Accurate Evaluation of Feature Contributions for Sentinel Lymph Node Status Classification in Breast Cancer
Abstract
1. Introduction
2. Materials
2.1. Data
2.2. Histological Features
3. Methods
3.1. Training Strategies
3.2. Classification Algorithms
3.2.1. Logistic LASSO
3.2.2. Random Forest
3.3. Performance Evaluation
- Accuracy
- Sensitivity
- Specificity
- Area Under the Receiver Operating Characteristics (ROC) Curve (AUC) that adjusts the decision threshold to plot sensitivity versus specificity.
3.4. Feature Ranking Analysis
4. Results and Discussion
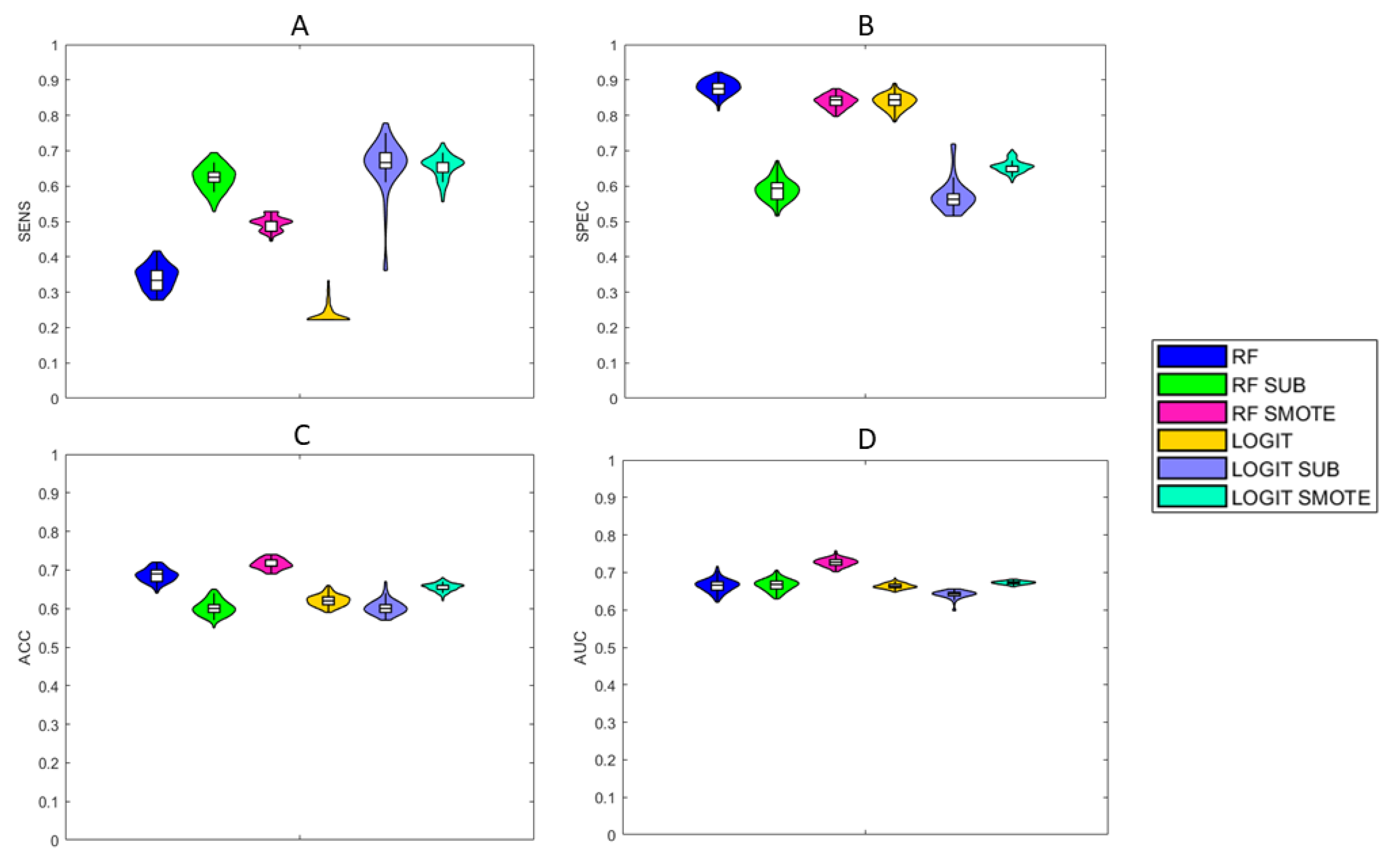
4.1. Performance
4.2. Feature Ranking Analysis
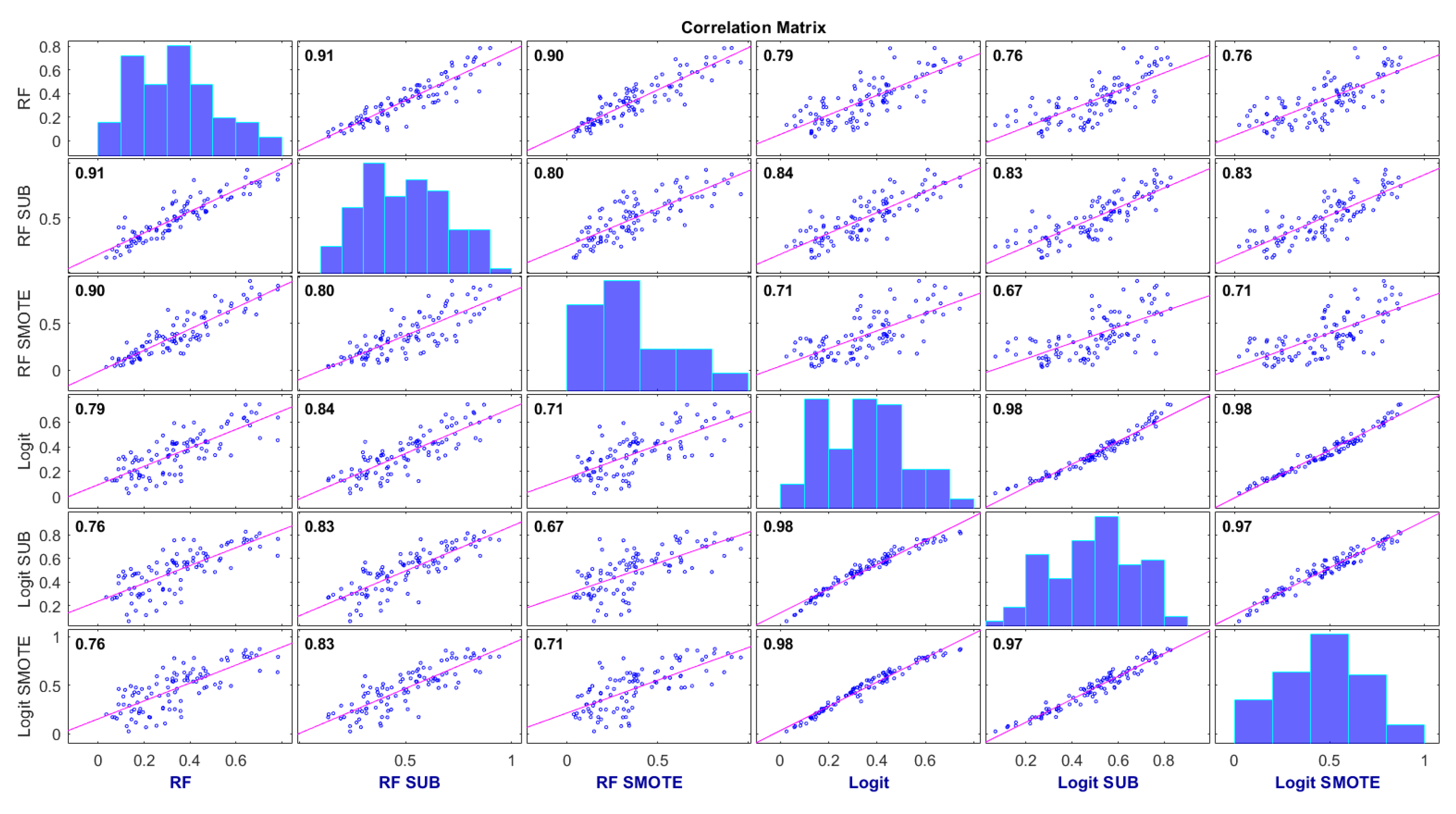
4.3. Consensus Degree of the Probability Scores
5. Landscape
6. Limitations and Future Perspectives
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mazo, C.; Kearns, C.; Mooney, C.; Gallagher, W.M. Clinical decision support systems in breast cancer: A systematic review. Cancers 2020, 12, 369. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Abdi, M.A.; Falkson, C. Axillary management in breast cancer patients: A comprehensive review of the key trials. Clin. Breast Cancer 2018, 18, e1251–e1259. [Google Scholar] [CrossRef] [PubMed]
- Cormier, J.N.; Askew, R.L.; Mungovan, K.S.; Xing, Y.; Ross, M.I.; Armer, J.M. Lymphedema beyond breast cancer: A systematic review and meta-analysis of cancer-related secondary lymphedema. Cancer 2010, 116, 5138–5149. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, A.E.; Ballman, K.V.; McCall, L.; Beitsch, P.D.; Brennan, M.B.; Kelemen, P.R.; Ollila, D.W.; Hansen, N.M.; Whitworth, P.W.; Blumencranz, P.W.; et al. Effect of axillary dissection vs no axillary dissection on 10-year overall survival among women with invasive breast cancer and sentinel node metastasis: The ACOSOG Z0011 (Alliance) randomized clinical trial. JAMA 2017, 318, 918–926. [Google Scholar] [CrossRef]
- Galimberti, V.; Fontana, S.R.; Maisonneuve, P.; Steccanella, F.; Vento, A.; Intra, M.; Naninato, P.; Caldarella, P.; Iorfida, M.; Colleoni, M.; et al. Sentinel node biopsy after neoadjuvant treatment in breast cancer: Five-year follow-up of patients with clinically node-negative or node-positive disease before treatment. Eur. J. Surg. Oncol. (EJSO) 2016, 42, 361–368. [Google Scholar] [CrossRef]
- Chen, K.; Liu, J.; Li, S.; Jacobs, L. Development of nomograms to predict axillary lymph node status in breast cancer patients. BMC Cancer 2017, 17, 1–10. [Google Scholar] [CrossRef]
- Houvenaeghel, G.; Lambaudie, E.; Classe, J.M.; Mazouni, C.; Giard, S.; Cohen, M.; Faure, C.; Charitansky, H.; Rouzier, R.; Daraï, E.; et al. Lymph node positivity in different early breast carcinoma phenotypes: A predictive model. BMC Cancer 2019, 19, 1–10. [Google Scholar] [CrossRef]
- Chen, J.Y.; Chen, J.J.; Yang, B.L.; Liu, Z.B.; Huang, X.Y.; Liu, G.Y.; Han, Q.X.; Yang, W.T.; Shen, Z.Z.; Shao, Z.M.; et al. Predicting sentinel lymph node metastasis in a Chinese breast cancer population: Assessment of an existing nomogram and a new predictive nomogram. Breast Cancer Res. Treat. 2012, 135, 839–848. [Google Scholar] [CrossRef]
- Okuno, J.; Miyake, T.; Sota, Y.; Tanei, T.; Kagara, N.; Naoi, Y.; Shimoda, M.; Shimazu, K.; Kim, S.J.; Noguchi, S. Development of prediction model including microRNA expression for sentinel lymph node metastasis in ER-positive and HER2-negative breast cancer. Ann. Surg. Oncol. 2021, 28, 310–319. [Google Scholar] [CrossRef]
- Fanizzi, A.; Pomarico, D.; Paradiso, A.; Bove, S.; Diotaiuti, S.; Didonna, V.; Giotta, F.; La Forgia, D.; Latorre, A.; Pastena, M.I.; et al. Predicting of sentinel lymph node status in breast cancer patients with clinically negative nodes: A Validation Study. Cancers 2021, 13, 352. [Google Scholar] [CrossRef]
- Estabrooks, A.; Jo, T.; Japkowicz, N. A multiple resampling method for learning from imbalanced data sets. Comput. Intell. 2004, 20, 18–36. [Google Scholar] [CrossRef]
- Weiss, G.M. Mining with rarity: A unifying framework. ACM Sigkdd Explor. Newsl. 2004, 6, 7–19. [Google Scholar] [CrossRef]
- Tang, Y.; Zhang, Y.Q.; Chawla, N.V.; Krasser, S. SVMs modeling for highly imbalanced classification. IEEE Trans. Syst. Man Cybern. Part B (Cybern.) 2008, 39, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Jo, T.; Japkowicz, N. Class imbalances versus small disjuncts. ACM Sigkdd Explor. Newsl. 2004, 6, 40–49. [Google Scholar] [CrossRef]
- Zhao, Y.; Wong, Z.S.Y.; Tsui, K.L. A framework of rebalancing imbalanced healthcare data for rare events’ classification: A case of look-alike sound-alike mix-up incident detection. J. Healthc. Eng. 2018, 2018, 6275435. [Google Scholar] [CrossRef] [PubMed]
- Muhamed Ali, A.; Zhuang, H.; Ibrahim, A.; Rehman, O.; Huang, M.; Wu, A. A machine learning approach for the classification of kidney cancer subtypes using mirna genome data. Appl. Sci. 2018, 8, 2422. [Google Scholar] [CrossRef]
- Jeong, B.; Cho, H.; Kim, J.; Kwon, S.K.; Hong, S.; Lee, C.; Kim, T.; Park, M.S.; Hong, S.; Heo, T.Y. Comparison between statistical models and machine learning methods on classification for highly imbalanced multiclass kidney data. Diagnostics 2020, 10, 415. [Google Scholar] [CrossRef]
- Barbieri, D.; Chawla, N.; Zaccagni, L.; Grgurinović, T.; Šarac, J.; Čoklo, M.; Missoni, S. Predicting cardiovascular risk in Athletes: Resampling improves classification performance. Int. J. Environ. Res. Public Health 2020, 17, 7923. [Google Scholar] [CrossRef]
- Tjoa, E.; Guan, C. A survey on explainable artificial intelligence (xai): Toward medical xai. IEEE Trans. Neural Netw. Learn. Syst. 2020, 32, 4793–4813. [Google Scholar] [CrossRef]
- Lombardi, A.; Diacono, D.; Amoroso, N.; Monaco, A.; Tavares, J.M.R.; Bellotti, R.; Tangaro, S. Explainable Deep Learning for Personalized Age Prediction With Brain Morphology. Front. Neurosci. 2021, 15, 578. [Google Scholar] [CrossRef]
- Szychta, P.; Westfal, B.; Maciejczyk, R.; Smolarz, B.; Romanowicz, H.; Krawczyk, T.; Zadrożny, M. Intraoperative diagnosis of sentinel lymph node metastases in breast cancer treatment with one-step nucleic acid amplification assay (OSNA). Arch. Med. Sci. AMS 2016, 12, 1239. [Google Scholar] [CrossRef] [PubMed]
- Egner, J.R. AJCC cancer staging manual. JAMA 2010, 304, 1726–1727. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- He, H.; Garcia, E.A. Learning from imbalanced data. IEEE Trans. Knowl. Data Eng. 2009, 21, 1263–1284. [Google Scholar]
- Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B (Methodol.) 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Wu, T.T.; Chen, Y.F.; Hastie, T.; Sobel, E.; Lange, K. Genome-wide association analysis by lasso penalized logistic regression. Bioinformatics 2009, 25, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Kim, Y.; Jeong, K.; Jeong, H.; Kim, J. Logistic LASSO regression for the diagnosis of breast cancer using clinical demographic data and the BI-RADS lexicon for ultrasonography. Ultrasonography 2018, 37, 36. [Google Scholar] [CrossRef]
- McEligot, A.J.; Poynor, V.; Sharma, R.; Panangadan, A. Logistic LASSO regression for dietary intakes and breast cancer. Nutrients 2020, 12, 2652. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Yamada, M.; Jitkrittum, W.; Sigal, L.; Xing, E.P.; Sugiyama, M. High-dimensional feature selection by feature-wise kernelized lasso. Neural Comput. 2014, 26, 185–207. [Google Scholar] [CrossRef] [PubMed]
- Grömping, U. Variable importance assessment in regression: Linear regression versus random forest. Am. Stat. 2009, 63, 308–319. [Google Scholar] [CrossRef]
- Bonett, D.G. Confidence interval for a coefficient of quartile variation. Comput. Stat. Data Anal. 2006, 50, 2953–2957. [Google Scholar] [CrossRef]
- Fanizzi, A.; Lorusso, V.; Biafora, A.; Bove, S.; Comes, M.C.; Cristofaro, C.; Digennaro, M.; Didonna, V.; Forgia, D.L.; Nardone, A.; et al. Sentinel Lymph Node Metastasis on Clinically Negative Patients: Preliminary Results of a Machine Learning Model Based on Histopathological Features. Appl. Sci. 2021, 11, 10372. [Google Scholar] [CrossRef]
- Dong, Y.; Feng, Q.; Yang, W.; Lu, Z.; Deng, C.; Zhang, L.; Lian, Z.; Liu, J.; Luo, X.; Pei, S.; et al. Preoperative prediction of sentinel lymph node metastasis in breast cancer based on radiomics of T2-weighted fat-suppression and diffusion-weighted MRI. Eur. Radiol. 2018, 28, 582–591. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Mao, N.; Ma, H.; Dong, J.; Zhang, K.; Che, K.; Duan, S.; Zhang, X.; Shi, Y.; Xie, H. Pharmacokinetic parameters and radiomics model based on dynamic contrast enhanced MRI for the preoperative prediction of sentinel lymph node metastasis in breast cancer. Cancer Imaging 2020, 20, 1–8. [Google Scholar] [CrossRef]
- Klar, M.; Foeldi, M.; Markert, S.; Gitsch, G.; Stickeler, E.; Watermann, D. Good prediction of the likelihood for sentinel lymph node metastasis by using the MSKCC nomogram in a German breast cancer population. Ann. Surg. Oncol. 2009, 16, 1136–1142. [Google Scholar] [CrossRef]
- Luo, J.; Ning, Z.; Zhang, S.; Feng, Q.; Zhang, Y. Bag of deep features for preoperative prediction of sentinel lymph node metastasis in breast cancer. Phys. Med. Biol. 2018, 63, 245014. [Google Scholar] [CrossRef]
- Sopik, V.; Narod, S.A. The relationship between tumour size, nodal status and distant metastases: On the origins of breast cancer. Breast Cancer Res. Treat. 2018, 170, 647–656. [Google Scholar] [CrossRef]
- Min, S.K.; Lee, S.K.; Woo, J.; Jung, S.M.; Ryu, J.M.; Yu, J.; Lee, J.E.; Kim, S.W.; Chae, B.J.; Nam, S.J. Relation between tumor size and lymph node metastasis according to subtypes of breast cancer. J. Breast Cancer 2021, 24, 75. [Google Scholar] [CrossRef]
- Lombardi, A.; Amoroso, N.; Diacono, D.; Monaco, A.; Logroscino, G.; De Blasi, R.; Bellotti, R.; Tangaro, S. Association between structural connectivity and generalized cognitive spectrum in Alzheimer’s disease. Brain Sci. 2020, 10, 879. [Google Scholar] [CrossRef] [PubMed]
- Awada, W.; Khoshgoftaar, T.M.; Dittman, D.; Wald, R.; Napolitano, A. A review of the stability of feature selection techniques for bioinformatics data. In Proceedings of the 2012 IEEE 13th International Conference on Information Reuse & Integration (IRI), Las Vegas, NV, USA, 8–10 August 2012; pp. 356–363. [Google Scholar]
- Nogueira, S.; Sechidis, K.; Brown, G. On the stability of feature selection algorithms. J. Mach. Learn. Res. 2017, 18, 6345–6398. [Google Scholar]
- Khaire, U.M.; Dhanalakshmi, R. Stability of feature selection algorithm: A review. J. King Saud-Univ.-Comput. Inf. Sci. 2019, 34, 1060–1073. [Google Scholar] [CrossRef]
- Bousquet, O.; Elisseeff, A. Stability and generalization. J. Mach. Learn. Res. 2002, 2, 499–526. [Google Scholar]
- Kernbach, J.M.; Staartjes, V.E. Foundations of Machine Learning-Based Clinical Prediction Modeling: Part II—Generalization and Overfitting. In Machine Learning in Clinical Neuroscience; Springer: Berlin/Heidelberg, Germany, 2022; pp. 15–21. [Google Scholar]
- Futoma, J.; Simons, M.; Doshi-Velez, F.; Kamaleswaran, R. Generalization in clinical prediction models: The blessing and curse of measurement indicator variables. Crit. Care Explor. 2021, 3, e0453. [Google Scholar] [CrossRef] [PubMed]
- Bonsang-Kitzis, H.; Mouttet-Boizat, D.; Guillot, E.; Feron, J.G.; Fourchotte, V.; Alran, S.; Pierga, J.Y.; Cottu, P.; Lerebours, F.; Stevens, D.; et al. Medico-economic impact of MSKCC non-sentinel node prediction nomogram for ER-positive HER2-negative breast cancers. PLoS ONE 2017, 12, e0169962. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.K.; Kim, M.K.; Kim, J.; Lee, E.; Yoo, T.K.; Lee, H.B.; Kang, Y.J.; Kim, J.; Moon, H.G.; Chang, J.M.; et al. Can we skip intraoperative evaluation of sentinel lymph nodes? Nomogram predicting involvement of three or more axillary lymph nodes before breast cancer surgery. Cancer Res. Treat. Off. J. Korean Cancer Assoc. 2017, 49, 1088–1096. [Google Scholar] [CrossRef]
- Bevilacqua, J.L.B.; Kattan, M.W.; Fey, J.V.; Cody III, H.S.; Borgen, P.I.; Van Zee, K.J. Doctor, what are my chances of having a positive sentinel node? A validated nomogram for risk estimation. J. Clin. Oncol. 2007, 25, 3670–3679. [Google Scholar] [CrossRef]
- Liu, J.; Sun, D.; Chen, L.; Fang, Z.; Song, W.; Guo, D.; Ni, T.; Liu, C.; Feng, L.; Xia, Y.; et al. Radiomics analysis of dynamic contrast-enhanced magnetic resonance imaging for the prediction of sentinel lymph node metastasis in breast cancer. Front. Oncol. 2019, 9, 980. [Google Scholar] [CrossRef]
- Bove, S.; Comes, M.C.; Lorusso, V.; Cristofaro, C.; Didonna, V.; Gatta, G.; Giotta, F.; La Forgia, D.; Latorre, A.; Pastena, M.I.; et al. A ultrasound-based radiomic approach to predict the nodal status in clinically negative breast cancer patients. Sci. Rep. 2022, 12, 1–10. [Google Scholar] [CrossRef]
- Ren, J.; He, T.; Li, Y.; Liu, S.; Du, Y.; Jiang, Y.; Wu, C. Network-based regularization for high dimensional SNP data in the case–control study of Type 2 diabetes. BMC Genet. 2017, 18, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Ma, S. A selective review of robust variable selection with applications in bioinformatics. Brief. Bioinform. 2015, 16, 873–883. [Google Scholar] [CrossRef] [PubMed]



| N. Patients | N. Positive/ N. Negative | |
|---|---|---|
| Overall | 635 | |
| Histologic Type | ||
| Ductal | 512 | |
| Lobular | 67 | |
| Special type | 56 | |
| Diameter | ||
| T1a | 31 | |
| T1b | 125 | |
| T1c | 281 | |
| T2 | 198 | |
| ER | ||
| Positive | 571 | |
| Negative | 64 | |
| Grading | ||
| G1 | 175 | |
| G2 | 287 | |
| G3 | 173 | |
| HER2 | ||
| 0 | 471 | |
| 1 | 78 | |
| 2 | 46 | |
| 3 | 39 | |
| Multifocality | ||
| Positive | 143 | |
| Negative | 492 | |
| In situ component | ||
| Positive | 369 | |
| Negative | 266 |
| Classification Scheme | SENS | SPEC | ACC | AUC |
|---|---|---|---|---|
| RF | ||||
| RF SUB | ||||
| RF SMOTE | ||||
| Logit Lasso | ||||
| Logit Lasso SUB | ||||
| Logit Lasso SMOTE |
| Classification Scheme | SENS | SPEC | ACC | AUC |
|---|---|---|---|---|
| RF | ||||
| RF SUB | ||||
| RF SMOTE | ||||
| Logit Lasso | ||||
| Logit Lasso SUB | ||||
| Logit Lasso SMOTE |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lombardi, A.; Amoroso, N.; Bellantuono, L.; Bove, S.; Comes, M.C.; Fanizzi, A.; La Forgia, D.; Lorusso, V.; Monaco, A.; Tangaro, S.; et al. Accurate Evaluation of Feature Contributions for Sentinel Lymph Node Status Classification in Breast Cancer. Appl. Sci. 2022, 12, 7227. https://doi.org/10.3390/app12147227
Lombardi A, Amoroso N, Bellantuono L, Bove S, Comes MC, Fanizzi A, La Forgia D, Lorusso V, Monaco A, Tangaro S, et al. Accurate Evaluation of Feature Contributions for Sentinel Lymph Node Status Classification in Breast Cancer. Applied Sciences. 2022; 12(14):7227. https://doi.org/10.3390/app12147227
Chicago/Turabian StyleLombardi, Angela, Nicola Amoroso, Loredana Bellantuono, Samantha Bove, Maria Colomba Comes, Annarita Fanizzi, Daniele La Forgia, Vito Lorusso, Alfonso Monaco, Sabina Tangaro, and et al. 2022. "Accurate Evaluation of Feature Contributions for Sentinel Lymph Node Status Classification in Breast Cancer" Applied Sciences 12, no. 14: 7227. https://doi.org/10.3390/app12147227
APA StyleLombardi, A., Amoroso, N., Bellantuono, L., Bove, S., Comes, M. C., Fanizzi, A., La Forgia, D., Lorusso, V., Monaco, A., Tangaro, S., Zito, F. A., Bellotti, R., & Massafra, R. (2022). Accurate Evaluation of Feature Contributions for Sentinel Lymph Node Status Classification in Breast Cancer. Applied Sciences, 12(14), 7227. https://doi.org/10.3390/app12147227













