Abstract
Exosomal nanoparticles (exosomes or nanovesicles) are biogenic membrane vesicles secreted by various cell types and represent a conservative mechanism of intercellular and interspecies communication in pro- and eukaryotic organisms. By transporting specific proteins, nucleic acids, and low molecular weight metabolites, the exosomes are involved in the regulation of developmental processes, activation of the immune system, and the development of a protective response to stress. Recently, the plant nanovesicles, due to an economical and affordable source of their production, have attracted a lot of attention in the biomedical field. Being a natural transport system, the plant exosomes represent a promising platform in biomedicine for the delivery of molecules of both endogenous and exogenous origin. This review presents current data on the biogenesis of plant exosomes and their composition, as well as mechanisms of their loading with various therapeutic compounds, which are determining factors for their possible practical use. We believe that further research in this area will significantly expand the potential of targeted therapy, particularly targeted gene regulation via the small RNAs, due to the use of plant exosomes in clinical practice.
1. Introduction
The discovery and development of new effective pharmaceuticals are considered as priority outcomes of biomedical science, aimed to improve the quality of human life. In recent years, significant advances have been made in this field, which have led to the development of many new drugs, including low-molecular-weight compounds of natural and synthetic origin, therapeutic proteins, and nucleic acids [1,2,3,4]. However, an important issue of therapy is the delivery effectiveness of these molecules to target cells or tissues. The targeted delivery makes it possible to create the required dose of an effector in the right location, protects it from degradation, reduces the toxic impact, and makes the therapy process more economical [5,6].
Many materials and approaches for delivery have been developed to date by the means of inorganic and organic platforms. Inorganic carriers include various nanoparticles, mainly based on metals and silica, as well as various combinations thereof [7,8]. The particles are functionalized with active groups, such as amine, carboxyl, and thiol, that allow them to chemically bind bioactive substances [9]. In particular, the mesoporous silica nanoparticles attract a lot of attention as promising carriers due to their high surface area and pore volume [10]. The carbon-based nanomaterials, especially graphene and multi-walled carbon nanotubes, also have a great potential for encapsulation and controlled release of bioactive compounds [11]. Still, limitations, such as particle aggregation in biological fluids and high cost of production, may restrict their use in practical applications.
Organic nanocarriers based on proteins (e.g., zein, gliadin, albumin) and polysaccharides (e.g., chitosan, alginate, cellulose) are promising candidates for targeted drug delivery [12]. The natural biopolymers used for their fabrication ensure the improved biological compatibility of such platforms, along with other advantages of nanosized materials. However, the low stability may limit their medical application in particular cases. Liposomes are simple vesicular structures and have been widely used as transporters for about 40 years [13]. Due to the use of physiological lipids as building blocks, liposomes have many advantages in transdermal, topical, and pulmonary applications [5]. However, the short half-life and production cost of such systems limit their therapeutic use [14]. The lipid nanoparticles represent a new delivery method, with improved properties compared to liposomes, including the ease of fabrication processes and stability [15]. They became particularly well known worldwide due to their use as the mRNA-containing vaccines for coronavirus (COVID-19) developed by Moderna and BioNTech/Pfizer. Such problems as the low drug-loading efficiency and lack of sufficient clinical observation should be solved to promote further the benefits of these carriers [16]. In addition to the above-mentioned delivery methods, the biological vectors derived from bacteriophages, mammalian, and plant viruses are also clinically used [17].
Exosomal nanoparticles from various biological objects, including mammals, plants, fungi, and bacteria have emerged as a new category of membrane vectors [17]. The practical application of plant exosomes requires a solid scientific basis and understanding of their biogenesis, molecular composition, physical characteristics, and biological properties as well as safety and effectiveness in disease treatment. Actually, thorough research in this field has only recently begun and thus many questions remain unsolved. In this review, we aimed to review critically the current achievements and challenges in terms of the possible use of plant exosomes as potential therapeutic agents and regulators of cellular networks.
2. General Characteristics
Membrane vesicles (exosomes) are biogenic nano-formations with a characteristic size of 30–200 nm, which are released from the cell by fusion of the multivesicular body with the plasma membrane [18]. Other varieties of extracellular membrane structures differ from the exosomes by their size, origin, and function. For example, the particles of 200–1000 nm, formed by budding of the plasma membrane, are related to microvesicles, while the structures of more than 1000 nm are related to apoptotic bodies, the products of the cell decay via programmed cell death [19]. The existence of nanovesicles in plants was questioned until numerous studies proving their presence in all plant organs were carried out in the last decade [20]. Morphologically, the plant nanovesicles have a rounded shape formed by a phospholipid bilayer with an average thickness of 5.3 nm [21]. The exosomal membrane protects their molecular contents from enzymatic degradation, as well as from environmental influences (e.g., high and low temperatures, extreme pH, high salinity, moisture, and sunlight) [22,23,24]. Any parts of plants can serve as a source of nanovesicles for biomedical purposes, with the most preferred being the leaves, fruits, and apoplastotic fluid. It should be noted that the different plant organs produce different amounts of nanovesicles with unique compositions and properties, which may reflect their specializations in intercellular communications [25].
3. Mechanism of Nanovesicles Formation
The mechanism of nanovesicle formation in plants is not yet completely studied, but according to the available data, it is generally similar to the biogenesis of mammalian exosomes (Figure 1). The formation of exosomes is tightly connected with the cellular membrane transport and is initiated by the formation of an early endosome through the invagination of the plasma membrane cooperating with the regulatory components of the Golgi trans-network [26]. The early endosome undergoes maturation accompanied by invagination of the endosomal membrane with the formation of intraluminal vesicles (ILVs) that selectively accumulate various intracellular biomolecules. In mammals, this process is controlled by a special endosomal sorting complex, required for their transport (ESCRT), consisting of four proteins, ESCRT-0, -I, -II, and -III, and vacuolar protein sorting 4 (VPS4) ATPase [26]. As a result of these processes, the formation of a multivesicular body (MVB) occurs, which is a late stage in the development of endosomes. Plants contain most of the ESCRT proteins, as well as VPS4/SKD1 homologs, suggesting the common functions for membrane modifications and endosomal trafficking [27]. However, a certain part of the exosomes has the potential to be formed in an ESCRT-independent way [28]. For example, the formation of some ILVs was dependent on sphingolipid ceramide produced by neutral sphingomyelinases [29]. The protein sorting in exosomes can be mediated by major histocompatibility complex class II enriched in the lipid microdomains [30]. In the case of plants, no alternative mechanisms for the exosome biogenesis are currently known. At the next stage of the biogenesis, MVB merges with the plasma membrane or with lysosomes. In the case of fusion with the plasma membrane, the nanovesicles are released into the extracellular space [31]. The fusion process is regulated itself by two protein complexes from the GTPase family Rab and soluble NSF attachment receptor (SNARE), which are localized on the outer MVB membrane and remain there after the fusion for the next MVB [32,33]. Upon the fusion with lysosomes, the enzymatic decomposition of MVB occurs [34].
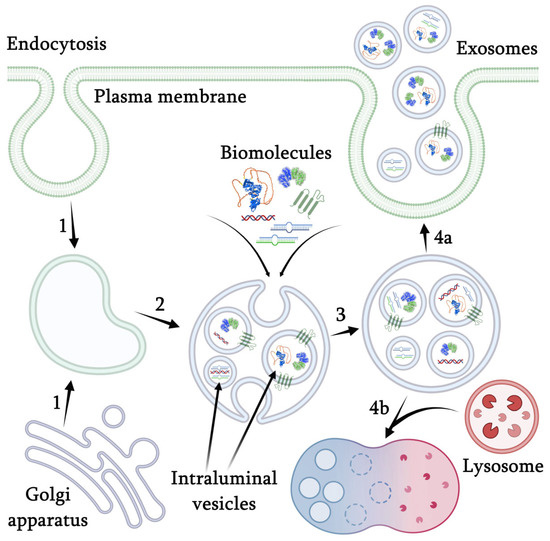
Figure 1.
Mechanism of nanovesicles formation. 1—early endosome formation through the invagination of the plasma membrane cooperating with the regulatory components of the Golgi trans-network; 2—endosome maturation accompanied by invagination of the endosomal membrane with the formation of intraluminal vesicles that selectively accumulate various intracellular biomolecules by a special endosomal sorting complex: ESCRT-0, -I, -II, -III, and vacuolar protein sorting 4 (VPS4) ATPase; 3—formation of a late endosome or multivesicular body (MBT); 4a—fusion with the plasma membrane accompanied by the release of exosomes, regulated by two protein complexes from the GTPase family Rab and soluble NSF attachment receptor; 4b—fusion with lysosome and further degradation.
4. Biochemical Characterization of Nanovesicles
The composition of nanovesicles varies depending on the source of their isolation. However, all of them contain specific proteins, messenger RNAs (mRNAs), microRNAs (miRNAs), and other small non-coding RNAs, as well as lipids and low-molecular-weight metabolites. Some established proteins and miRNAs frequently found in the exosomes are given in Table 1 and Table 2.
The nanovesicles contain two main groups of lipids: phospholipids and glycolipids, which comprise their membrane and are necessary for proper formation, release, and targeted delivery. Many nanovesicles contain phosphatidic acid that modulates membrane fission and fusion [35]. The plant nanovesicles are also enriched in transmembrane proteins, such as tetraspanins (TETs), which are an integral part of the membrane and involved in the movement and recognition of the nanovesicles. TETs are analogues of animal protein markers of exosomes (CD9, CD63, CD81) [36]. In addition, the nanovesicles contain the plant-specific secretory syntaxin called penetration 1 (PEN1 = SYP121). The plant-specific protein PEN1, an important player in plant cell wall biogenesis and modification, is required for fungal pathogen resistance [37,38]. PEN1, together with the GTPase Rab, forms complexes in the exosomes that stimulate membrane fusion and regulate their transport [39]. The vesicles often contain proteins associated with the plant defense system, namely chitinases, peroxidases, β-glucosidases, and others [40]. Interestingly, a negative regulator of the plant immunity (the RPM1-interacting protein RIN4) which inhibits the development of the cell response towards the pathogen-associated molecular patterns (PAMP) has been also found in the nanovesicles [41]. Apart from the proteins listed above, the exosomes may contain a large number of unclassified proteins that have probably a random origin. Thus, more than 200 proteins were found in the nanovesicles from the model plant Arabidopsis thaliana, but most of them were present in trace amounts [35]. It is evident that the plant species, tissue, organ, physiological state, and other factors could influence both protein composition and high the variabilities of the published data (Table 1). In addition, the method used to isolate the nanovesicles influences their contents greatly. It is known that the highly purified fractions of the exosomes from the human placenta contain a relatively small amount of the main proteins, therefore other reported samples have rather impurities associated with their preparation [42]. Considering the fact that the study of plant exosomes is still at the initial stage, the number of proteins found in them can be greatly exaggerated.

Table 1.
Protein components of plant nanovesicles.
Table 1.
Protein components of plant nanovesicles.
| Plant Species | Name of Protein | Protein Function | References |
|---|---|---|---|
| Thale cress (Arabidopsis thaliana) | ABC transporter G family member 36 (PEN3) | Defense responses, transmembrane transport | [35] |
| Thale cress (A. thaliana) Kenya violet (Craterostigma plantagineum) orange (Citrus sinensis) lemon (C. limon), grapefruit (C. paradise) bitter orange (C. aurantium) | Tetraspanin-8 (TET8) | Signaling pathway, defense response to bacterium | [35,43,44,45] |
| Thale cress (A. thaliana), Kenya violet (C. plantagineum), orange (Citrus sinensis) lemon (C. limon) grapefruit (C. paradise), bitter orange (C. aurantium) | Annexin D1 and D5 (ANN1, ANN5) | Phospholipids and calcium ions transmembrane transport | [35,43,45] |
| Thale cress (A. thaliana) | RPM1-interacting protein 4 (RIN4) | Defense responses, regulation of plant-type hypersensitive response | [35] |
| Thale cress (A. thaliana), Kenya violet (C. plantagineum), orange (Citrus sinensis) lemon (C. limon) grapefruit (C. paradise), bitter orange (C. aurantium) | Patellin 1 and 3 (PATL1, PATL3) | Cell division cycle | [35,43,45] |
| Thale cress (A. thaliana) | Syntaxin-122 (SYP122) | Defense responses, exocytosis, intracellular protein transport, vesicle fusion | [35] |
| Thale cress (A. thaliana), orange (Citrus sinensis) lemon (C. limon) grapefruit (C. paradise), bitter orange (C. aurantium) | H(+)-ATPase 1 and 10 (AHA1, AHA10) | Ion transmembrane transport, regulation of intracellular pH | [35,45] |
| Thale cress (A. thaliana), orange (Citrus sinensis), lemon (C. limon), grapefruit (C. paradise), bitter orange (C. aurantium) | Phospholipase D alpha and delta (PLDα, PLDδ) | Regulation of abscisic acid-activated signaling pathway, programmed cell death | [35,45] |
| Thale cress (A. thaliana) | GDSL esterase/lipase ESM1 (ESM1) | Glucosinolate and lipid catabolism | [35] |
| Thale cress (A. thaliana) | Protein NRT1/PTR (NPF2.10) | Glucosinolate transport | [35] |
| Thale cress (A. thaliana), Kenya violet (C. plantagineum), orange (Citrus sinensis), lemon (C. limon), grapefruit (C. paradise), bitter orange (C. aurantium) | Heat shock 70 kDa protein 3 (HSP70-3) | Chaperone cofactor-dependent protein refolding, stress responses | [35,43,45] |
| Thale cress (A. thaliana) | L-ascorbate peroxidase 1 (APX1) | Responses to oxidative stress, lignin and phenylpropanoid biosynthesis | [35] |
| Kenya violet (C. plantagineum) | Beta-galactosidase 3 (BGal3) | Carbohydrate metabolism in plants | [43] |
| Kenya violet (C. plantagineum) | Peptidylprolyl isomerase (PPIase) | Chaperone-dependent protein refolding | [43] |
| Kenya violet (C. plantagineum), orange (Citrus sinensis) lemon (C. limon) grapefruit (C. paradise), bitter orange (C. aurantium) | Coatomer protein complex (COP) subunits α1, β1, β2, γ | Membrane transport in the Golgi apparatus trans-network | [43,45] |
Moreover, the plant nanovesicles contain a significant amount of genetic material, represented mainly by small RNA patterns (Table 2). MicroRNAs (miRNAs) are of particular interest as important epigenetic regulators of cellular processes that can specifically suppress the work of genes by degradation of target mRNAs [18,46]. The miRNAs are frequently found in the extracellular fluids of plants in both freeform and protein-miRNA complexes due to the passive diffusion or active secretion [47]. The free miRNA molecules are unstable and susceptible to destructive effects, while their packaging inside the plant nanovesicles allows them to avoid these consequences and even to be transported to the external environment. Currently, several hundred representatives of the exosomal miRNAs from various plant species are known. However, the majority of these miRNAs were probably copurified with the exosomes associated with the RNA-binding proteins, and only a limited fraction were factually encapsulated inside the vesicles [48]. Thus, in Arabidopsis, only seven miRNAs, namely: miR157c, miR167a, miR168a, miR168b, miR169a, miR172e, and miR8175, were primarily accumulated in the nanovesicles, while the other 55 miRNAs were located outside [48]. This observation means that, as in the case of proteins, the content of plant exosomal miRNAs (and probably from other sources) could also be greatly overestimated. Moreover, the plant exosomes are enriched in the “tiny” RNAs of 10 to 17 nucleotides in the length, but their origin and functions still remain unknown [49].

Table 2.
Examples of miRNAs in plant nanovesicles.
Table 2.
Examples of miRNAs in plant nanovesicles.
| Plant Species | miRNA Name | miRNA Function | References |
|---|---|---|---|
| Thale cress (Arabidopsis thaliana) | miR396 | Reduces stomata density, regulates leaf and flower development | [50] |
| miR156 | Increases the accumulation of anthocyanin | [51] | |
| miR398 | Regulates the expression of superoxide dismutases | [52] | |
| Honeysuckle (Lonicera xylosteum), Thale cress (A. thaliana) | miR2911 | Suppresses some viral infections | [53] |
| Ginger (Zingiber officinale) | miR1078 | Acts on the leptin gene, which is associated with lipopolysaccharide-induced expression of IL-6 | [54] |
| miR7267 | Modulates immunity by suppression of the Lactobacillus rhamnosus monooxygenase expression in the gut microbiome | [55] | |
| Ginger (Z. officinale), grapefruit (Citrus paradisi) | miR-2911 | Suppresses influenza virus (H5N1) infection, and suppresses SARS-CoV-2 virus replication | [55] |
| Cabbage (Brassica oleracea) | miR172 | Promotes flowering by inhibiting the function of the APETALA2 family genes | [56] |
| Grapefruit (C. paradisi) | miR17 | Suppresses the expression of the histocompatibility complex, thereby inhibiting tumor growth | [57] |
| Blueberry (Vaccinium spp.)coconut (Cocos nucifera) ginger (Z. officinale), grapefruit (C. paradisi) melon (Cucumis melo) kiwi (Actinidia chinensis), orange (Citrus reticulata) pear (Pyrus communis) soybean (Glycine max) tomato (Solanum lycopersicum) | miR168 | Regulates the functions of the AGO2 protein complex | [51,58] |
| miR319 | Acts on the transcription factor TCP, which controls leaf development | [51] | |
| Soybean (G. max) ginger (Z. officinale), grapefruit (C. paradisi) tomato (S. lycopersicum) pear (Pyrus communis) | miR530 | Participates in defense reactions, circadian rhythm and secondary metabolism | [59] |
| Watermelon (Citrullus lanatus) walnut (Juglans regia) | miR156 | Regulates the mammalian TNF-α signaling pathway in adipocytes; found in human blood following oral administration | [59,60] |
The composition of secondary metabolites in the plant nanovesicles is still not well understood, but it is assumed that their presence may be related to the hydrophobicity of the vesicles themselves [61]. In dependence on the biochemical characteristics of the source, they contain the flavonoids, such as naringin and naringenin, ascorbic acid, and other compounds [62]. In addition, several proteins whose function is associated with the secondary metabolism of plants have been identified in the nanovesicles (Table 1), thus indicating that plants can modify the biosynthesis of these compounds in the target cells via exosomal signalling. The nanovesicles have been also found to contain considerable amounts of carbohydrates (glucose, fructose, sucrose, etc.) and amino acids (alanine, asparagine, isoleucine, threonine, leucine, etc.), the concentration of which also varies and depends on the source of the material [26]. It is probable that these small molecules are encapsulated into the exosomes unspecifically via passive diffusion.
5. Functions of Plant Nanovesicles
The extracellular vesicles were previously believed to be mainly needed for cellular waste removal and do not have any important function. However, recent studies have shown that exosomes mediate intercellular communication by transporting various biologically active molecules in the organism. This issue is currently actively investigated in various animal and human models. Their exosomes play important roles in the regulation of developmental processes, activation of the immune system, and protective mechanisms in response to stresses, maintaining the pluripotency of embryonic stem cells, and in many other functions [63].
The functions of nanovesicles in plants are mostly considered in terms of their protective function in the system of plant-pathogen interactions. The ability of plants to quickly respond to various pathogens is essential for their survival. Restructuring the signaling pathways, cytoskeleton, and cell wall, as well as increased synthesis of the defense compounds, lead to the formation of physical (modification of plant cell wall) and biochemical (defense-related molecules) barriers that are designed to resist infection [64]. Such changes can be carried out through the rapid and targeted delivery of the necessary molecules by vesicles. It has been shown that the fungal infection enhances the rapid accumulation of exosomes between the plasma membrane and cell wall in plant cells, indicating their important role in the immune response [41,65]. For example, the vesicular structures containing polyphenolic metabolites and hydrogen peroxide prevented infection of the barley leaves by the powdery mildew fungus Blumeria graminis [66]. During the infection of Arabidopsis by a biotrophic fungus Golovinomyces orontii, the PEN1 and PEN3 proteins were transported by the exosomes and incorporated into the cell wall, acting as a protective barrier [67]. The transfer of miRNAs from the host plant to the pathogen, causing silencing of virulence genes, has also been described in the plant pathosystems, such as cotton/Verticillium dahliae and wheat/Fusarium graminearum [57,58]. At the same time, pathogens can transport their own miRNAs in the invaded area, contributing to the suppression of the immune response and defense systems of their host plant [39]. The grain yellow rust pathogen Puccinia striiformis produces miRNA-like RNA that suppresses the expression of defense genes in wheat [68]. The transfer of specific miRNAs of Hyaloperonospora arabidopsidis and Botrytis cinerea into the cells of Arabidopsis thaliana suppresses the expression of AGO1 protein, thus disrupting the loading of the plant nanovesicles with protective miRNAs [69]. Notably, the plant pathogenic fungi also widely utilize the extracellular vesicles to transport the effectors through host barriers aiming to suppress the plant immunity [40].
6. Isolation and Purification of Plant Nanovesicles
Various parts of plants are used to isolate the nanovesicles, mainly the fruits, roots, and apoplastotic fluid from their leaves. Currently, five approaches are applicable for plant nanovesicle isolation, namely differential and gradient density ultracentrifugation, precipitation using highly hydrophilic polymers (e.g., protamine, dextran, polyethylene glycol), size exclusion chromatography, and immunoprecipitation (Figure 2), with preference of the methods based on ultracentrifugation [70]. The methods based on ultracentrifugation are preferred due to resulting in a larger amount of the material [42]. Differential ultracentrifugation includes series of centrifugation cycles with different centrifugal forces and durations, providing differences in the densities and sizes of nanovesicles and other cellular components, including organelles, microvesicles, and apoplastic bodies [71]. Typically, the biological fluids are successively centrifuged at 5000×, 10,000×, and 20,000× g, followed by the ultracentrifugation step. For ultracentrifugation, the speed in the range of 100,000× g to 120,000× g is sufficient to pellet nanovesicles [72]. However, for the study on extracellular vesicles from the apoplastic fluids of A. thaliana leaves, recovery efficiency and quality were maximally improved by the ultracentrifugation at 40,000× g that allowed them not to use the speed of 100,000× g [35]. Later, the research of Huang et al. demonstrated the opposite results for the same model plant [72]. In another study, the nanovesicles were isolated from Nicotiana tabacum L., Vinca minor L., and Viscum album L. by differential ultracentrifugation at 50,000× g (the optimal speed for the nanovesicles sedimentation), followed by agarose gel electrophoresis purification to increase their purity [73]. Additionally, the method was improved by adding a thin pad (Optiprep™) to the bottom of the centrifuge tube that prevented the damage of nanovesicles [74]. This means that the issue of the most suitable ultracentrifugation conditions is still open and they should obviously be optimized for each plant species and type of the biofluids as the starting material. Moreover, the differential ultracentrifugation method should be used in combination with the additional purification methods, since the disadvantage of this method is the contamination of the resulting vesicles with protein molecules similar in density and size to the nanovesicles [42].
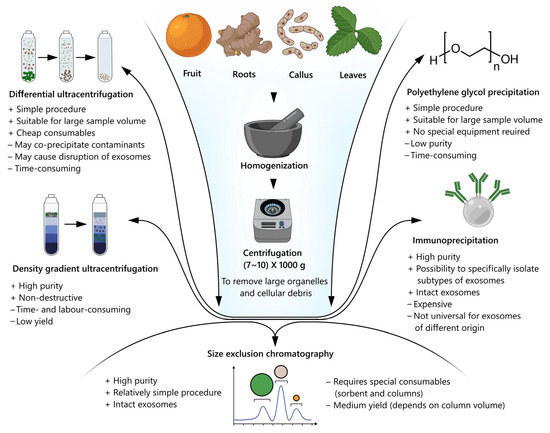
Figure 2.
Schematic flow diagram of the isolation and purification of plant nanovesicles.
Density gradient ultracentrifugation is based on the separation of nanovesicles depending on their size, mass, and density in a centrifuge tube with a pre-designed medium with a gradually decreasing gradient. The sample is subjected to a long cycle of ultracentrifugation, whereby the solutes of the sample, including the nanovesicles, move as separate zones through the medium with a density gradient to the bottom, each with its own specific sedimentation rate [71]. The samples of nanovesicles isolated by this method are much more homogeneous and of less foreign biomolecules. When both methods of ultracentrifugation are applied together, the yield of the purified nanovesicles is much higher. Thus, the ginger nanovesicles were isolated firstly by differential centrifugation and then purified in a sucrose gradient [75]. The nanovesicles from the plant Asparagus cochinchinensis were also obtained by a combination of differential and gradient ultracentrifugation methods [76]. The disadvantage of density gradient ultracentrifugation is the comparatively lower yield and contamination of the isolated nanovesicles, but their quality is higher than that of isolation by differential ultracentrifugation.
The polyethylene glycol (PEG) method is based on changing the solubility or dispersibility of nanovesicles using water-exclusive polymers, such as PEG, in the range of a molecular weight of more than 8000 [71]. This displacement agent can create a network-like structure, trapping nanovesicles before their precipitation. By adjusting the concentration of PEG from 8%, 10%, 12% to 15%, the nanoparticles of 365 nm, 304 nm, 256 nm, and 252 nm are able to be obtained, respectively, while the ultracentrifugation method results in the formation of nanoparticles of 403 nm [77]. The highest yield of nanovesicles from ginger using PEG with the different molecular weights (4000, 6000, and 8000) was observed with the use of PEG4000, with its residual concentration at the end of preparation remaining acceptable for consumption [78]. In general, this method results in high yield but low-quality isolation due to its non-specific mechanism. However, combining the PEG-based isolation with other isolation methods is an effective strategy to avoid the disadvantages associated with using PEG alone. There are many benefits of such a strategy providing the processing of multiple samples simultaneously: it is easier, faster, and is relatively low cost, without damaging the nanovesicles. However, lengthy sample preparation and purification procedures are needed, while the proper selective isolation mechanism is absent. These factors inevitably compromise the purity of the isolated nanovesicles, thereby impairing subsequent analysis [79].
Size exclusion chromatography (SEC) is based on the size separation of biomolecules. Sample components with a small hydrodynamic radius may pass through the pores, resulting in a late elution. The components with a larger hydrodynamic radius, including nanovesicles, cannot enter the pores, thereby eluting by the first washing solution volume [71]. For example, in one study, the nanovesicles were isolated from two types of cabbage using three methods, including SEC, and it was concluded that the nanovesicles isolated by SEC were the most homogeneous and purified from protein impurities, as well as retaining their biological activity [80]. This method of isolation can be used as an additional method for the purification of nanovesicles obtained by ultracentrifugation. Thus, the nanovesicles obtained from Cucumis sativus by differential ultracentrifugation was further successfully purified by SEC to remove the protein impurities [81]. The SEC isolation method has a great potential to produce highly-purified nanovesicles whose integrity and biological activity are largely preserved, probably because SEC relies on the use of gravity rather than centrifugal force reaching high values. Additionally, there are plenty of pre-packed commercial columns, specially designed for the isolation of exosomes from various biofluids. Nevertheless, its long runtime limits the SEC application in any large-scale production [79].
The method of immunoprecipitation or immunoaffinity consists of coating magnetic beads with the antibodies that target the proteins present on the surface of exosomes [71]. This method is based on the surface biomarkers that are uniquely expressed in nanovesicles, allowing the isolation of specific nanovesicles. Despite its high potential, the immunoaffinity method has not been extensively studied for the isolation of plant nanovesicles, probably due to the lack of extensive knowledge of the surface composition and antibody-antigen interactions that could be used to isolate them. However, in a study on the isolation of the nanovesicles from Arabidopsis thaliana, this method was used in conjunction with ultracentrifugation and, as a result, the nanovesicles were separated from the TET8 proteins mix by adding the appropriate antibodies, then passed through electrophoresis and isolated from the gel [82]. Although it provides high purity isolation, the overall yield of nanovesicles is reduced as only those recognized by the antibodies are captured. In addition, if the antibodies cannot be easily removed from the vesicles after the precipitation, this may disrupt their integrity. The specificity and quality of antibody is another issue that limits the use of this technique, since most antibodies commercially available for immunoprecipitation are non-specific. In general, the immunoaffinity method is one of the most expensive methods for isolating nanovesicles from a large sample volume since it requires a large number of antibody-conjugated vesicles which can limit its application. Therefore, it may only be suitable for studies with small sample volumes, which creates a barrier to any potential therapeutic use [79].
7. Characterization of Exosomes
The characterization of plant nanovesicles includes a wide spectrum of morphological, physical, and biochemical analytical approaches. One of the most common imaging techniques used is scanning and transmission electronic microscopy (SEM and TEM, respectively) [83]. SEM provides information about the three-dimensional structure of the exosome surface, however, as soon as drying is applied during sample preparation, their natural morphology could be changed, leading to the formation of cup-shaped structures [84]. In turn, TEM is a more accurate method, as nanovesicles are not deformed during processing. Moreover, staining with heavy metals, such as osmium tetroxide and uranyl acetate, creates lipid membrane contrasts allowing distinguishing exosomes from impurities [85]. Cryo-SEM, or cryomicroscopy, implies freezing and sample analysis at a very low temperature (below −100 °C) [86]. It is used to assess the morphology of nanovesicles in a state close to the native one due to their perfect preservation, as well as the absence of pre-fixation operations or the addition of heavy metals [21,83]. Atomic force microscopy can also be used as an additional indirect method for assessing the morphological and physical properties (e.g., adhesion and stiffness) of nanovesicles [87]. The method does not require extensive sample preparation: exosomes could be rapidly adsorbed and dried on glass or mica surfaces.
The hydrodynamic sizes of exosomes are evaluated by using dynamic light scattering (DLS) or nanoparticle tracking analysis (NTA). DLS measures the volumetric scattered light from nanovesicles when illuminated with a monochromatic light source. Since the particles are in Brownian motion, scattered light from all particles interferes and intensity fluctuates with time, and information about the particles is obtained from the autocorrelation of the oscillation intensity recorded during the experiment [88]. At the same time, NTA is based on the use of a concentrated beam of light to illuminate the particles in the sample. As the particles scatter light and undergo Brownian motion, the camera records the path of each individual particle to determine the average speed and size [85]. As the measurement of nanoparticles using the latter method is more accurate when analyzing polydispersed samples [89,90], NTA is now considered a gold standard for exosome characterization [91].
The identification of specific protein markers is one of the most important biochemical characteristics of exosomes. Usually, Western blotting with antibodies specific to such proteins as PEN3, TET8, and HSP70 indicates the plant-derived nanovesicle fraction [92]. However, in the case of exosomes from the non-model plants, the commercially available antibodies for Arabidopsis thaliana may have a low affinity to the proteins from evolutionarily distant species. Another versatile way to identify proteins, including marker ones, is mass spectrometry [93]. In addition to the purification, the peptide fractionation prior to mass spectrometric analysis is considered an important prerequisite for the identification of vesicular proteins with high confidence. In terms of detection sensitivity, mass spectrometry is not as sensitive as antibody-based methods, but it allows analysis of a whole spectrum of exosomal proteins at once [94]. As mentioned above, the purity of the exosomes must be checked very strictly, since the identification of many hundreds of proteins in exosomes is probably due to contamination [42]. Mass spectrometry can also be used to analyze the low molecular weight components, such as secondary metabolites, fatty acids, and sugars [95,96].
New generation sequencing (NGS) methods are used to identify the nucleic acid fractions, primarily the small RNAs [97]. Taking into account that a huge fraction of small RNAs is coprecipitated with exosomes in the complexes with proteins, protease trypsin digestion should precede RNase treatment to ensure degradation of RNAs located outside vesicles [48]. For low-scale analysis, quantitative real-time PCR, applying a stem-loop approach [98] or poly(A)-tailing [99] could be used to verify the individual miRNA representatives. In this term, an interesting question is whether there are inherent miRNAs that can be used as markers along with protein components.
8. Methods for Loading Biomolecules into Nanovesicles
For a long time, the presence in plant cells of a molecular mechanism that regulates the specific composition of the exosomes remained an unresolved issue. It was found that the profile of miRNAs in the nanovesicles is strikingly different from the profile of all miRNAs in the cells [44], indicating the selective nature of their transport. It turned out that the TET proteins fraction, associated with nanovesicles, contains several RNA-binding proteins: argonaute (AGO1), helicases, and annexins. Moreover, AGO1 is the only representative of this family of proteins that is secreted by nanovesicles and binds precisely to the exosomal, rather than cellular, miRNAs [82]. On the contrary, the annexins demonstrate a nonspecific binding to miRNAs, but play an important role in their stabilization in the exosomes. The ESCRT complex is simultaneously involved in the loading of intraluminal vesicles with both ubiquitinylated proteins, which are further broken down by lysosomes (Figure 1, 4b), and those secreted as a part of the exosomes (Figure 1, 4a). However, the exact mechanism of protein selection for one or another pathway still remains not fully understood [27]. An ESCRT-independent transport pathway has so far been described for a relatively small number of proteins and is likely to be involved in certain highly specialized processes [31] (Figure 3).
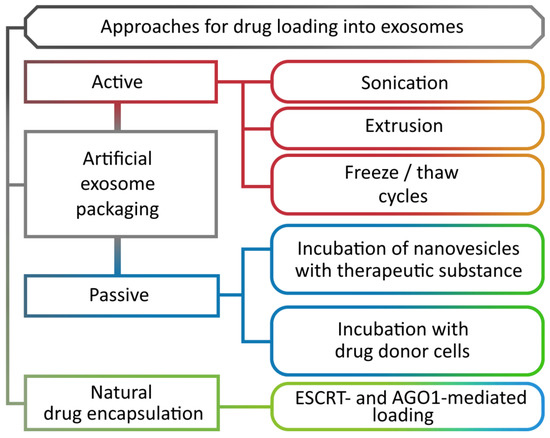
Figure 3.
Methods for loading biomolecules into nanovesicles.
Currently, the largest number of studies is devoted to the use of native exosomes containing such biomolecules as proteins, miRNAs, and secondary metabolites inherent in certain plant species [62]. At the same time, the therapeutic potential of native nanovesicles is determined mainly by the plant species used for the isolation [26]. It should be noted that there is no information in the literature about any adverse reaction on the part of the recipient organism induced by the introduction of plant nanovesicles that indicates their safety and biocompatibility.
The composition of exosomes can also be artificially altered by introducing both high- and low-molecular-weight target molecules (Figure 3). There are two main methods of loading nanovesicles: active and passive. Active loading includes such methods as sonication, extrusion, and freeze and thaw cycles that temporarily disrupt the nanovesicle membrane, allowing various compounds to diffuse inwards, after which the membrane is repaired [100]. Passive loading is an incubation process during which the loading takes place. The incubation is carried out by two methods: incubation of nanovesicles with a compound [101], and incubation with donor cells of target molecules [102].
9. Routes of Exosome Administration
Plant nanovesicles have a number of advantages in the delivery of substances in comparison with existing analogues. Firstly, their small size, negative charge, affinity for the plasma membrane, and pronounced physicochemical stability at various pH values and temperatures allow the nanovesicles to penetrate effectively the target cells. At the same time, the phospholipid bilayer protects the content of nanovesicles from enzymatic degradation by proteinases and nucleases. Secondly, the nanovesicles have a high potential for targeted delivery of therapeutic agents, which reduces the probability of adverse reactions in clinical use. Thirdly, the nanovesicles successfully diffuse through the blood-brain barrier (BBB), avoiding an inflammatory response, unlike artificially made liposomes and animal exosomes [82]. Currently, the main obstacles for the use of plant nanovesicles are insufficient standardization levels of respective technologies and limited practical experience. Particularly, there is no universally recommended isolation protocol to date, while the existing ones still have their own drawbacks, such as high cost, the possibility of introducing contaminants, and the instability of exosomes during processing [103]. There is still a need for further development of the methods for precise quantification of the molecular and biochemical parameters of exosomes. The main disadvantage for promoting exosomes as therapeutics is the lack of a way to obtain a drug with a single potency, or at least containing a specific set of bioactive molecules. Effective technologies for obtaining exosomes in the amounts sufficient for a therapeutic application on humans are yet to be developed. Furthermore, the lack of a clear regulatory procedure (EMA/FDA) that is needed to move research into the production and commercialization phases, presents certain barriers to the development of exosome-based therapies.
Nevertheless, there are four main ways of introducing nanovesicles into the body at present, namely oral, transdermal, intranasal, and intravenous. The choice of a specific method depends on the purpose of therapy and the administered substance, as well as the target organ of the patient.
Oral administration is the simplest method in comparison with others. When nanovesicles are not cleaved by food enzymes, they can be preserved in the recipient’s body for a long time. This method is preferred for the targeted therapy of the stomach, colon, upper ileum, and liver [104]. However, the surface properties of exosomes, particularly their membrane proteins, could be partially altered by interactions with other substances in the digestive tract [105]. Moreover, it is difficult to select the exact dose of the drug, since oral absorption is variable and incomplete. The oral route of administration is not applicable in emergency situations because the process of absorption is time consuming and the patient must be able and compliant to take the drug.
The transdermal method is used primarily for delivery to the skin and circulatory system. There are two routes of nanovesicle penetration during transdermal application: using physical channels of the skin with a sufficient micrometer diameter to pass to the vessels and through the stratum corneum. It is assumed that after applying nanovesicles to the skin, they penetrate through the surface of lipid-rich channels on the hair follicles. These nanovesicles can reach the hair shafts by entering the hair matrix cells and moving further by cell differentiation or by the direct penetration into the hair shafts from the hair tip. Another method of penetration into the dermis is through the stratum corneum. Nanovesicles are able to penetrate the skin through trans-follicular pathways, since their surface has a bilayer flexible structure [106]. Formulations for the transdermal route should avoid the risk of irritation and inflammation. Despite the universal penetration mechanisms, a significant proportion of the drug will remain inaccessible due to the barrier function of the skin, which varies from patient to patient by different parts of the body and the patient’s age, which also creates problems for the dosage selection [107].
Intranasal delivery requires lower doses and leads to a faster effect due to the absorption of a therapeutic agent through the nasal mucosa, cumulating in a high concentration of nanovesicles in the system and avoiding the first-pass effect of the drug through the liver, which, in turn, leads to using lower concentrations of the substance [108]. This route of injection is predominantly chosen for targeted therapy of the lungs and brain. For the nasal route, however, only a very limited volume (25–200 μL) can be administered to a relatively small absorption area for a short period of time due to the mucociliary clearance process [109], requiring a stable formulation with a high concentration of exosomes. In addition, this method may not be available in the case of certain respiratory diseases like the common cold, for example. While the single application is sufficiently safe, the continuous intranasal route may be associated with additional risks to the health of the nasal mucosa.
The least common way to inject nanovesicles is intravenous. This type of injection allows you to quickly create the required concentration of the drug in the blood and the target organ, but at the same time carries a certain risk of side effects [110]. Another disadvantage of this method is the rapid clearance of a drug from the blood after its administration [111,112].
10. Therapeutic Effects of Plant Nanovesicles
In the last decade, the attention of researchers has been attracted by the possibility of using plant exosomes as a drug delivery system. The plant origin and high biocompatibility, as well as the universal therapeutic potential of the exosomes, make them advantageous over synthetic liposomes used as nanocarriers.
It was found that the plant exosomes participate in the regulation of the immune response and have a pronounced anti-inflammatory effect. Due to the interspecies translocalization ability of exosomes, the molecules they contain can regulate the interaction between the intestinal microbiota and the host immune system, resulting in a homeostatic balance [113]. In a recurrent inflammatory bowel disease, such as colitis, the intestinal macrophages lose their tolerogenic abilities. Wang and co-authors showed that grapefruit-derived exosomes have a beneficial effect on the gut immune homeostasis by enhancing the anti-inflammatory capacity of intestinal macrophages, ultimately alleviating colitis in mice. After exosome capture by the intestinal macrophages, these nanocarriers increased the expression of heme oxygenase-1 and interleukin (IL)-10, but at the same time suppressed IL-6, IL-1b, and tumor necrosis factor (TNF) [45]. In addition, the nanovesicles delivered naringin, a key flavanone in grapefruit. Upon release, naringin is hydrolyzed by the gut microbiota to its active metabolite naringenin. The exosomes have been shown to exhibit antitumor activity in a mouse model of dextran sodium sulfate-induced colitis [101]. It has also been reported that miRNAs of the exosomes isolated from ginger and grapefruit specifically downregulated some genes of the intestinal probiotic Lactobacillus rhamnosus in mice [67].
A great number of studies are devoted to the nanovesicles isolated from ginger [74,82,102,104,110]. When ginger nanovesicles are loaded with doxorubicin for delivery to colon tumor cells, they effectively invade colon tumors and suppress their growth [82]. Probably, the nanovesicles release doxorubicin at the acidic pH of the tumor extracellular microenvironment, reducing the side effects of the drug [82]. Moreover, the ginger exosomes have shown promising results in reducing colorectal tumorigenesis in mice by decreasing the level of pro-inflammatory cytokines, as well as suppressing the intestinal epithelial cells proliferation and apoptosis by reducing the cyclin D1 expression, which is a marker in the early stages of cancer development [102]. Along with the above effects of ginger exosomes, successful suppression of tumor growth has been shown by reducing the level of survival genes expression using intravenous nanovesicular microRNA delivery [104]. The grape nanovesicles also exhibited an antitumor activity. The effect was achieved by increasing the expression of the Lgr5 and BMI1 genes, which serve as the intestinal stem cell markers and are the genes that regulate the growth and proliferation of stem cells [106]. Other researchers have found that the lemon juice nanovesicles are able to inhibit the proliferation of the A549 (human lung carcinoma), LAMA84 (chronic myeloid leukemia), and human colorectal adenocarcinoma cell lines. The inhibition of proliferation occurred due to the activation of apoptotic cell death mediated by an apoptosis-inducing ligand associated with TNF [108].
It should be noted that in addition to the antitumor effects, the plant exosomes are known to be used in regenerative medicine. It was shown that the grape nanovesicles are able to penetrate into intestinal stem cells, induce their proliferation by regulating the expression of genes responsible for pluripotency (Oct4, SOX2, and Klf4), and thereby regenerate epithelial tissue [110]. At the same time, the ginger nanovesicles exhibited the hepatoprotective activity by reducing the generation of reactive oxygen species in alcohol-damaged mouse livers. Due to their bioactive components, the exosomes can influence the nuclear factor NRF2. The activation of NRF2 is known to increase the expression of liver detoxification genes and antioxidants, which together promote hepatoprotection [82].
In addition, in a similar study, the nanovesicles isolated from broccoli were shown to have a preventive and therapeutic effect on acute and chronic colitis by increasing the level of anti-inflammatory cytokines. The exosomes preserved the intestinal environment with minimal adverse reactions due to the regulation of AMP-activated kinase, which controls the cell energy balance [100]. The ginseng root nanovesicles exerted the anti-aging and anti-pigmentation effects on the ultraviolet-treated human dermal fibroblasts by inhibiting the activity of age-related β-galactosidase and melanogenesis proteins [110]. The nanovesicles have also been isolated from wheat to study their effect on the skin regeneration using primary human dermal fibroblast (HDF), human keratinocyte (HaCaT), and human umbilical vein endothelial cells (HUVEC) in in vitro studies. The results of the study showed an increase in the expression level of type 1 collagen. In addition to their proliferative and migratory effects, the nanovesicles are pro-angiogenic in nature, causing the formation of a tubular structure in human HUVEC lineage. Probably, the nanovesicles have the ability to induce the vascular formation during wound healing [113].
Grapefruit nanovesicles loaded with methotrexate have been used to study immunological responses. When administered orally, these vesicles have been shown to effectively target the mouse F4/80 macrophages located in the gut via micropinocytosis and clathrin-dependent cellular uptake pathways. After the application of nanovesicles, weight loss and reduction in the length of the large intestine stopped. Along with the apparent anti-inflammatory response, the production of pro-inflammatory cytokines TNF-a, IL-1b, and IL-6 decreased. The side effects of methotrexate in the composition of nanovesicles were significantly reduced [45]. In addition, miRNAs in the ginger- and grapefruit-derived nanovesicles targeted the genes of the mice intestinal probiotic L. rhamnosus, increasing its growth and stimulating the antimicrobial immunity to promote the gut microbiota well-being [114]. Similar studies with nanovesicles from broccoli, grapes, and carrots are also known, confirming the significant potential of nanovesicles as modulators of immunological reactions [67]. Some examples of the therapeutic effect of plant nanovesicles are summarized in Table 3.

Table 3.
Therapeutic effects of plant-derived nanovesicles.
11. Targeted Gene Regulation via Engineered Exosomes
The most promising direction might be the investigation of the potential for the use of exosomes in the targeted delivery of genetic material into the cells, particularly when the exosomal vesicles, loaded with the specific microRNA or CRISPR/Cas9, are delivered to regulate the expression of target genes (Figure 4).
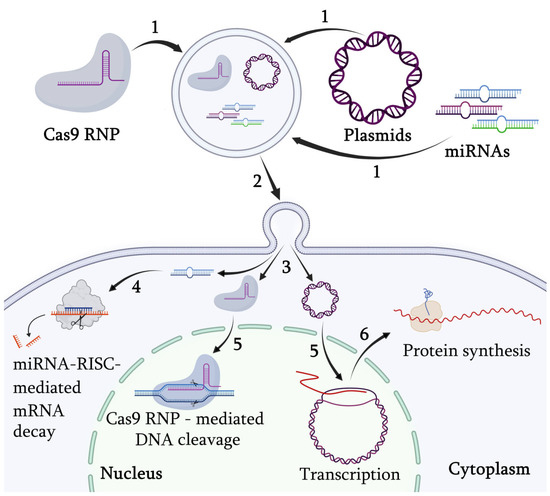
Figure 4.
The use of exosomal vesicles for genetic material delivery. 1—exosome loading with therapeutic macromolecules; 2—exosome delivery; 3—genetic material release upon fusion of the exosome with the cell plasma membrane; 4—miRNA—mediated regulation of gene expression through activation of the RNA-induced silencing complex (RISC); 5—nuclear import of Cas9—ribonucleoprotein (RNP) and plasmid DNA; 6—mRNA nuclear export for translation.
Although the native miRNAs contained in the plant vesicles are mainly delivered, the exogenous miRNAs and siRNAs (small interfering RNAs) have already been reported to be used for loading exosomes and targeted delivery. Thus, the isolated grapefruit exosomes, passively loaded with siRNAs, are directed against the luciferase reporter gene stably expressed in the GL26-Luc and A549-Luc cell cultures. It has been found that the luciferase siRNAs carried by the grapefruit exosomes effectively inhibited the gene expression compared to the native plant exosomes and free miRNAs of luciferase itself [120]. In a similar study, the targeting was improved for the grapefruit exosomes by dimethyl sulfoxide [121]. The exosomes were loaded by incubation with therapeutic miRNA17 (miR17), whose genomic target is antigenic peptide MHC-1 expressed on tumor cells. They were injected intranasally into a mouse with a brain tumor GL-26. As a result, mouse survival was increased, probably due to the selective uptake of exosomes by the GL-26 cells, subsequently inhibiting the MHC-1 expression that caused natural killer cells to become activated to kill tumor cells [121]. In another study, exosomes isolated from ginger were loaded with siRNA-CD98 by sonication and administered orally. The exosomes accumulated in the colon and ileum, while free siRNAs as a control were retained only in the stomach, proving the effective targeting of the exosomes to the intestine. In addition, the delivered siRNAs specifically reduced the expression of CD98 gene, the excess accumulation of which causes inflammatory bowel disease, attenuated the inflammatory response, and effectively alleviated colitis and cancer associated with colitis [122]. Thus, studies in the field of targeted delivery of miRNAs and siRNAs prove the promise of using the exosomal plant vesicles for this purpose. For mammalian exosomes, the studies on the delivery of the CRISPR/Cas9 system are prevalent. For example, the exosomes isolated from the tumor cells SKOV3 were used as the CRISPR/Cas9 delivery platform, which was loaded by electroporation to inhibit the cancer cell proliferation by activating the apoptotic pathway. More specifically, the exosomes loaded with CRISPR/Cas9 could inhibit polymerase-1 (PARP-1) expression and induce apoptosis in ovarian cancer cells. In addition, the inhibition of PARP-1 by CRISPR/Cas9, mediated by genome editing, increased the cell sensitivity to cisplatin (an oncocytotoxic drug) [123]. In another study, the exosomes from the HEK 293T cells, previously transfected with the plasmids containing sgRNA and spCAS9, and the exosomes with the fused exosomal membrane protein CD63 with GFP, which can bind to the GFP antibody fused with Cas9 protein, were isolated. The A549 cells were then treated with exosomes, and the efficiency of their delivery was assessed by fluorescent signals (the reporter gene-GFP), thereby confirming that the exosomes were taken up by the recipient cells. This loading of the CRISPR-Cas9 components contributed to efficient genome editing in the reporter cells [124]. Although the data on the plant exosomal vesicles are still absent, the opportunity of their use in the similar research should also be realized.
Exosome-mediated methods are advantageous compared to the alternative methods for genetic material delivery (mainly liposomes and viral vectors) for several reasons. The first factor is that exosomes have extremely low immunogenicity, which increases the effectiveness of therapy and, most importantly, makes repetition possible [125]. Secondly, exosomes are able to penetrate the blood-brain barrier, as well as other biological barriers, opening up the possibility of targeting hard-to-reach disease cases [126]. Thirdly, the half-life of exosomes in the blood is longer than that of artificial liposomes, which is achieved due to the presence of various transmembrane and membrane-bound proteins on the exosome surface, allowing them to avoid phagocytic clearance [127]. The effective targeting by exome-based vectors may be possible in the future by engineering the composition of specific proteins on their surfaces. For the purposes of genome editing technology, exosomes can be loaded not only with a plasmid containing the CRISPR/Cas9 system, but also in a more advanced way, namely Cas9 ribonucleoprotein (RNP), which allows avoidance of the undesirable consequences of bacterial gene expression in patient cells [124]. All of these advantages will allow the creation of more effective and safer drugs for patients.
12. Conclusions
In the last two decades, there has been an exponential increase in the number of studies devoted to the biological characterization of nanovesicles (the most important carriers of biological information) but significant interest in plant-derived nanovesicles has only appeared in recent years. The endogenous nature of plant nanovesicles formation is their natural and unique advantage for intercellular communication and a wide range of biological activity. Nanovesicles can be taken up selectively by both nearby and distant cells, reprogramming them with their biologically active contents, primarily due to the proteins and ribonucleic acids.
The regulated formation of nanovesicles, the possibility of imparting to them specific biological activity and cell targeting, as well as the potential of their scale production, are of great interest for practical use. Along with the fact that the use of human exosomes in some cases is on a par with stem cell therapy in terms of its effectiveness, there are already a number of successful examples of medical application of plant nanovesicles. However, since the properties and composition of nanovesicles from many plant species remain incompletely understood, further studies are required to standardize protocols for their isolation and characterization, to develop storage technologies and efficient loading, and to improve the targeting to certain cell types.
Currently, new information about the biogenesis of exosomes of the plant origin is required due to the relative lack of knowledge of the processes underlying their formation in different species. A deeper understanding of the mechanisms of nanovesicles formation in plants, as well as environmental factors affecting their number and the content of their biologically active molecules, is necessary to optimize the production of drugs based on plant exosomes. Further study of regulation of the plant nanovesicles properties, using the methods of metabolic and genetic engineering of the plant producers, is of particular relevance, in view of the possibility of controlling the amount and composition of such natural components of exosomes as low-molecular-weight biologically active substances and small RNAs. Since the oral administration of exosomes would require huge amounts of purified preparation, the technology for the production of various types of exosome preparations should be optimized as much as possible. For the same reasons, it is extremely important to confirm the stability and safety of natural nanovesicles in numerous experiments when using them as delivery vehicles in various food matrices.
In general, the plant vesicles are characterized by low immunogenicity, lack of cytotoxicity, natural anti-inflammatory activity, and high biocompatibility. All together, these qualities will enable the plant-derived nanovesicles to make important contributions to the development of nanomedicine therapeutics.
Author Contributions
Conceptualization, Y.S. and T.R.; methodology, Z.T.; software, A.D.; validation, Y.S., A.D. and Y.Y.; formal analysis, L.B.; investigation, Y.S.; resources, Y.Y.; data curation, Y.S.; writing—original draft preparation, Z.T.; writing—review and editing, Y.S.; visualization, A.D.; supervision, V.B.; project administration, Y.S.; funding acquisition, T.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Russian Science Foundation grant number 22-24-00091 (T.V. Rusapetova).
Acknowledgments
Financial support was provided by the Russian Science Foundation, Grant no. 22-24-00091 (T.V. Rusapetova). The experiments described in this work were performed using equipment from the Instrumental Centre for Biotechnology and Gene Engineering at the Federal Scientific Centre of East Asia Terrestrial Biodiversity of the Far East Branch of the Russian Academy of Sciences.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sridharan, K.; Gogtay, N.J. Therapeutic nucleic acids: Current clinical status. Br. J. Clin. Pharmacol. 2016, 82, 659–672. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, N.; Zhang, W.; Cheng, X.; Yan, Z.; Shao, G.; Wang, X.; Wang, R.; Fu, C. Therapeutic peptides: Current applications and future directions. Signal Transduct. Target. Ther. 2022, 7, 48. [Google Scholar] [CrossRef] [PubMed]
- Dzobo, K. The role of natural products as sources of therapeutic agents for innovative drug discovery. Compr. Pharmacol. 2022, 408–422. [Google Scholar] [CrossRef]
- Gerry, C.; Schreiber, S. Chemical probes and drug leads from advances in synthetic planning and methodology. Nat. Rev. Drug Discov. 2018, 17, 333–352. [Google Scholar] [CrossRef]
- Tewabe, A.; Abate, A.; Tamrie, M.; Seyfu, A.; Abdela Siraj, E. Targeted drug delivery—From magic bullet to nanomedicine: Principles, challenges, and future perspectives. J. Multidiscip. Healthc. 2021, 5, 1711–1724. [Google Scholar] [CrossRef] [PubMed]
- Manzari, M.T.; Shamay, Y.; Kiguchi, H.; Rosen, N.; Scaltriti, M.; Heller, D.A. Targeted drug delivery strategies for precision medicines. Nat. Rev. Mater. 2021, 6, 351–370. [Google Scholar] [CrossRef] [PubMed]
- Desai, N.; Momin, M.; Khan, T.; Gharat, S.; Ningthoujam, R.S.; Omri, A. Metallic nanoparticles as drug delivery system for the treatment of cancer. Expert Opin. Drug Deliv. 2021, 18, 1261–1290. [Google Scholar] [CrossRef]
- De Oliveira, L.F.; Bouchmella, K.; De Almeida Gonçalves, K.; Bettini, J.; Kobarg, J.; Borba Cardoso, M. Functionalized silica nanoparticles as an alternative platform for targeted drug-delivery of water insoluble drugs. Langmuir 2016, 32, 3217–3225. [Google Scholar] [CrossRef]
- Seidu, T.A.; Kutoka, P.T.; Asante, D.O.; Farooq, M.A.; Alolga, R.N.; Bo, W. Functionalization of nanoparticulate drug delivery systems and its influence in cancer therapy. Pharmaceuticals 2022, 14, 1113. [Google Scholar] [CrossRef]
- Bharti, C.; Nagaich, U.; Pal, A.K.; Gulati, N. Mesoporous silica nanoparticles in target drug delivery system: A review. Int. J. Pharm. Investig. 2015, 5, 124–133. [Google Scholar] [CrossRef]
- Maiti, D.; Tong, X.; Mou, X.; Yang, K. Carbon-based nanomaterials for biomedical applications: A recent study. Front. Pharmacol. 2019, 9, 1401. [Google Scholar] [CrossRef] [PubMed]
- Patra, J.K.; Das, G.; Fraceto, L.F.; Campos, E.V.R.; del Pilar Rodriguez-Torres, M.; Acosta-Torres, L.S.; Diaz-Torres, L.A.; Grillo, R.; Swamy, M.K.; Sharma, S.; et al. Nano based drug delivery systems: Recent developments and future prospects. J. Nanobiotechnol. 2018, 16, 71. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Chen, G.; Zhang, J. A review of liposomes as a drug delivery system: Current status of approved products, regulatory environments, and future perspectives. Molecules 2022, 27, 1372. [Google Scholar] [CrossRef]
- Sercombe, L.; Veerati, T.; Moheimani, F.; Wu, S.Y.; Sood, A.K.; Hua, S. Advances and challenges of liposome assisted drug delivery. Front. Pharmacol. 2015, 6, 286. [Google Scholar] [CrossRef] [PubMed]
- Dhiman, N.; Awasthi, R.; Sharma, B.; Kharkwal, H.; Kulkarni, G.T. Lipid nanoparticles as carriers for bioactive delivery. Front. Chem. 2021, 9, 580118. [Google Scholar] [CrossRef]
- Ghasemiyeh, P.; Mohammadi-Samani, S. Solid lipid nanoparticles and nanostructured lipid carriers as novel drug delivery systems: Applications, advantages and disadvantages. Res. Pharm. Sci. 2018, 13, 288–303. [Google Scholar] [CrossRef]
- Chen, L.; Hong, W.; Ren, W.; Xu, T.; Qian, Z.; He, Z. Recent progress in targeted delivery vectors based on biomimetic nanoparticles. Signal Transduct. Target. Ther. 2021, 6, 225. [Google Scholar] [CrossRef]
- Yang, M.; Liu, X.; Luo, Q.; Xu, L.; Chen, F. An efficient method to isolate lemon derived extracellular vesicles for gastric cancer therapy. J. Nanobiotechnol. 2020, 18, 100. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.; Heuser, J.; Stahl, P. Endocytosis and intracellular processing of transferrin and colloidal gold-transferrin in rat reticulocytes: Demonstration of a pathway for receptor shedding. Eur. J. Cell Biol. 1984, 35, 256–263. [Google Scholar]
- An, Q.; van Bel, A.J.; Huckelhoven, R. Do plant cells secrete exosomes derived from multivesicular bodies? Plant Signal. Behav. 2007, 2, 4–7. [Google Scholar] [CrossRef]
- Garaeva, L.; Kamyshinsky, R.; Kil, Y.; Varfolomeeva, E.; Verlov, N.; Komarova, E.; Garmay, Y.; Landa, S.; Burdakov, V.; Myasnikov, A.; et al. Delivery of functional exogenous proteins by plant-derived vesicles to human cells in vitro. Sci. Rep. 2021, 11, 6489. [Google Scholar] [CrossRef] [PubMed]
- Perez-Bermudez, P.; Blesa, J.; Soriano, J.M.; Marcilla, A. Extracellular vesicles in food: Experimental evidence of their secretion in grape fruits. Eur. J. Pharm. Sci. 2017, 98, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Park, J.; Sohn, Y.; Oh, C.E.; Park, J.H.; Yuk, J.M.; Yeon, J.H. Stability of Plant Leaf-Derived Extracellular Vesicles According to Preservative and Storage Temperature. Pharmaceutics 2022, 14, 457. [Google Scholar] [CrossRef]
- Yuan, F.; Li, Y.M.; Wang, Z. Preserving extracellular vesicles for biomedical applications: Consideration of storage stability before and after isolation. Drug Deliv. 2021, 28, 1501–1509. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.A.; Rhee, W.J. Exosomes: Biogenesis, Composition, Functions, and Their Role in Pre-metastatic Niche Formation. Biotechnol. Bioprocess Eng. 2019, 24, 689–701. [Google Scholar] [CrossRef]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef]
- Gao, C.; Zhuang, X.; Shen, J.; Jiang, L. Plant ESCRT Complexes: Moving Beyond Endosomal Sorting. Trends Plant Sci. 2017, 22, 986–998. [Google Scholar] [CrossRef]
- Stuffers, S.; Sem Wegner, C.; Stenmark, H.; Brech, A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 2009, 10, 925–937. [Google Scholar] [CrossRef]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- De Gassart, A.; Geminard, C.; Fevrier, B.; Raposo, G.; Vidal, M. Lipid raft-associated protein sorting in exosomes. Blood 2003, 102, 4336–4344. [Google Scholar] [CrossRef]
- Van der Pol, E.; Boing, A.N.; Harrison, P.; Sturk, A.; Nieuwland, R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol. Rev. 2012, 64, 676–705. [Google Scholar] [CrossRef] [PubMed]
- Collins, N.C.; Thordal-Christensen, H.; Lipka, V.; Bau, S.; Kombrink, E.; Qiu, J.L.; Hückelhoven, R.; Stein, M.; Freialdenhoven, A.; Somerville, S.C.; et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature 2003, 425, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Noriko, I.; Shigeyuki, B.; Takashi, L.S.; Kashio, E.; Emi, I.; Natsumaro, K.; Seiichiro, H.; Yoshitaka, T.; Hiroo, F.; Akihiko, N.; et al. Modulation of Plant RAB GTPase-Mediated Membrane Trafficking Pathway at the Interface between Plants and Obligate Biotrophic Pathogens. Plant Cell 2016, 57, 1854–1864. [Google Scholar] [CrossRef]
- Katsiarimpa, A.; Kalinowska, K.; Anzenberger, F.; Weis, C.; Ostertag, M.; Tsutsumi, C.; Schwechheimer, C.; Brunner, F.; Hückelhoven, R.; Isono, E. The deubiquitinating enzyme AMSH1 and the ESCRT-III subunit VPS2.1 are required for autophagic degradation in Arabidopsis. Plant Cell 2013, 25, 2236–2252. [Google Scholar] [CrossRef]
- Rutter, B.D.; Innes, R.W. Extracellular Vesicles Isolated from the Leaf Apoplast Carry Stress-Response Proteins. Plant Physiol. 2017, 173, 728–741. [Google Scholar] [CrossRef] [PubMed]
- Boavida, L.C.; Qin, P.; Broz, M.; Becker, J.D.; McCormick, S. Arabidopsis tetraspanins are confined to discrete expression domains and cell types in reproductive tissues and form homo- and heterodimers when expressed in yeast. Plant Physiol. 2013, 163, 696–712. [Google Scholar] [CrossRef]
- Waghmare, S.; Lileikyte, E.; Karnik, R.; Goodman, J.K.; Blatt, M.R.; Jones, A.M.E. SNAREs SYP121 and SYP122 Mediate the Secretion of Distinct Cargo Subsets. Plant Physiol. 2018, 178, 1679–1688. [Google Scholar] [CrossRef]
- Assaad, F.F.; Qiu, J.L.; Youngs, H.; Ehrhardt, D.; Zimmerli, L.; Kalde, M.; Wanner, G.; Peck, S.C.; Edwards, H.; Ramonell, K.; et al. The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell 2004, 15, 5118–5129. [Google Scholar] [CrossRef]
- Nielsen, M.E.; Feechan, A.; Bohlenius, H.; Ueda, T.; Thordal-Christensen, H. Arabidopsis ARF-GTP exchange factor, GNOM, mediates transport required for innate immunity and focal accumulation of syntaxin PEN1. Proc. Natl. Acad. Sci. USA 2012, 109, 11443–11448. [Google Scholar] [CrossRef]
- Samuel, M.; Bleackley, M.; Anderson, M.; Mathivanan, S. Extracellular vesicles including exosomes in cross kingdom regulation: A viewpoint from plant-fungal interactions. Front. Plant Sci. 2015, 6, 766. [Google Scholar] [CrossRef]
- An, Q.; Ehlers, K.; Kogel, K.H.; Van Bel, A.J.E.; Huckelhoven, R. Multivesicular compartments proliferate in susceptible and resistant MLA12-barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytol. 2006, 172, 563–576. [Google Scholar] [CrossRef] [PubMed]
- Burkova, E.E.; Grigoreva, A.E.; Bulgakov, D.V.; Dmitrenok, P.S.; Vlassov, V.V.; Ryabchikova, E.I.; Sedykh, S.E.; Nevinsky, G.A. Extra Purified Exosomes from Human Placenta Contain an Unpredictable Small Number of Different Major Proteins. Int. J. Mol. Sci. 2019, 20, 2434. [Google Scholar] [CrossRef] [PubMed]
- Woith, E.; Guerriero, G.; Hausman, J.F.; Renaut, J.; Leclercq, C.C.; Weise, C.; Legay, S.; Weng, A.; Melzig, M.F. Plant Extracellular Vesicles and Nanovesicles: Focus on Secondary Metabolites, Proteins and Lipids with Perspectives on Their Potential and Sources. Int. J. Mol. Sci. 2021, 22, 3719. [Google Scholar] [CrossRef]
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.M.; Palmquist, J.; Huang, S.D.; Jin, H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018, 360, 1126–1129. [Google Scholar] [CrossRef] [PubMed]
- Pocsfalvi, G.; Turiak, L.; Ambrosone, A.; Del Gaudio, P.; Puska, G.; Fiume, I.; Silvestre, T.; Vekey, K. Physiochemical and protein datasets related to citrus juice sac cells-derived nanovesicles and microvesicles. Data Brief 2018, 22, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Jimenez, S.; Hashimoto, K.; Santana, O.; Aguirre, J.; Kuchitsu, K.; Cardenas, L. Emerging roles of tetraspanins in plant inter-cellular and inter-kingdom communication. Plant Signal. Behav. 2019, 14, e1581559. [Google Scholar] [CrossRef]
- Caillaud, M.C.; Wirthmueller, L.; Sklenar, J.; Findlay, K.; Piquerez, S.J.; Jones, A.M.; Robatzek, S.; Jones, J.D.; Faulkner, C. The plasmodesmal protein PDLP1 localises to haustoria-associated membranes during downy mildew infection and regulates callose deposition. PLoS Pathog. 2014, 10, e1004496. [Google Scholar] [CrossRef]
- Zand Karimi, H.; Baldrich, P.; Rutter, B.D.; Borniego, L.; Zajt, K.K.; Meyers, B.C.; Innes, R.W. Arabidopsis apoplastic fluid contains sRNA- and circular RNA–protein complexes that are located outside extracellular vesicles. Plant Cell 2022, 34, 1863–1881. [Google Scholar] [CrossRef]
- Baldrich, P.; Rutter, B.D.; Karimi, H.Z.; Podicheti, R.; Meyers, B.C.; Innes, R.W. Plant Extracellular Vesicles Contain Diverse Small RNA Species and Are Enriched in 10- to 17-Nucleotide “Tiny” RNAs. Plant Cell 2019, 31, 315–324. [Google Scholar] [CrossRef]
- Liu, D.; Song, Y.; Chen, Z.; Yu, D. Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol. Plant 2009, 136, 223–236. [Google Scholar] [CrossRef]
- Xiao, J.; Feng, S.; Wang, X.; Long, K.; Luo, Y.; Wang, Y.; Ma, J.; Tang, Q.; Jin, L.; Li, X.; et al. Identification of exosome-like nanoparticle-derived microRNAs from 11 edible fruits and vegetables. PeerJ 2018, 6, e5186. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational Identification of Plant MicroRNAs and Their Targets, Including a Stress-Induced miRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Li, X.; Liu, J.; Dong, L.; Chen, Q.; Liu, J.; Kong, H.; Zhang, Q.; Qi, X.; Hou, D.; et al. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. 2015, 25, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Fairfax, B.P.; Vannberg, F.O.; Radhakrishnan, J.; Hakonarson, H.; Keating, B.J.; Hill, A.V.; Knight, J.C. An integrated expression phenotype mapping approach defines common variants in LEP, ALOX15 and CAPNS1 associated with induction of IL-6. Hum. Mol. Genet. 2010, 19, 720–730. [Google Scholar] [CrossRef]
- Kalarikkal, S.P.; Sundaram, G.M. Edible plant-derived exosomal microRNAs: Exploiting a cross-kingdom regulatory mechanism for targeting SARS-CoV-2. Toxicol. Appl. Pharmacol. 2021, 414, 115425. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Zhu, Y.; Sun, B.; Shao, Y.; Jing, A.; Wang, J.; Xiao, Z. Assessing the survival of exogenous plant microRNA in mice. Food Sci. Nutr. 2014, 2, 380–388. [Google Scholar] [CrossRef]
- Wang, B.; Zhuang, X.; Deng, Z.B.; Jiang, H.; Mu, J.; Wang, Q.; Xiang, X.; Guo, H.; Zhang, L.; Dryden, G.; et al. Targeted drug delivery to intestinal macrophages by bioactive nanovesicles released from grapefruit. Mol. Ther. 2014, 22, 522–534. [Google Scholar] [CrossRef]
- Campo, S.; Peris-Peris, C.; Sire, C.; Moreno, A.B.; Donaire, L.; Zytnicki, M.; Notredame, C.; Llave, C.; San Segundo, B. Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol. 2013, 199, 212–227. [Google Scholar] [CrossRef]
- Kalarikkal, S.P.; Sundaram, G.M. Inter-kingdom regulation of human transcriptome by dietary microRNAs: Emerging bioactives from edible plants to treat human diseases? Trends Food Sci. Technol. 2021, 118A, 723–734. [Google Scholar] [CrossRef]
- Liang, H.; Zhang, S.; Fu, Z.; Wang, Y.; Wang, N.; Liu, Y.; Zhao, C.; Wu, J.; Hu, Y.; Zhang, J.; et al. Effective detection and quantification of dietetically absorbed plant microRNAs in human plasma. J. Nutr. Biochem. 2015, 26, 505–512. [Google Scholar] [CrossRef]
- Hansen, L.L.; Nielsen, M.E. Plant exosomes: Using an unconventional exit to prevent pathogen entry? J. Exp. Bot. 2017, 69, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Dad, H.A.; Gu, T.W.; Zhu, A.Q.; Huang, L.Q.; Peng, L.H. Plant Exosome-like Nanovesicles: Emerging Therapeutics and Drug Delivery Nanoplatforms. Mol. Ther. 2021, 29, 13–31. [Google Scholar] [CrossRef] [PubMed]
- Tkach, M.; Thery, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Frei dit Frey, N.; Robatzek, S. Trafficking vesicles: Pro or contra pathogens? Curr. Opin. Plant Biol. 2009, 12, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Shang, Y.; Fan, B.; Yu, J.Q.; Chen, Z. Arabidopsis LIP5, a positive regulator of multivesicular body biogenesis, is a critical target of pathogen-responsive MAPK cascade in plant basal defense. PLoS Pathog. 2014, 10, e1004243. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, G.; Khong, N.G.; Tisserant, B.; Randoux, B.; Fontaine, J.; Magnin-Robert, M.; Reignault, P.; Sahraoui, A.L. Defence mechanisms associated with mycorrhiza-induced resistance in wheat against powdery mildew. Funct. Plant Biol. 2017, 44, 443–454. [Google Scholar] [CrossRef]
- Meyer, D.; Pajonk, S.; Micali, C.; O’Connell, R.; Schulze-Lefert, P. Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J. 2009, 57, 986–999. [Google Scholar] [CrossRef]
- Wang, B.; Sun, Y.; Song, N.; Zhao, M.; Liu, R.; Feng, H.; Wang, X.; Kang, Z. Puccinia striiformis f. sp. tritici microRNA-like RNA 1 (Pst-milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesis-related 2 gene. New Phytol. 2017, 215, 338–350. [Google Scholar] [CrossRef]
- Weiberg, A.; Wang, M.; Lin, F.M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.D.; Jin, H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef]
- Nakai, W.; Yoshida, T.; Diez, D.; Miyatake, Y.; Nishibu, T.; Imawaka, N.; Naruse, K.; Sadamura, Y.; Hanayama, R. A novel affinity-based method for the isolation of highly purified extracellular vesicles. Sci. Rep. 2016, 6, 33935. [Google Scholar] [CrossRef]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in Exosome Isolation Techniques. Theranostics 2017, 7, 789–804. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, S.; Cai, Q.; Jin, H. Methodological guidelines for isolation and purification of plant extracellular vesicles. bioRxiv 2021, 458648. [Google Scholar] [CrossRef]
- Woith, E.; Melzig, M.F. Extracellular Vesicles from Fresh and Dried Plants-Simultaneous Purification and Visualization Using Gel Electrophoresis. Int. J. Mol. Sci. 2019, 20, 357. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, H.; Yin, H.; Bennett, C.; Zhang, H.G.; Guo, P. Arrowtail RNA for Ligand Display on Ginger Exosome-like Nanovesicles to Systemic Deliver siRNA for Cancer Suppression. Sci. Rep. 2018, 8, 14644. [Google Scholar] [CrossRef]
- Teng, Y.; Xu, F.; Zhang, X.; Mu, J.; Sayed, M.; Hu, X.; Lei, C.; Sriwastva, M.; Kumar, A.; Sundaram, K.; et al. Plant-derived exosomal microRNAs inhibit lung inflammation induced by exosomes SARS-CoV-2 Nsp12. Mol. Ther. 2021, 29, 2424–2440. [Google Scholar] [CrossRef]
- Zhang, L.; He, F.; Gao, L.; Cong, M.; Sun, J.; Xu, J.; Wang, Y.; Hu, Y.; Asghar, S.; Hu, L.; et al. Engineering Exosome-Like Nanovesicles Derived from Asparagus cochinchinensis Can Inhibit the Proliferation of Hepatocellular Carcinoma Cells with Better Safety Profile. Int. J. Nanomed. 2021, 16, 1575–1586. [Google Scholar] [CrossRef]
- Suharta, S.; Barlian, A.; Hidajah, A.C.; Notobroto, H.B.; Ana, I.D.; Indariani, S.; Wungu, T.D.K.; Wijaya, C.H. Plant-derived exosome-like nanoparticles: A concise review on its extraction methods, content, bioactivities, and potential as functional food ingredient. J. Food Sci. 2021, 86, 2838–2850. [Google Scholar] [CrossRef]
- Kalarikkal, S.P.; Prasad, D.; Kasiappan, R.; Chaudhari, S.R.; Sundaram, G.M. A cost-effective polyethylene glycol-based method for the isolation of functional edible nanoparticles from ginger rhizomes. Sci. Rep. 2020, 10, 4456. [Google Scholar] [CrossRef]
- Sidhom, K.; Obi, P.O.; Saleem, A. A Review of Exosomal Isolation Methods: Is Size Exclusion Chromatography the Best Option? Int. J. Mol. Sci. 2020, 21, 6466. [Google Scholar] [CrossRef]
- You, J.Y.; Kang, S.J.; Rhee, W.J. Isolation of cabbage exosome-like nanovesicles and investigation of their biological activities in human cells. Bioact. Mater. 2021, 6, 4321–4332. [Google Scholar] [CrossRef]
- Abraham, A.M.; Wiemann, S.; Ambreen, G.; Zhou, J.; Engelhardt, K.; Brubler, J.; Bakowsky, U.; Li, S.M.; Mandic, R.; Pocsfalvi, G.; et al. Cucumber-Derived Exosome-like Vesicles and PlantCrystals for Improved Dermal Drug Delivery. Pharmaceutics 2022, 14, 476. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Cai, Q.; Qiao, L.; Huang, C.Y.; Wang, S.; Miao, W.; Ha, T.; Wang, Y.; Jin, H. RNA-binding proteins contribute to small RNA loading in plant extracellular vesicles. Nat. Plants 2021, 7, 342–352. [Google Scholar] [CrossRef] [PubMed]
- Echlin, P. Low-temperature scanning electron microscopy. In Low-Temperature Microscopy and Analysis; Plenum Press: New York, NY, USA, 1992; pp. 349–411. [Google Scholar] [CrossRef]
- Noble, J.M.; Roberts, L.D.M.; Vidavsky, N.; Chiou, A.E.; Fischbach, C.; Paszek, M.J.; Estroff, L.A.; Kourkoutis, L.F. Direct comparison of optical and electron microscopy methods for structural characterization of extracellular vesicles. J. Struct. Biol. 2020, 210, 107474. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Chung, J.; Balaj, L.; Charest, A.; Bigner, D.D.; Carter, B.S.; Hochberg, F.H.; Breakefield, X.O.; Weissleder, R.; Lee, H. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat. Med. 2012, 18, 1835–1840. [Google Scholar] [CrossRef]
- Tatischeff, I.; Larquet, E.; Falcon-Perez, J.M.; Turpin, P.Y.; Kruglik, S.G. Fast characterisation of cell-derived extracellular vesicles by nanoparticles tracking analysis, cryo-electron microscopy, and raman tweezers microspectroscopy. J. Extracell. Vesicles 2012, 1, 19179. [Google Scholar] [CrossRef]
- Sharma, S.; Rasool, H.I.; Palanisamy, V.; Mathisen, C.; Schmidt, M.; Wong, D.T.; Gimzewski, J.K. Structural-mechanical characterization of nanoparticle exosomes in human saliva, using correlative afm, fesem, and force spectroscopy. ACS Nano 2010, 4, 1921–1926. [Google Scholar] [CrossRef]
- Sitar, S.; Kejzar, A.; Pahovnik, D.; Kogej, K.; Tusek-Znidaric, M.; Lenassi, M.; Zagar, E. Size characterization and quantification of exosomes by asymmetrical-flow field-flow fractionation. Anal. Chem. 2015, 87, 9225–9233. [Google Scholar] [CrossRef]
- Hou, J.; Ci, H.; Wang, P.; Wang, C.; Lv, B.; Miao, L.; You, G. Nanoparticle tracking analysis versus dynamic light scattering: Case study on the effect of Ca2+ and alginate on the aggregation of cerium oxide nanoparticles. J. Hazard. Mater. 2018, 360, 319–328. [Google Scholar] [CrossRef]
- Filipe, V.; Hawe, A.; Jiskoot, W. Critical evaluation of nanoparticle tracking analysis (nta) by nanosight for the measurement of nanoparticles and protein aggregates. Pharm. Res. 2010, 27, 796–810. [Google Scholar] [CrossRef]
- Szatanek, R.; Baj-Krzyworzeka, M.; Zimoch, J.; Lekka, M.; Siedlar, M.; Baran, J. The methods of choice for extracellular vesicles (evs) characterization. Int. J. Mol. Sci. 2017, 18, 1153. [Google Scholar] [CrossRef]
- Rutter, B.D.; Innes, R.W. Growing pains: Addressing the pitfalls of plant extracellular vesicle research. New Phytol. 2020, 228, 1505–1510. [Google Scholar] [CrossRef]
- Thongboonkerd, V. Practical points in urinary proteomics. J. Proteome Res. 2007, 6, 3881–3890. [Google Scholar] [CrossRef] [PubMed]
- Kreimer, S.; Belov, A.M.; Ghiran, I.; Murthy, S.K.; Frank, D.A.; Ivanov, A.R. Mass-spectrometry-based molecular characterization of extracellular vesicles: Lipidomics and proteomics. J. Proteome Res. 2015, 14, 2367–2384. [Google Scholar] [CrossRef] [PubMed]
- Kedjouar, B.; De Medina, P.; Oulad-Abdelghani, M.; Payre, B.; Silvente-Poirot, S.; Favre, G.; Faye, J.C.; Poirot, M. Molecular characterization of the microsomal tamoxifen binding site. J. Biol. Chem. 2004, 279, 34048–34061. [Google Scholar] [CrossRef]
- Mayr, M.; Grainger, D.; Mayr, U.; Leroyer, A.S.; Leseche, G.; Sidibe, A.; Herbin, O.; Yin, X.; Gomes, A.; Madhu, B.; et al. Proteomics, metabolomics, and immunomics on microparticles derived from human atherosclerotic plaques. Circ. Cardiovasc. Genet. 2009, 2, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Yagi, Y.; Ohkubo, T.; Kawaji, H.; Machida, A.; Miyata, H.; Goda, S.; Roy, S.; Hayashizaki, Y.; Suzuki, H.; Yokota, T. Next-generation sequencing-based small RNA profiling of cerebrospinal fluid exosomes. Neurosci. Lett. 2017, 636, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Mao, W.; Shu, Y.; Lin, F.; Liu, S.; Shen, H.; Gao, W.; Li, S.; Shen, D. A cluster of specified microRNAs in peripheral blood as biomarkers for metastatic non-small-cell lung cancer by stem-loop RT-PCR. J. Cancer Res. Clin. Oncol. 2012, 138, 85–93. [Google Scholar] [CrossRef]
- Li, X.; Chen, C.; Wang, Z.; Liu, J.; Sun, W.; Shen, K.; Lv, Y.; Zhu, S.; Zhan, P.; Lv, T.; et al. Elevated exosome-derived miRNAs predict osimertinib resistance in non-small cell lung cancer. Cancer Cell Int. 2021, 21, 428. [Google Scholar] [CrossRef]
- Fuhrmann, G.; Serio, A.; Mazo, M.; Nair, R.; Stevens, M.M. Active loading into extracellular vesicles significantly improves the cellular uptake and photodynamic effect of porphyrins. J. Control. Release 2015, 205, 35–44. [Google Scholar] [CrossRef]
- Vashisht, M.; Rani, P.; Onteru, S.K.; Singh, D. Curcumin Encapsulated in Milk Exosomes Resists Human Digestion and Possesses Enhanced Intestinal Permeability In Vitro. Appl. Biochem. Biotechnol. 2017, 183, 993–1007. [Google Scholar] [CrossRef]
- Zhang, M.; Viennois, E.; Prasad, M.; Zhang, Y.; Wang, L.; Zhang, Z.; Han, M.K.; Xiao, B.; Xu, C.; Srinivasan, S.; et al. Edible ginger-derived nanoparticles: A novel therapeutic approach for the prevention and treatment of inflammatory bowel disease and colitis-associated cancer. Biomaterials 2016, 101, 321–340. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, N.; Whiteside, T.L.; Reichert, T.E. Challenges in exosome isolation and analysis in health and disease. Int. J. Mol. Sci. 2019, 20, 4684. [Google Scholar] [CrossRef]
- Mu, J.; Zhuang, X.; Wang, Q.; Jiang, H.; Deng, Z.B.; Wang, B.; Zhang, L.; Kakar, S.; Jun, Y.; Miller, D.; et al. Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol. Nutr. Food Res. 2014, 58, 1561–1573. [Google Scholar] [CrossRef]
- Munir, J.; Lee, M.; Ryu, S. Exosomes in food: Health benefits and clinical relevance in diseases. Adv. Nutr. 2020, 11, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Gu, T.W.; Wang, M.Z.; Niu, J.; Chu, Y.; Guo, K.R.; Peng, L.H. Outer membrane vesicles derived from E. coli as novel vehicles for transdermal and tumor targeting delivery. Nanoscale 2020, 12, 18965–18977. [Google Scholar] [CrossRef]
- Alkilani, A.Z.; McCrudden, M.T.; Donnelly, R.F. Transdermal drug delivery: Innovative pharmaceutical developments based on disruption of the barrier properties of the stratum corneum. Pharmaceuticals 2015, 7, 438–470. [Google Scholar] [CrossRef]
- Moss, L.D.; Sode, D.; Patel, R.; Lui, A.; Hudson, C.; Patel, N.A.; Bickford, P.C. Intranasal delivery of exosomes from human adipose derived stem cells at forty-eight hours post injury reduces motor and cognitive impairments following traumatic brain injury. Neurochem. Int. 2021, 150, 105173. [Google Scholar] [CrossRef] [PubMed]
- Ghadiri, M.; Young, P.M.; Traini, D. Strategies to enhance drug absorption via nasal and pulmonary routes. Pharmaceuticals 2019, 11, 113. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Deng, Z.B.; Mu, J.; Zhang, L.; Yan, J.; Miller, D.; Feng, W.; McClain, C.J.; Zhang, H.G. Ginger-derived nanoparticles protect against alcohol-induced liver damage. J. Extracell. Vesicles 2015, 4, 28713. [Google Scholar] [CrossRef]
- Takahashi, Y.; Nishikawa, M.; Shinotsuka, H.; Matsui, Y.; Ohara, S.; Imai, T. Visualization and in vivo tracking of the exosomes of murine melanoma B16-BL6 cells in mice after intravenous injection. J. Biotechnol. 2013, 165, 77–84. [Google Scholar] [CrossRef]
- Smyth, T.; Kullberg, M.; Malik, N.; Smith-Jones, P.; Graner, M.W.; Anchordoquy, T.J. Biodistribution and delivery efficiency of unmodified tumor-derived exosomes. J. Control. Release 2015, 199, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Rong, Y.; Teng, Y.; Mu, J.; Zhuang, X.; Tseng, M.; Samykutty, A.; Zhang, L.; Yan, J.; Miller, D.; et al. Broccoli-Derived Nanoparticle Inhibits Mouse Colitis by Activating Dendritic Cell AMP-Activated Protein Kinase. Mol. Ther. 2017, 25, 1641–1654. [Google Scholar] [CrossRef]
- Amaro, M.I.; Rocha, J.; Vila-Real, H.; Eduardo-Figueira, M.; Mota-Filipe, H.; Sepodes, B.; Ribeiro, M.H. Anti-inflammatory activity of naringin and the biosynthesised naringenin by naringinase immobilized in microstructured materials in a model of DSS-induced colitis in mice. Food Res. Int. 2009, 42, 1010–1017. [Google Scholar] [CrossRef]
- Raimondo, S.; Naselli, F.; Fontana, S.; Monteleone, F.; Lo Dico, A.; Saieva, L.; Zito, G.; Flugy, A.; Manno, M.; Di Bella, M.A.; et al. Citrus limon-derived nanovesicles inhibit cancer cell proliferation and suppress CML xenograft growth by inducing TRAIL-mediated cell death. Oncotarget 2015, 6, 19514–19527. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Viennois, E.; Xu, C.; Merlin, D. Plant derived edible nanoparticles as a new therapeutic approach against diseases. Tissue Barriers 2016, 4, e1134415. [Google Scholar] [CrossRef] [PubMed]
- Ju, S.; Mu, J.; Dokland, T.; Zhuang, X.; Wang, Q.; Jiang, H.; Xiang, X.; Deng, Z.B.; Wang, B.; Zhang, L.; et al. Grape exosome-like nanoparticles induce intestinal stem cells and protect mice from DSS-induced colitis. Mol. Ther. 2013, 21, 1345–1357. [Google Scholar] [CrossRef] [PubMed]
- Sahin, F.; Kocak, P.; Gunes, M.Y.; Ozkan, I.; Yildirim, E.; Kala, E.Y. In Vitro Wound Healing Activity of Wheat-Derived Nanovesicles. Appl. Biochem. Biotechnol. 2019, 188, 381–394. [Google Scholar] [CrossRef]
- Cho, E.G.; Choi, S.Y.; Kim, H.; Choi, E.J.; Lee, E.J.; Park, P.J.; Ko, J.; Kim, K.P.; Baek, H.S. Panax ginseng-Derived Extracellular Vesicles Facilitate Anti-Senescence Effects in Human Skin Cells: An Eco-Friendly and Sustainable Way to Use Ginseng Substances. Cells 2021, 10, 486. [Google Scholar] [CrossRef]
- Wang, Q.; Zhuang, X.; Mu, J.; Deng, Z.B.; Jiang, H.; Zhang, L.; Xiang, X.; Wang, B.; Yan, J.; Miller, D.; et al. Delivery of therapeutic agents by nanoparticles made of grapefruit-derived lipids. Nat. Commun. 2013, 4, 1867. [Google Scholar] [CrossRef]
- Zhuang, X.; Teng, Y.; Samykutty, A.; Mu, J.; Deng, Z.; Zhang, L.; Cao, P.; Rong, Y.; Yan, J.; Miller, D.; et al. Grapefruit-derived Nanovectors Delivering Therapeutic miR17 through an Intranasal Route Inhibit Brain Tumor Progression. Mol. Ther. 2016, 24, 96–105. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, X.; Han, M.K.; Collins, J.F.; Merlin, D. Oral administration of ginger-derived nanolipids loaded with siRNA as a novel approach for efficient siRNA drug delivery to treat ulcerative colitis. Nanomedicine 2017, 12, 1927–1943. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Yang, Y.; Oh, S.J.; Hong, Y.; Seo, M.; Jang, M. Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting. J. Control. Release 2017, 266, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Zhang, X.; Xie, F.; Xu, B.; Xie, P.; Yang, T.; Shi, Q.; Zhang, C.Y.; Zhang, Y.; Chen, J.; et al. An engineered exosome for delivering sgRNA:Cas9 ribonucleoprotein complex and genome editing in recipient cells. Biomater. Sci. 2020, 8, 2966–2976. [Google Scholar] [CrossRef] [PubMed]
- Mendt, M.; Kamerkar, S.; Sugimoto, H.; McAndrews, K.M.; Wu, C.C.; Gagea, M.; Yang, S.; Blanko, E.V.R.; Peng, Q.; Ma, X.; et al. Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI Insight 2018, 3, e99263. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Kamerkar, S.; LeBleu, V.S.; Sugimoto, H.; Yang, S.; Ruivo, C.F.; Melo, S.A.; Lee, J.J.; Kalluri, R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017, 546, 498–503. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).