training procedure. Eighteen photos were chosen at random for validation during each round. The models in the validation dataset with the best performance, or those with the highest PSNR, were chosen for further independent testing.
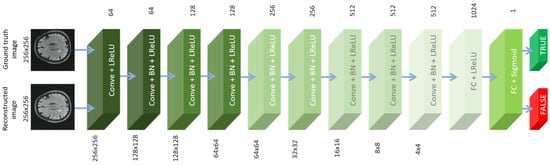
Figure 4.
Discriminator architecture.
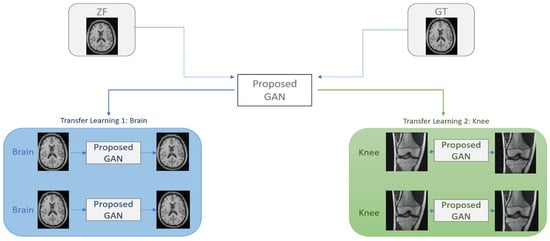
Figure 5.
Transfer learning for a GAN reconstruction model proposed for under-sampled MRI reconstruction.
Using random sampling trajectories for AF = 2 and AF = 4, retroactively, all fully sampled k-space data were undersampled. We experimented with different filter sizes, changing the filter sizes according to the pool of our data sample. Figure 3 and Figure 4 show the details of the architecture. The networks were trained using the Adam [43] optimizer and the various hyperparameters. The model was trained with an initial learning rate of 10−3, filter size 3 × 3, Xavier initialization and an 8-batch size with a monotonically decreasing learning rate over 500 epochs.
3. Results and Discussion
The experiments were carried out in Python3 with the TensorFlow backend. On a workstation equipped with an NVIDIA GV100GL graphics processor unit (GPU), the reconstruction methods were executed. PSNR and SSIM were used to evaluate the acquired reconstruction outcomes.
We experimented with several filter sizes in order to determine the best filter size, and we ultimately chose the filter size that produced the greatest results on our samples of public and private data. Three distinct filter sizes, including 3 × 3, 5 × 5 and 7 × 7, were used. In comparison to other filter sizes, the 3 × 3 filters produced greater results. The experimental findings on the private brain and public knee datasets with various filter sizes are shown in Figure 6.
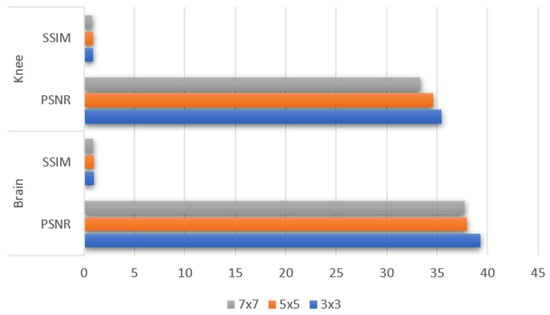
Figure 6.
The graph of the PSNR/SSIM values obtained as a result of experiments by using different filter size (3 × 3, 5 × 5 and 7 × 7) on our private brain and public dataset.
The model produced excellent reconstruction results (PSNR, 37.98; SSIM, 0.97). Then, using a test dataset made up of just a few hundred images from various domains, we applied our model to it. The test results of several brain and knee image reconstruction methods on a private and public dataset are shown in Figure 7, Figure 8, Figure 9 and Figure 10. As demonstrated by brain images, the findings of Directly Trained (PSNR, 35.78; SSIM, 0.95) were marginally superior to those of the Calgary Model (PSNR, 34.73; SSIM, 0.94) and Image Net (PSNR, 34.25, SSIM, 0.92), which had artifacts.
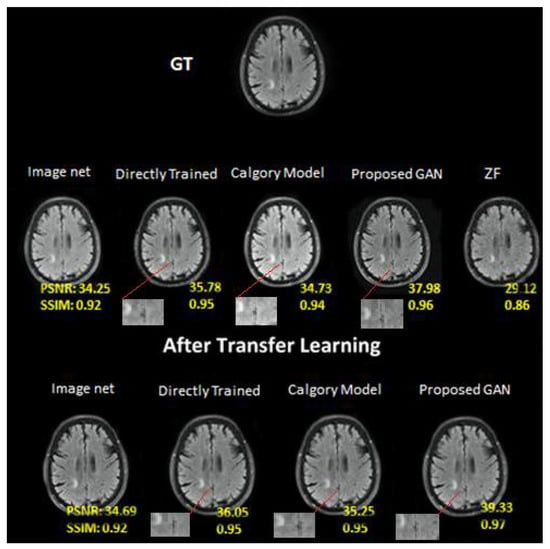
Figure 7.
At AF = 2, Reconstruction results on private brain images. From left to right: (i) Image Net (IN); (ii) Directly trained (DT); (iii) Calgary Model (CM); (iv) Proposed GAN; (v) ZF. In the second row: (i) Image net; (ii) Directly Trained; (iii) Calgary Model; (iv) Proposed GAN results of same slices from brain image dataset after applying transfer learning.
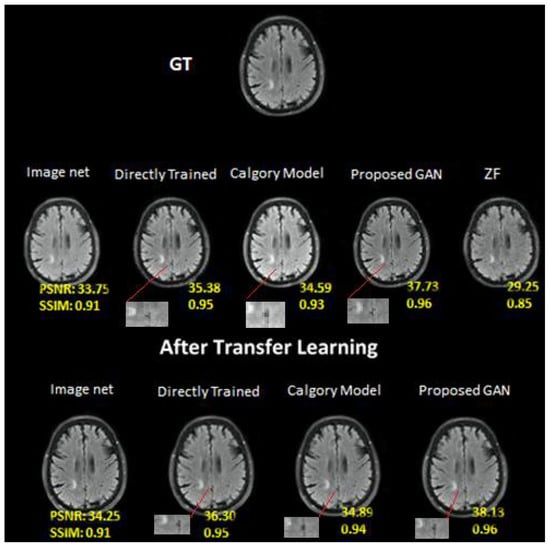
Figure 8.
At AF = 4, Reconstruction results on private brain images. From left to right: (i) Image Net; (ii) Directly trained; (iii) Calgary Model; (iv) Proposed GAN; (v) ZF. In the second row: (i) Image net; (ii) Directly Trained; (iii) Calgary Model; (iv) Proposed GAN results of same slices from brain image dataset after applying transfer learning.
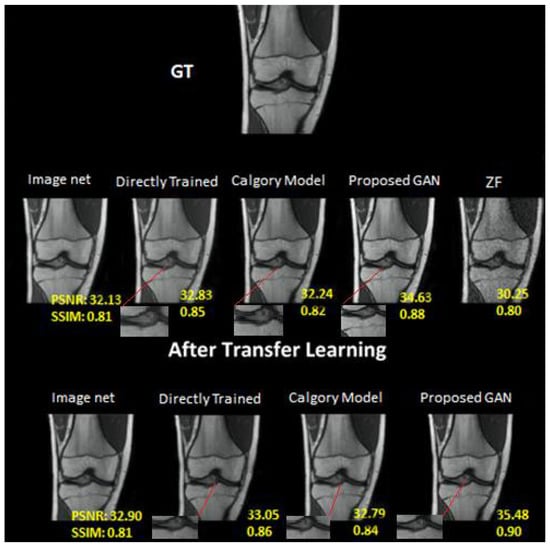
Figure 9.
At AF = 2, Reconstruction results on knee dataset. From left to right: (i) Image Net; (ii) Directly trained; (iii) Calgary Model; (iv) Proposed GAN; (v) ZF. In the second row: (i) Image net; (ii) Directly Trained; (iii) Calgary Model; (iv) Proposed GAN results of same slices from knee image dataset after applying transfer learning.
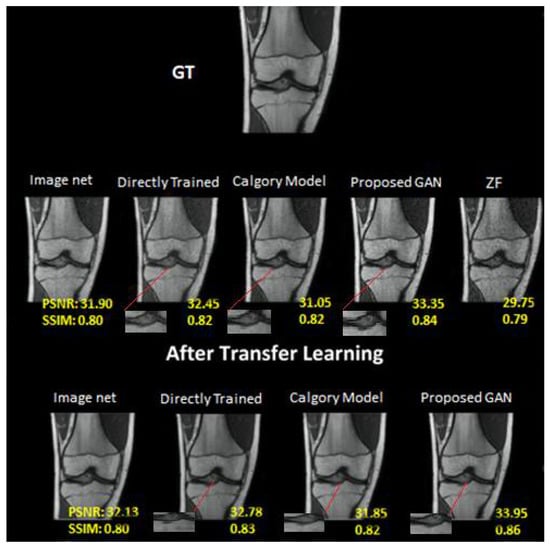
Figure 10.
At AF = 4, Reconstruction results on knee dataset. Moving left to right: (i) Image Net; (ii) Directly trained; (iii) Calgary Model; (iv) Proposed GAN; (v) ZF. In the second row: (i) Image net; (ii) Directly Trained; (iii) Calgary Model; (iv) Proposed GAN results of same slices from knee image dataset after applying transfer learning.
The outcomes of our proposed GAN model (PSNR, 37.98; SSIM, 0.97) were superior to those of the other models examined. Our proposed transfer learning model beat other reconstruction techniques (PSNR, 39.33; SSIM, 0.95). Due to a short training dataset, the DT image (PSNR = 36.05; SSIM = 0.95) produced artifacts in the associated brain images. The image net model (PSNR, 34.69; SSIM, 0.92) and CM image (PSNR, 35.25; SSIM, 0.95) had worse results than the other models.
The results of our proposed GAN model at AF 4 were 37.73 PSNR and 0.96 SSIM. Image net (PSNR, 34.25; SSIM, 0.92), CM (PSNR, 34.59; SSIM, 0.93) and DT (PSNR, 35.38; SSIM, 0.95) at AF 4. The image net model performed the worst, the category model performed marginally better and the directly trained model performed better than both. Similarly, the performance was enhanced when transfer learning was applied to the same dataset at AF 4. On the knee dataset, the performance of our proposed GAN was also better than other models. As compared to the brain image dataset, the knee dataset had the lowest accuracy of all mentioned models at AF2 and AF 4. On the knee dataset, our proposed GAN model at AF 2 was 34.63 PSNR and 0.88 SSIM. Image net (PSNR, 32.13; SSIM, 0.81), Calgary Model (PSNR, 32.24; SSIM, 0.82) and Directly Trained (PSNR, 32.83; SSIM, 0.85) at AF 2. The Calgary model outperformed the ImageNet model, whereas the directly trained model outperformed both. The performance was enhanced when transfer learning was applied to the same dataset at AF 2.
There were the following results after applying transfer learning: Directly Trained (PSNR, 33.05; SSIM, 0.86), Image net (PSNR, 32.90; SSIM, 0.81), Calgary Model (PSNR, 32.79; SSIM, 0.84) and proposed GAN model (PSNR, 35.48: SSIM, 0.90).
This study’s key contribution is the development of a transfer learning enhanced GAN technique for reconstructing numerous previously unreported multi-channel MR datasets. The findings indicate that TL from our proposed method may be able to lessen variation in image contrast, anatomy and AF between the training and test datasets. With the brain tumor dataset, reconstruction image performance was better.
This demonstrates that the best method might be to generate training and test data with the same contrasts because brain data were initially used to train the proposed model. When the distributions of the training and test datasets were similar, the reconstruction performance was good. The PSNR and SSIM of the images were significantly enhanced after applying transfer learning. This demonstrates that the extra information provided by these reconstructions makes fine-tuning more efficient when data are replicated across domains.
Table 2 shows the comparison of different models’ reconstruction results under AF = 2 for brain and knee images. The directly trained model for brain and knee images performed better than the Image Net and Calgary model. Our proposed GAN models beat all other compared methods, having PSNR (37.98) and SSIM (0.96) for brain images and PSNR (34.63) and SSIM (0.88) for knee images. All compared models perform slightly less on knee images than on brain images.

Table 2.
Quantitative analysis of PSNR and SSIM values acquired from brain and knee test images using various reconstruction techniques.
Figure 11 displays the outcomes of reconstruction for knee and brain images at an AF of 2. The x-axis shows various models, and the y-axis shows the value of PSNR. The blue color legend depicts the brain images, and the brown color legend depicts the knee. The proposed GAN model had the highest accuracy (PSNR, 37.98 and 34.63) on brain and knee images, respectively. ZF and image net had the least PSNR compared to other models. Figure 12 displays the outcomes of reconstruction for knee and brain images at an AF of 2. There are multiple model counts on the x-axis, and the SSIM value is displayed on the y-axis. The knee is represented by the brown color legend, and the brain by the blue color legend. In images of the knee and the brain, the proposed GAN model had the highest SSIM (0.96 and 0.88, respectively). When compared to other models, ZF and image net had the lowest SSIM.
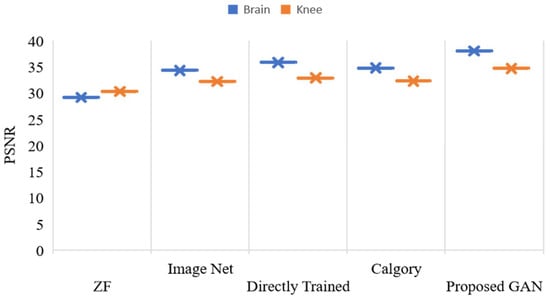
Figure 11.
PSNR, values of reconstructed images of the knee and brain using various models, with AF = 2.
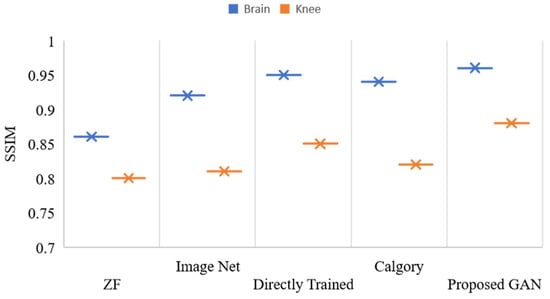
Figure 12.
SSIM, values of reconstructed images of the knee and brain using various models, with AF = 2.
Figure 13 and Figure 14 show the reconstruction results at AF 4 for brain and knee images. Our proposed GAN model showed promising results as compared to the other model. Results at AF 4, PSNR and SSIM of all compared models in Figure 13 and Figure 14 were slightly decreased. Furthermore, if we discuss the performance difference at AF 2 and AF 4, the Proposed GAN model improved PSNR by 1.06% and SSIM by 1.01% at AF 2, as compared to AF 4.
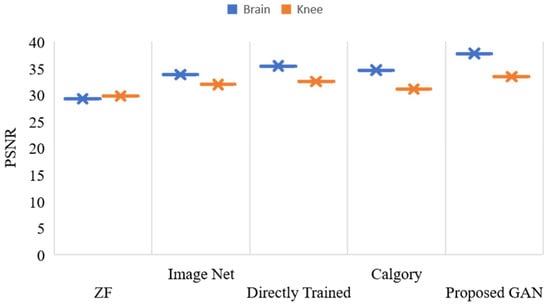
Figure 13.
PSNR, values of reconstructed images of the knee and brain using various models, with AF = 4.
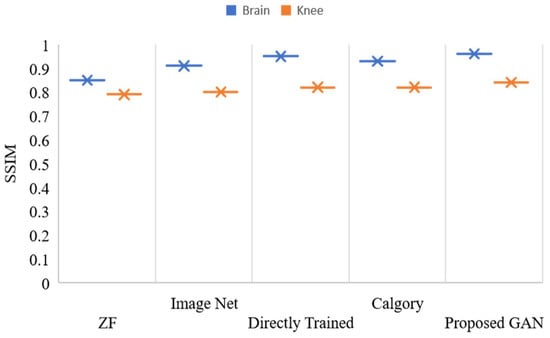
Figure 14.
SSIM, values of reconstructed images of the knee and brain using various models, with AF = 4.
After using transfer learning, Table 3 compares the reconstruction outcomes of several models for pictures of the knee and the brain with AF = 2. Our proposed GAN models beat all other compared methods by improved PSNR (39.33) and SSIM (0.97) for brain images and PSNR (35.48) and SSIM (0.90) for knee images. The directly trained model for brain and knee image performance improved by PSNR (36.05), SSIM (0.95) and PSNR (33.05), SSIM (0.86) better than the Image Net and Calgary model. All compared models’ performance increased on knee images than brain images after applying transfer learning.

Table 3.
PSNR and SSIM quantitatively evaluated values for the brain and knee test images acquired using various reconstruction models after using transfer learning.
Reconstruction results for images of the knee and brain at AF = 2 after TL are shown in Figure 15 and Figure 16. Figure 15 displays the PSNR value-based outcomes of reconstruction, while Figure 16 shows the SSIM value-based results of reconstruction after applying TL. The blue color legend depicts the brain images, and the brown color legend depicts the knee. Proposed GAN model PSNR (37.98 and 34.63), SSIM (0.97 and 0.90), on brain and knee images, respectively. Calgary model performance PSNR (35.25 and 32.79) and SSIM (0.95 and 0.84) were slightly better compared to the image net model. Directly trained model PSNR (36.05 and 33.05) and SSIM (0.95 and 0.84) results for brain and knee images, respectively. Performance increased by 3.0 % on brain images compared to knee images.
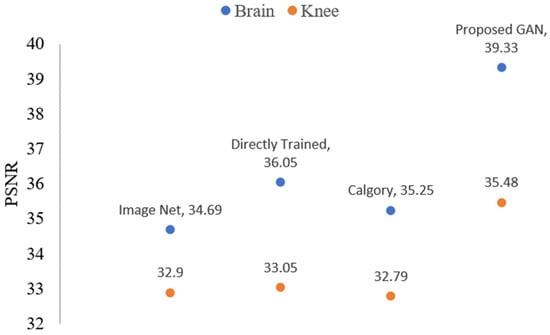
Figure 15.
The blue dots represent brain data PSNR values while orange dots represent knee data PSNR values of the reconstructed images when AF = 2 following TL.
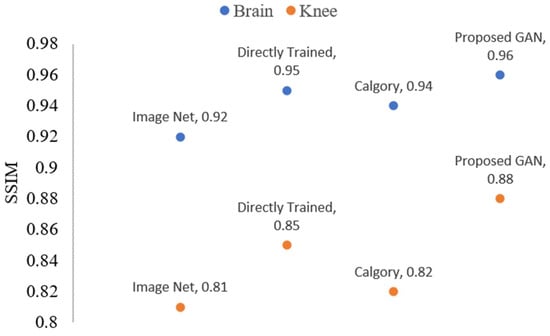
Figure 16.
The blue dots represent brain data SSIM values, while orange dots represent knee data SSIM values of the reconstructed images when AF = 2 following TL.
The reconstruction outcomes for images of the knee and brain at AF 4 are shown in Figure 17 and Figure 18. In comparison to the other model, the proposed GAN model demonstrated good results. Additionally, if we compare the performance between AF 2 and AF 4, the proposed GAN model enhanced PSNR and SSIM at AF 2 compared to AF 4 by 1.20 and 2% percent, respectively.
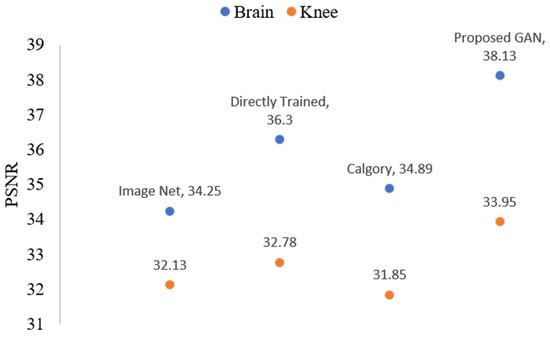
Figure 17.
The blue dots represent brain data PSNR values, while orange dots represent knee data PSNR values of the reconstructed images when AF = 4 following TL.
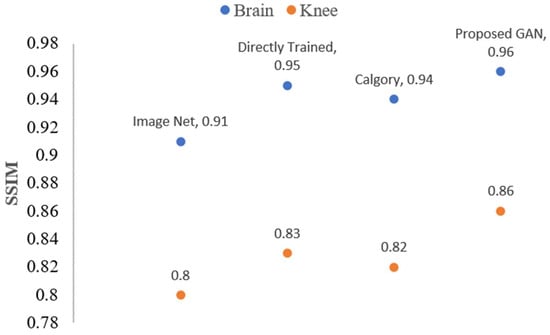
Figure 18.
The blue dots represent brain data SSIM values, while orange dots represent knee data SSIM values of the reconstructed images when AF = 4 following TL.
Our research shows that when compared to other techniques, the reconstruction’s image’s distribution produced by transfer learning is more similar to the distribution of the completely sampled image, which can help with the segmentation and diagnosis of cancerous tumors. We can also successfully use transferred learning across a range of anatomies. We discovered that brain tumor samples converged faster than knee datasets. This might be because only a few transfer learning steps were necessary to achieve the best results because the brain tumor data were located at similar anatomical locations as the training data.
As an alternative, we used a fixed training set and a range of iterations to test model-reconstructed image performance after fine-tuning. It makes sense to draw the conclusion that performance increases with dataset size. Given that there are essentially no data that can be collected, we think the current study is more realistic. The outcomes of reconstruction will be better than using a tiny portion of its own data for training as long as fine-tuning is carried out, regardless of whether the AF is more or less than its own under-sampling AF. A model with a low AF should be chosen for TL because both brain and knee data show that AF = 2 is ideal for fine-tuning. In the future, we will evaluate our transfer learning method’s reconstruction performance compared to that of existing unsupervised learning methodologies. The proposed method would facilitate the application of future MRI reconstruction models without requiring the collection of sizable imaging datasets.
4. Conclusions
This work examines the generalization capabilities of a learned proposed GAN model for under-sampled multi-channel MR images in terms of the differences across training and test datasets. Our research demonstrates that the proposed GAN model was used to analyze private brain images, knee images and images with varying AF while utilizing TL and a small tuning dataset. As compared to the results of transfer learning for the brain and knee, fewer training data being used produced superior results, with acceleration factor (AF) 2 (for brain PSNR (39.33) and SSIM (0.97); for knee PSNR (35.48) and SSIM (0.90)) and AF 4 (for brain PSNR (38.13) and SSIM (0.95); for knee PSNR (33.95) and SSIM (0.86)). The proposed method would facilitate the application of future MRI reconstruction models without requiring the collection of sizable imaging datasets.
Author Contributions
Conceptualization, M.Y. and F.J.; methodology, M.Y.; software, M.Y.; validation, F.J., M.Y. and S.A.; formal analysis, M.Y.; investigation, F.J.; resources, M.A.B.; data curation, K.A.; writing—original draft preparation, M.Y.; writing—review and editing, F.J.; visualization, M.P.A.; supervision, F.J. and M.S.Z.; project administration, F.J.; funding acquisition, F.J. All authors have read and agreed to the published version of the manuscript.
Funding
This work was sponsored by the National Science Foundation of China under Grant No. 81871394 and the Beijing Laboratory of Advanced Information Networks.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to thank sponsored organizations.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chen, Y.; Schönlieb, C.-B.; Liò, P.; Leiner, T.; Dragotti, P.L.; Wang, G.; Rueckert, D.; Firmin, D.; Yang, G.J. AI-based reconstruction for fast MRI—A systematic review and meta-analysis. Proc. IEEE 2022, 110, 224–245. [Google Scholar] [CrossRef]
- Feng, C.-M.; Yang, Z.; Chen, G.; Xu, Y.; Shao, L. Dual-octave convolution for accelerated parallel MR image reconstruction. In Proceedings of the AAAI Conference on Artificial Intelligence, Vancouver, BC, Canada, 2–9 February 2021; pp. 116–124. [Google Scholar]
- Yang, C.; Liao, X.; Wang, Y.; Zhang, M.; Liu, Q. Virtual Coil Augmentation Technology for MRI via Deep Learning. arXiv 2022, arXiv:2201.07540. [Google Scholar]
- Shan, S.; Gao, Y.; Liu, P.Z.; Whelan, B.; Sun, H.; Dong, B.; Liu, F.; Waddington, D.E.J. Distortion-Corrected Image Reconstruction with Deep Learning on an MRI-Linac. arXiv 2022, arXiv:2205.10993. [Google Scholar]
- Hollingsworth, K.G. Reducing acquisition time in clinical MRI by data undersampling and compressed sensing reconstruction. Phys. Med. Biol. 2015, 60, R297. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Kang, J.; Oh, S.-H.; Ye, D.H. Multi-Domain Neumann Network with Sensitivity Maps for Parallel MRI Reconstruction. Sensors 2022, 22, 3943. [Google Scholar] [CrossRef] [PubMed]
- Scott, A.D.; Wylezinska, M.; Birch, M.J.; Miquel, M.E. Speech MRI: Morphology and function. Phys. Med. 2014, 30, 604–618. [Google Scholar] [CrossRef] [PubMed]
- Oostveen, L.J.; Meijer, F.J.; de Lange, F.; Smit, E.J.; Pegge, S.A.; Steens, S.C.; van Amerongen, M.J.; Prokop, M.; Sechopoulos, I. Deep learning-based reconstruction may improve non-contrast cerebral CT imaging compared to other current reconstruction algorithms. Eur. Radiol. 2021, 31, 5498–5506. [Google Scholar] [CrossRef]
- Lebel, R.M. Performance characterization of a novel deep learning-based MR image reconstruction pipeline. arXiv 2020, arXiv:2008.06559. [Google Scholar]
- Lv, J.; Wang, C.; Yang, G.J.D. PIC-GAN: A parallel imaging coupled generative adversarial network for accelerated multi-channel MRI reconstruction. Diagnostics 2021, 11, 61. [Google Scholar] [CrossRef]
- Schlemper, J.; Caballero, J.; Hajnal, J.V.; Price, A.; Rueckert, D. A Deep Cascade of Convolutional Neural Networks for MR image Reconstruction. In Information Processing in Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Jiang, M.; Yuan, Z.; Yang, X.; Zhang, J.; Gong, Y.; Xia, L.; Li, T. Accelerating CS-MRI reconstruction with fine-tuning Wasserstein generative adversarial network. IEEE Access 2019, 7, 152347–152357. [Google Scholar] [CrossRef]
- Mardani, M.; Gong, E.; Cheng, J.Y.; Vasanawala, S.S.; Zaharchuk, G.; Xing, L.; Pauly, J.M. Deep generative adversarial neural networks for compressive sensing MRI. IEEE Trans. Med. Imaging 2018, 38, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Sandilya, M.; Nirmala, S.; Saikia, N. Compressed Sensing MRI Reconstruction Using Generative Adversarial Network with Rician De-noising. Appl. Magn. Reson. 2021, 52, 1635–1656. [Google Scholar] [CrossRef]
- Wu, Y.; Ma, Y.; Liu, J.; Du, J.; Xing, L. Self-attention convolutional neural network for improved MR image reconstruction. Inf. Sci. 2019, 490, 317–328. [Google Scholar] [CrossRef] [Green Version]
- Rempe, M.; Mentzel, F.; Pomykala, K.L.; Haubold, J.; Nensa, F.; Kröninger, K.; Egger, J.; Kleesiek, J. k-strip: A novel segmentation algorithm in k-space for the application of skull stripping. arXiv 2022, arXiv:2205.09706. [Google Scholar]
- Bydder, M.; Larkman, D.; Hajnal, J. Combination of signals from array coils using image-based estimation of coil sensitivity profiles. Magn. Reson. Med. 2002, 47, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Shitrit, O.; Riklin Raviv, T. Accelerated Magnetic Resonance Imaging by Adversarial Neural Network. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Berlin/Heidelberg, Germany, 2017; pp. 30–38. [Google Scholar]
- Pruessmann, K.P.; Weiger, M.; Scheidegger, M.B.; Boesiger, P. SENSE: Sensitivity encoding for fast MRI. Magn. Reson. Med. 1999, 42, 952–962. [Google Scholar] [CrossRef]
- HashemizadehKolowri, S.; Chen, R.-R.; Adluru, G.; Dean, D.C.; Wilde, E.A.; Alexander, A.L.; DiBella, E.V. Simultaneous multi-slice image reconstruction using regularized image domain split slice-GRAPPA for diffusion MRI. Med. Image Anal. 2021, 70, 102000. [Google Scholar] [CrossRef]
- Candès, E.J. Compressive sampling. In Proceedings of the International Congress of Mathematicians, Madrid, Spain, 22–30 August 2006. [Google Scholar]
- Liu, B.; Zou, Y.M.; Ying, L. SparseSENSE: Application of compressed sensing in parallel MRI. In Proceedings of the 2008 International Conference on Information Technology and Applications in Biomedicine, Shenzhen, China, 30–31 May 2008. [Google Scholar]
- Wen, B.; Ravishankar, S.; Bresler, Y. Structured overcomplete sparsifying transform learning with convergence guarantees and applications. Int. J. Comput. Vis. 2015, 114, 137–167. [Google Scholar] [CrossRef]
- Qin, C.; Schlemper, J.; Caballero, J.; Price, A.N.; Hajnal, J.V.; Rueckert, D. Convolutional recurrent neural networks for dynamic MR image reconstruction. IEEE Trans. Med. Imaging 2018, 38, 280–290. [Google Scholar] [CrossRef]
- Ruijsink, B.; Puyol-Antón, E.; Usman, M.; van Amerom, J.; Duong, P.; Forte, M.N.V.; Pushparajah, K.; Frigiola, A.; Nordsletten, D.A.; King, A.P.; et al. Semi-automatic Cardiac and Respiratory Gated MRI for Cardiac Assessment during Exercise. In Molecular Imaging, Reconstruction and Analysis of Moving Body Organs, and Stroke Imaging and Treatment; Springer: Berlin/Heidelberg, Germany, 2017; pp. 86–95. [Google Scholar]
- Bhatia, K.K.; Caballero, J.; Price, A.N.; Sun, Y.; Hajnal, J.V.; Rueckert, D. Fast reconstruction of accelerated dynamic MRI using manifold kernel regression. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 510–518. [Google Scholar]
- Hammernik, K.; Klatzer, T.; Kobler, E.; Recht, M.P.; Sodickson, D.K.; Pock, T.; Knoll, F. Learning a variational network for reconstruction of accelerated MRI data. Magn. Reson. Med. 2018, 79, 3055–3071. [Google Scholar] [CrossRef]
- Aggarwal, H.K.; Mani, M.P.; Jacob, M. MoDL: Model-based deep learning architecture for inverse problems. IEEE Trans. Med. Imaging 2018, 38, 394–405. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Han, F.; Ghodrati, V.; Gao, Y.; Yin, W.; Yang, Y.; Hu, P. Parallel imaging and convolutional neural network combined fast MR image reconstruction: Applications in low-latency accelerated real-time imaging. Med. Phys. 2019, 46, 3399–3413. [Google Scholar] [CrossRef] [PubMed]
- Du, T.; Zhang, H.; Li, Y.; Pickup, S.; Rosen, M.; Zhou, R.; Song, H.K.; Fan, Y. Adaptive convolutional neural networks for accelerating magnetic resonance imaging via k-space data interpolation. Med. Image Anal. 2021, 72, 102098. [Google Scholar] [CrossRef] [PubMed]
- Schlemper, J.; Qin, C.; Duan, J.; Summers, R.M.; Hammernik, K. Σ-net: Ensembled Iterative Deep Neural Networks for Accelerated Parallel MR Image Reconstruction. arXiv 2019, arXiv:1912.05480. [Google Scholar]
- Lv, J.; Wang, P.; Tong, X.; Wang, C. Parallel imaging with a combination of sensitivity encoding and generative adversarial networks. Quant. Imaging Med. Surg. 2020, 10, 2260. [Google Scholar] [CrossRef]
- Arvinte, M.; Vishwanath, S.; Tewfik, A.H.; Tamir, J.I. Deep J-Sense: Accelerated MRI Reconstruction via Unrolled Alternating Optimization. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021. [Google Scholar]
- Souza, R.; Bento, M.; Nogovitsyn, N.; Chung, K.J.; Loos, W.; Lebel, R.M.; Frayne, R. Dual-domain cascade of U-nets for multi-channel magnetic resonance image reconstruction. Magn. Reson. Imaging 2020, 71, 140–153. [Google Scholar] [CrossRef]
- Li, Z.; Sun, N.; Gao, H.; Qin, N.; Li, Z. Adaptive subtraction based on U-Net for removing seismic multiples. IEEE Trans. Geosci. Remote Sens. 2021, 59, 9796–9812. [Google Scholar] [CrossRef]
- Chen, Y.; Firmin, D.; Yang, G. Wavelet improved GAN for MRI reconstruction. In Medical Imaging 2021: Physics of Medical Imaging; SPIE: Bellingham, WA, USA, 2021; Volume 11595, pp. 285–295. [Google Scholar]
- Zhang, K.; Zuo, W.; Gu, S.; Zhang, L. Learning deep CNN denoiser prior for image restoration. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Kulkarni, K.; Lohit, S.; Turaga, P.; Kerviche, R.; Ashok, A. Reconnet: Non-iterative reconstruction of images from compressively sensed measurements. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Jin, K.H.; McCann, M.T.; Froustey, E.; Unser, M. Deep convolutional neural network for inverse problems in imaging. IEEE Trans. Image Process. 2017, 26, 4509–4522. [Google Scholar] [CrossRef]
- Song, P.; Weizman, L.; Mota, J.F.; Eldar, Y.C.; Rodrigues, M.R.D. Coupled dictionary learning for multi-contrast MRI reconstruction. IEEE Trans. Med. Imaging 2019, 39, 621–633. [Google Scholar] [CrossRef]
- Adler, J.; Öktem, O. Learned primal-dual reconstruction. IEEE Trans. Med. Imaging 2018, 37, 1322–1332. [Google Scholar] [CrossRef]
- Putzky, P.; Welling, M. Recurrent inference machines for solving inverse problems. arXiv 2017, arXiv:1706.04008. [Google Scholar]
- Sajjad, M.; Khan, S.; Muhammad, K.; Wu, W.; Ullah, A.; Baik, S.W. Multi-grade brain tumor classification using deep CNN with extensive data augmentation. J. Comput. Sci. 2019, 30, 174–182. [Google Scholar] [CrossRef]
- Afshar, P.; Mohammadi, A.; Plataniotis, K.N. Brain tumor type classification via capsule networks. In Proceedings of the 2018 25th IEEE International Conference on Image Processing (ICIP), Athens, Greece, 7–10 October 2018. [Google Scholar]
- Zhang, J.; Xie, Y.; Wu, Q.; Xia, Y. Medical image classification using synergic deep learning. Med. Image Anal. 2019, 54, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Isensee, F.; Kickingereder, P.; Wick, W.; Bendszus, M.; Maier-Hein, K.H. Brain tumor segmentation and radiomics survival prediction: Contribution to the brats 2017 challenge. In Proceedings of the International MICCAI Brainlesion Workshop, Quebec City, QC, Canada, 14 September 2017. [Google Scholar]
- Khan, H.; Shah, P.M.; Shah, M.A.; ul Islam, S.; Rodrigues, J.J.P.C. Cascading handcrafted features and Convolutional Neural Network for IoT-enabled brain tumor segmentation. Comput. Commun. 2020, 153, 196–207. [Google Scholar] [CrossRef]
- Han, Y.; Yoo, J.; Kim, H.H.; Shin, H.J.; Sung, K.; Ye, J.C. Deep learning with domain adaptation for accelerated projection-reconstruction MR. Magn. Reson. Med. 2018, 80, 1189–1205. [Google Scholar] [CrossRef] [PubMed]
- Healy, J.J.; Curran, K.M.; Serifovic Trbalic, A. Deep Learning for Magnetic Resonance Images of Gliomas. In Deep Learning for Cancer Diagnosis; Springer: Berlin/Heidelberg, Germany, 2021; pp. 269–300. [Google Scholar]
- Shabbir, A.; Ali, N.; Ahmed, J.; Zafar, B.; Rasheed, A.; Sajid, M.; Ahmed, A.; Dar, S.H. Satellite and scene image classification based on transfer learning and fine tuning of ResNet50. Math. Probl. Eng. 2021, 2021, 5843816. [Google Scholar] [CrossRef]
- Waddington, D.E.; Hindley, N.; Koonjoo, N.; Chiu, C.; Reynolds, T.; Liu, P.Z.; Zhu, B.; Bhutto, D.; Paganelli, C.; Keall, P.J.J.a.p.a. On Real-time Image Reconstruction with Neural Networks for MRI-guided Radiotherapy. arXiv 2022, arXiv:2202.05267. [Google Scholar]
- Guo, P.; Wang, P.; Zhou, J.; Jiang, S.; Patel, V.M. Multi-institutional collaborations for improving deep learning-based magnetic resonance image reconstruction using federated learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Montreal, QC, Canada, 11–17 October 2021. [Google Scholar]
- Yiasemis, G.; Sonke, J.-J.; Sánchez, C.; Teuwen, J. Recurrent Variational Network: A Deep Learning Inverse Problem Solver applied to the task of Accelerated MRI Reconstruction. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 19–24 June 2022. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).