Development of Novel Markers and Creation of Non-Anthocyanin and Anthocyanin-Rich Broccoli (Brassica oleracea var. italica) Cultivars
Abstract
:1. Introduction
2. Materials and Methods
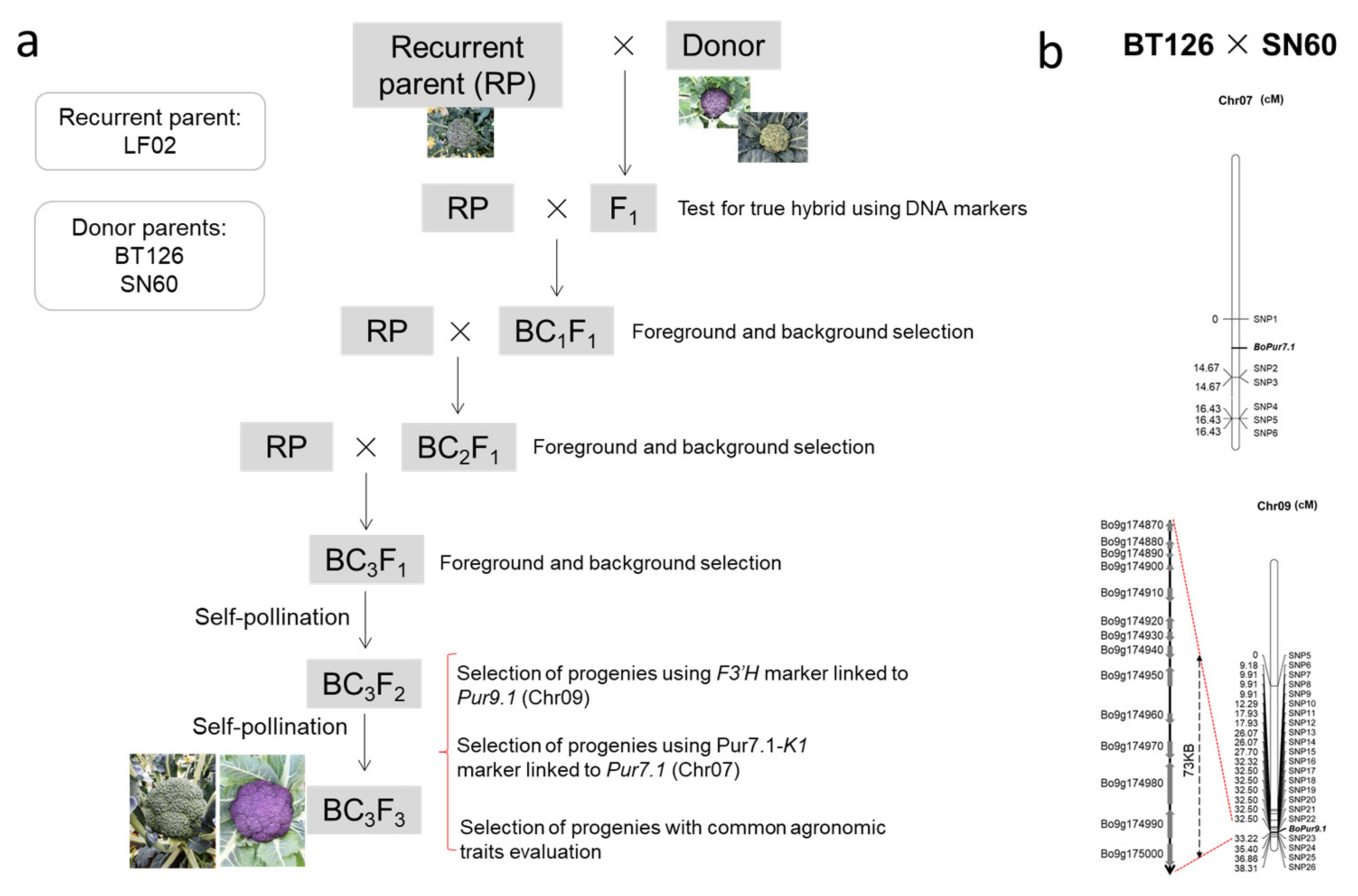
2.1. Materials and Breeding Procedures
2.2. DNA Extraction
2.3. Primer Synthesis and PCR Detection
2.4. Anthocyanin Content Identification
2.5. Field Planting and Agronomic Trait Examination
3. Results
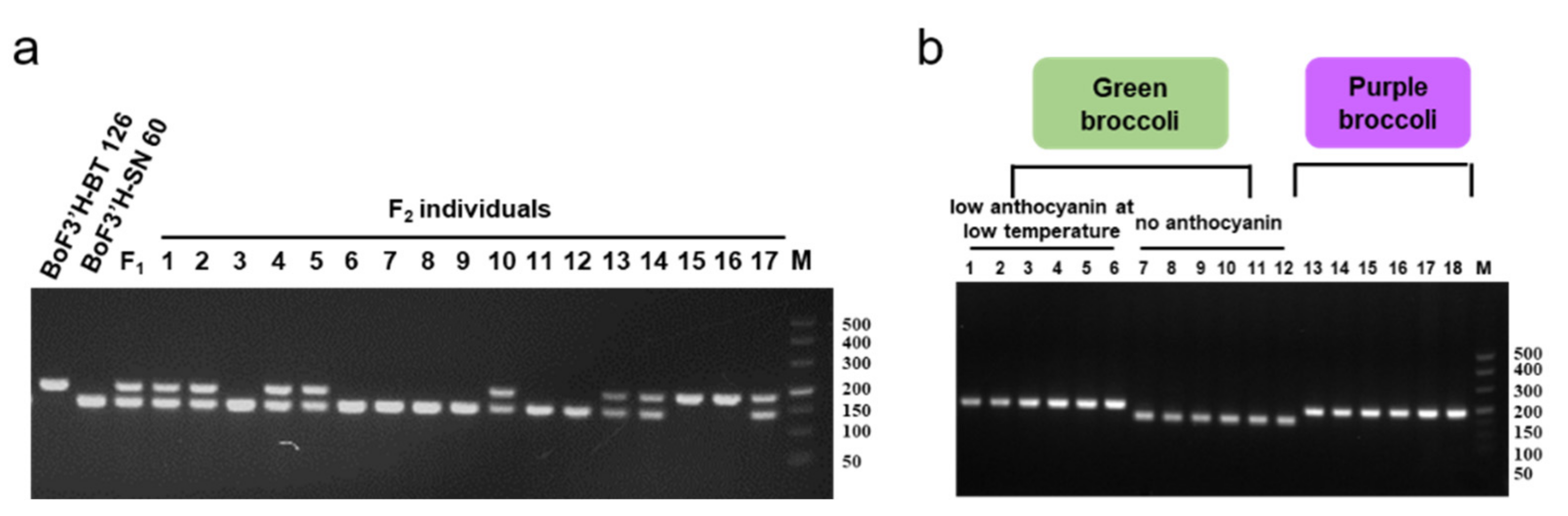
3.1. Analysis of F3′H Homologous Sequences and Development of Specific Markers
3.2. Crossbreeding and Backcross Transfer
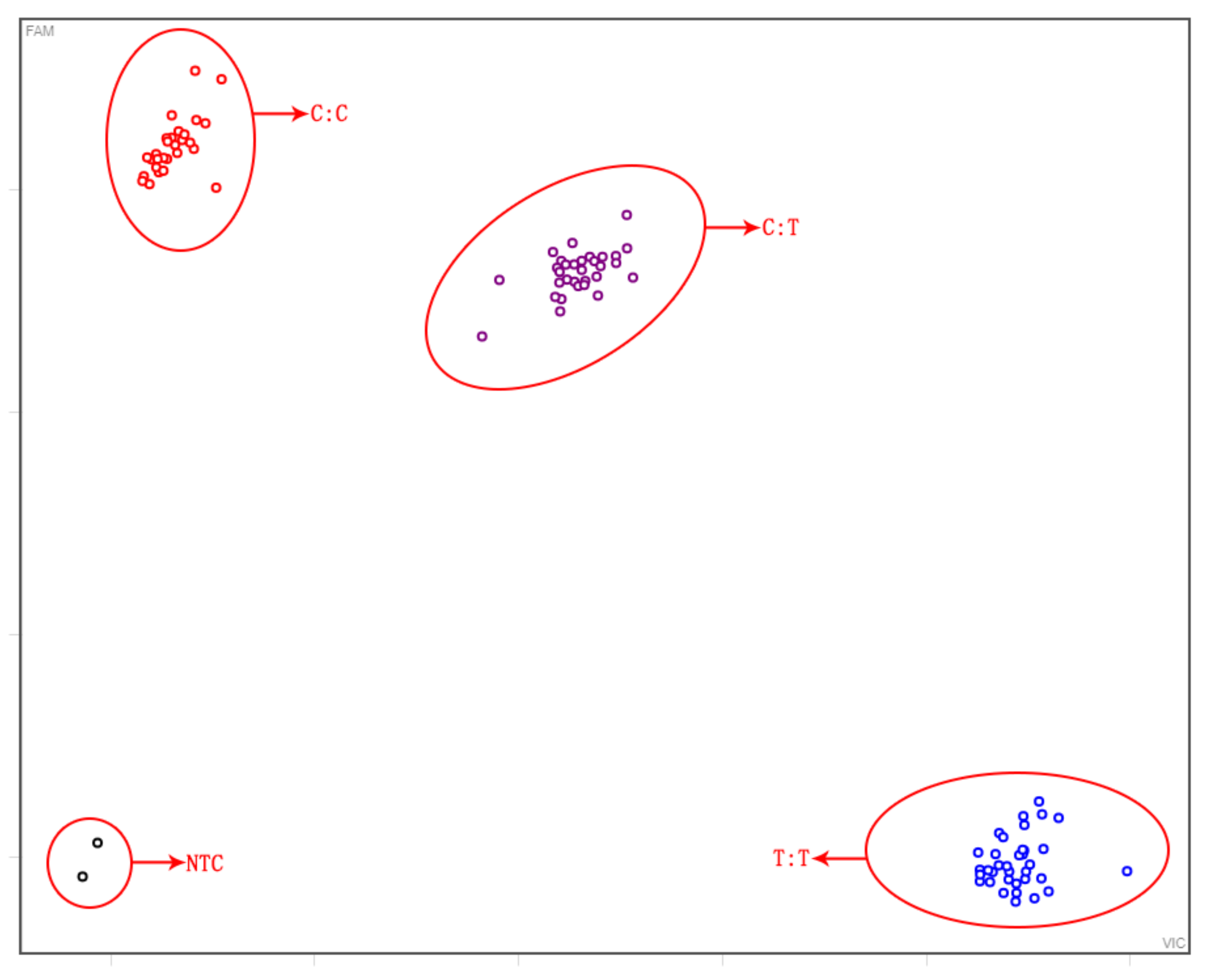
3.3. Screening of Foreground and Background Selection Primers
3.4. Agronomic Traits and Yield Performance in the Field
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Adefegha, S.A. Functional foods and nutraceuticals as dietary intervention in chronic diseases; novel perspectives for health promotion and disease prevention. J. Diet. Suppl. 2018, 15, 977–1009. [Google Scholar] [CrossRef] [PubMed]
- Gould, K.S.; Markham, K.R.; Smith, R.H.; Goris, J.J. Functional role of anthocyanins in the leaves of Quintinia serrata A. Cunn. J. Exp. Bot. 2000, 51, 1107–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalker-Scott, L. Environmental significance of anthocyanins in plant stress responses. Photochem. Photobiol. 1999, 70, 1–9. [Google Scholar] [CrossRef]
- Abdull Razis, A.F.; Iori, R.; Ioannides, C. The natural chemopreventive phytochemical R-sulforaphane is a far more potent inducer of the carcinogen-detoxifying enzyme systems in rat liver and lung than the S-isomer. Int. J. Cancer 2011, 128, 2775–2782. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-R.; Chen, M.; Lee, J.D.; Zhang, J.; Lin, S.-Y.; Fu, T.-M.; Chen, H.; Ishikawa, T.; Chiang, S.-Y.; Katon, J. Reactivation of PTEN tumor suppressor for cancer treatment through inhibition of a MYC-WWP1 inhibitory pathway. Science 2019, 364, eaau0159. [Google Scholar] [CrossRef] [PubMed]
- Crisp, P.; Gray, A.; James, H.; Ives, S.; Angell, S. Improving purple sprouting broccoli by breeding. J. Hortic. Sci. 1985, 60, 325–333. [Google Scholar] [CrossRef]
- Rodríguez-Hernández, M.d.C.; Moreno, D.A.; Carvajal, M.; García-Viguera, C.; Martínez-Ballesta, M.d.C. Natural antioxidants in purple sprouting broccoli under Mediterranean climate. J. Food Sci. 2012, 77, C1058–C1063. [Google Scholar] [CrossRef]
- Porter, Y. Antioxidant properties of green broccoli and purple-sprouting broccoli under different cooking conditions. Bioscience Horizons. Int. J. Stud. Res. 2012, 5, hzs004. [Google Scholar]
- Chaudhary, A.; Choudhary, S.; Sharma, U.; Vig, A.P.; Singh, B.; Arora, S. Purple head broccoli (Brassica oleracea L. var. italica Plenck), a functional food crop for antioxidant and anticancer potential. J. Food Sci. Technol. 2018, 55, 1806–1815. [Google Scholar]
- Moreno, D.A.; Pérez-Balibrea, S.; Ferreres, F.; Gil-Izquierdo, Á.; García-Viguera, C. Acylated anthocyanins in broccoli sprouts. Food Chem. 2010, 123, 358–363. [Google Scholar] [CrossRef]
- Liu, C.; Yao, X.; Li, G.; Huang, L.; Xie, Z. Transcriptomic profiling of purple broccoli reveals light-induced anthocyanin biosynthetic signaling and structural genes. PeerJ 2020, 8, e8870. [Google Scholar] [CrossRef] [PubMed]
- Clifford, M.N. Anthocyanins–nature, occurrence and dietary burden. J. Sci. Food Agric. 2000, 80, 1063–1072. [Google Scholar] [CrossRef]
- Fleschhut, J.; Kratzer, F.; Rechkemmer, G.; Kulling, S.E. Stability and biotransformation of various dietary anthocyanins in vitro. Eur. J. Nutr. 2006, 45, 7–18. [Google Scholar] [CrossRef]
- Ballaré, C.L. Stress under the sun: Spotlight on ultraviolet-B responses. Am. Soc. Plant Biol. 2003, 132, 1725–1727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chao, P.Y.; Huang, W.Y.; Hu, S.P.; Lo, H.F.; Lin, K.H.; Huang, M.Y.; Chang, T.R.; Yang, C.M. Indigenous purple vegetable extracts protect against hydrogen peroxide-induced DNA damage in human lymphocytes. Food Nutr. Sci. 2013, 4, 35258. [Google Scholar] [CrossRef] [Green Version]
- Tajalli, F.; Saeedi, M.; Malekabadi, A.V. Anticancer and antioxidant effects of red cabbage on three cancerous cell lines and comparison with a normal cell line (HFF-3). J. Genes Cells 2020, 6, 12–20. [Google Scholar] [CrossRef]
- Houghton, A.; Appelhagen, I.; Martin, C. Natural blues: Structure meets function in anthocyanins. Plants 2021, 10, 726. [Google Scholar] [CrossRef]
- Wu, M.; Lv, X.; Zhou, Y.; Zeng, Y.; Liu, D. High anthocyanin accumulation in an Arabidopsis mutant defective in chloroplast biogenesis. Plant Growth Regul. 2019, 87, 433–444. [Google Scholar] [CrossRef]
- Paulsmeyer, M.N.; Brown, P.J.; Juvik, J.A. Discovery of anthocyanin acyltransferase1 (AAT1) in maize using genotyping-by-sequencing (GBS). G3: Genes Genomes Genet. 2018, 8, 3669–3678. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Xu, X.; Huang, R.; Yang, S.; Li, M.; Guo, Y. CRISPR/Cas9-mediated targeted mutation reveals a role for AN4 rather than DPL in regulating venation formation in the corolla tube of Petunia hybrida. Hortic. Res. 2021, 8, 116. [Google Scholar] [CrossRef]
- He, F.; Mu, L.; Yan, G.L.; Liang, N.N.; Pan, Q.H.; Wang, J.; Reeves, M.J.; Duan, C.Q. Biosynthesis of anthocyanins and their regulation in colored grapes. Molecules 2010, 15, 9057–9091. [Google Scholar] [CrossRef] [Green Version]
- Mushtaq, M.A.; Pan, Q.; Chen, D.; Zhang, Q.; Ge, X.; Li, Z. Comparative leaves transcriptome analysis emphasizing on accumulation of anthocyanins in Brassica: Molecular regulation and potential interaction with photosynthesis. Front. Plant Sci. 2016, 7, 311. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.F.; Chen, Y.Z.; Kawabata, S.; Li, Y.H.; Wang, Y. Identification of light-independent anthocyanin biosynthesis mutants induced by ethyl methane sulfonate in Turnip “Tsuda”(Brassica rapa). Int. J. Mol. Sci. 2017, 18, 1288. [Google Scholar] [CrossRef] [Green Version]
- Jin, S.W.; Rahim, M.A.; Kim, H.T.; Park, J.I.; Kang, J.G.; Nou, I.S. Molecular analysis of anthocyanin-related genes in ornamental cabbage. Genome 2018, 61, 111–120. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Chen, G.; Dong, T.; Pan, Y.; Zhao, Z.; Tian, S.; Hu, Z. Anthocyanin accumulation and transcriptional regulation of anthocyanin biosynthesis in purple bok choy (Brassica rapa var. chinensis). J. Agric. Food Chem. 2014, 62, 12366–12376. [Google Scholar] [CrossRef]
- Chiu, L.W.; Zhou, X.; Burke, S.; Wu, X.; Prior, R.L.; Li, L. The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiol. 2010, 154, 1470–1480. [Google Scholar] [CrossRef] [Green Version]
- Holton, T.A.; Cornish, E.C. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 1995, 7, 1071. [Google Scholar] [CrossRef]
- Chiu, L.W.; Li, L. Characterization of the regulatory network of BoMYB2 in controlling anthocyanin biosynthesis in purple cauliflower. Planta 2012, 236, 1153–1164. [Google Scholar] [CrossRef]
- Song, H.; Yi, H.; Lee, M.; Han, C.T.; Lee, J.; Kim, H.; Park, J.I.; Nou, I.S.; Kim, S.J.; Hur, Y. Purple Brassica oleracea var. capitata F. rubra is due to the loss of BoMYBL2–1 expression. BMC Plant Biol. 2018, 18, 82. [Google Scholar]
- Wang, Y.; Wang, H.; Gao, M.; Fan, Z.; Chen, Y.; Jin, Y. Overexpression of kale (Brassica oleracea L. var. acephala) BoMYB increases anthocyanin content in Arabidopsis thaliana. Biotechnol. Biotechnol. Equip. 2019, 33, 902–910. [Google Scholar]
- Schoenbohm, C.; Martens, S.; Eder, C.; Forkmann, G.; Weisshaar, B. Identification of the Arabidopsis thaliana flavonoid 3′-hydroxylase gene and functional expression of the encoded P450 enzyme. Biol. Chem. 2000, 381, 749–753. [Google Scholar] [CrossRef] [PubMed]
- Vikhorev, A.V.; Strygina, K.V.; Khlestkina, E.K. Duplicated flavonoid 3′-hydroxylase and flavonoid 3′, 5′-hydroxylase genes in barley genome. PeerJ 2019, 7, e6266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, J.; Li, S.; Wang, Q.; Yuan, Y.; Ma, J.; Xu, M.; Yang, Y.; Zhang, C.; Chen, L.; Sun, Y. Construction of a high-density genetic map based on specific-locus amplified fragment sequencing and identification of loci controlling anthocyanin pigmentation in Yunnan red radish. Hortic. Res. 2022, 9, uhab031. [Google Scholar] [CrossRef]
- Fang, H.; Dong, Y.; Yue, X.; Hu, J.; Jiang, S.; Xu, H.; Wang, Y.; Su, M.; Zhang, J.; Zhang, Z. The B-box zinc finger protein MdBBX20 integrates anthocyanin accumulation in response to ultraviolet radiation and low temperature. Plant Cell Environ. 2019, 42, 2090–2104. [Google Scholar] [CrossRef]
- Nguyen, N.H. HY5, an integrator of light and temperature signals in the regulation of anthocyanins biosynthesis in Arabidopsis. AIMS Mol. Sci. 2020, 7, 70–81. [Google Scholar] [CrossRef]
- Guo, J.; Han, W.; Wang, M. Ultraviolet and environmental stresses involved in the induction and regulation of anthocyanin biosynthesis: A review. Afr. J. Biotechnol. 2008, 7, 25. [Google Scholar] [CrossRef]
- He, Q.; Ren, Y.; Zhao, W.; Li, R.; Zhang, L. Low temperature promotes anthocyanin biosynthesis and related gene expression in the seedlings of purple head Chinese cabbage (Brassica rapa L.). Genes 2020, 11, 81. [Google Scholar] [CrossRef] [Green Version]
- Gould, K.S.; Kuhn, D.N.; Lee, D.W.; Oberbauer, S.F. Why leaves are sometimes red. Nature 1995, 378, 241–242. [Google Scholar] [CrossRef]
- Pietrini, F.; Iannelli, M.; Massacci, A. Anthocyanin accumulation in the illuminated surface of maize leaves enhances protection from photo-inhibitory risks at low temperature, without further limitation to photosynthesis. Plant Cell Environ. 2002, 25, 1251–1259. [Google Scholar] [CrossRef]
- Liu, C.Q.; Li, G.Q.; Yao, X.Q.; Huang, L.; Wu, X.Y.; Xie, Z.J. Characterization of Ogura CMS fertility-restored interspecific hybrids and backcross progenies from crosses between broccoli and rapeseed. Euphytica 2020, 216, 194. [Google Scholar] [CrossRef]
- Yu, H.; Wang, J.; Sheng, X.; Zhao, Z.; Shen, Y.; Branca, F.; Gu, H. Construction of a high-density genetic map and identification of loci controlling purple sepal trait of flower head in Brassica oleracea L. italica. BMC Plant Biol. 2019, 19, 228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahim, M.A.; Afrin, K.S.; Jung, H.J.; Kim, H.T.; Park, J.I.; Hur, Y.; Nou, I.S. Molecular analysis of anthocyanin biosynthesis-related genes reveal BoTT8 associated with purple hypocotyl of broccoli (Brassica oleracea var. italica L.). Genome 2019, 62, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Hospital, F.; Chevalet, C.; Mulsant, P. Using markers in gene introgression breeding programs. Genetics 1992, 132, 1199–1210. [Google Scholar] [CrossRef]
- Nadeem, M.A.; Nawaz, M.A.; Shahid, M.Q.; Doğan, Y.; Comertpay, G.; Yıldız, M.; Hatipoğlu, R.; Ahmad, F.; Alsaleh, A.; Labhane, N. DNA molecular markers in plant breeding: Current status and recent advancements in genomic selection and genome editing. Biotechnol. Biotechnol. Equip. 2018, 32, 261–285. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Kalia, P.; Meena, R.K.; Mangal, M.; Islam, S.; Saha, S.; Tomar, B.S. Genetics and expression analysis of anthocyanin accumulation in curd portion of Sicilian purple to facilitate biofortification of Indian cauliflower. Front. Plant Sci. 2020, 10, 1766. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Yao, X.; Li, G.; Huang, L.; Wu, X.; Xie, Z. Identification of Major Loci and Candidate Genes for Anthocyanin Biosynthesis in Broccoli Using QTL-Seq. Horticulturae 2021, 7, 246. [Google Scholar] [CrossRef]
- Kidwell, K.K.; Osborn, T.C. Simple plant DNA isolation procedures. In Plant Genomes: Methods for Genetic and Physical Mapping; Springer: Berlin, Germany, 1992; pp. 1–13. [Google Scholar]
- Li, H.; Chen, X.; Yang, Y.; Xu, J.; Gu, J.; Fu, J.; Qian, X.; Zhang, S.; Wu, J.; Liu, K. Development and genetic mapping of microsatellite markers from whole genome shotgun sequences in Brassica oleracea. Mol. Breed. 2011, 28, 585–596. [Google Scholar] [CrossRef]
- Li, H.; Younas, M.; Wang, X.; Li, X.; Chen, L.; Zhao, B.; Chen, X.; Xu, J.; Hou, F.; Hong, B. Development of a core set of single-locus SSR markers for allotetraploid rapeseed (Brassica napus L.). Appl. Genet. 2013, 126, 937–947. [Google Scholar] [CrossRef]
- Lee, J.; Durst, R.W.; Wrolstad, R.E. Determination of total monomeric anthocyanin pigment content of fruit juices, beverages, natural colorants, and wines by the pH differential method: Collaborative study. J AOAC Int. 2005, 88, 1269–1278. [Google Scholar] [CrossRef] [Green Version]
- Welch, C.R.; Wu, Q.; Simon, J.E. Recent advances in anthocyanin analysis and characterization. Curr. Anal. Chem. 2008, 4, 75–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, N.; Cheng, F.; Wu, J.; Liu, B.; Zheng, S.; Liang, J.; Wang, X. Anthocyanin biosynthetic genes in Brassica rapa. BMC Genom. 2014, 15, 426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konczak, I.; Zhang, W. Anthocyanins—More than nature’s colours. J. Biomed. Biotechnol. 2004, 2004, 239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordeeva, T.; Sheldon, K.; Sychev, O. Linking academic performance to optimistic attributional style: Attributions following positive events matter most. Eur. J. Psychol. Educ. 2020, 35, 21–48. [Google Scholar] [CrossRef]
- Xu, Y.; Crouch, J.H. Marker-assisted selection in plant breeding: From publications to practice. Crop Sci. 2008, 48, 391–407. [Google Scholar] [CrossRef] [Green Version]
- Masukawa, T.; Cheon, K.S.; Mizuta, D.; Nakatsuka, A.; Kobayashi, N. Insertion of a Retrotransposon into a Flavonoid 3′-hydroxylase Homolog Confers the Red Root Character in the Radish (Raphanus sativus L. var. longipinnatus LH Bailey). Hortic. J. 2018, 87, 89–96. [Google Scholar] [CrossRef] [Green Version]
- Varshney, R.K.; Mohan, S.M.; Gaur, P.M.; Gangarao, N.; Pandey, M.K.; Bohra, A.; Sawargaonkar, S.L.; Chitikineni, A.; Kimurto, P.K.; Janila, P. Achievements and prospects of genomics-assisted breeding in three legume crops of the semi-arid tropics. Biotechnol. Adv. 2013, 31, 1120–1134. [Google Scholar] [CrossRef] [Green Version]
- Visscher, P.M.; Haley, C.S.; Thompson, R. Marker-assisted introgression in backcross breeding programs. Genetics 1996, 144, 1923–1932. [Google Scholar] [CrossRef]
- Miah, G.; Rafii, M.Y.; Ismail, M.R.; Puteh, A.B.; Rahim, H.A.; Latif, M.A. Recurrent parent genome recovery analysis in a marker-assisted backcrossing program of rice (Oryza sativa L.). C. R. Biol. 2015, 338, 83–94. [Google Scholar] [CrossRef]
- Agnola, B.; Silue, D.; Boury, S. Identification of RAPD markers of downy mildew (Peronospora parasitica) resistance gene in broccoli (Brassica oleracea var. italica). In Proceedings of the 3rd ISHS international symposium on Brassicas, Horticulture Research International, Wellesbourne, UK, 5–9 September 2000; pp. 5–9. [Google Scholar]
- Farinho, M.; Coelho, P.; Monteiro, A.; Leitao, J. RAPD and AFLP markers linked to Peronospora parasitica resistance genes in broccoli. In Proceedings of the 3rd ISHS International Symposium on Brassicas, Horticulture Research International, Wellesbourne, UK, 5–9 September 2000; p. 75. [Google Scholar]
- Han, F.; Liu, Y.; Fang, Z.; Yang, L.; Zhuang, M.; Zhang, Y.; Lv, H.; Wang, Y.; Ji, J.; Li, Z. Advances in Genetics and Molecular Breeding of Broccoli. Horticulturae 2021, 7, 280. [Google Scholar] [CrossRef]



| Line | Color | Anthocyanin Content | Characterization |
|---|---|---|---|
| BT126 | Red | High anthocyanin | Late ripening |
| SN60 | Green | No anthocyanin | Early ripening |
| LF02 | Green | Low anthocyanin at low temperatures | Medium ripening and good head quality |
| Marker Name | Primer Sequence | Linked to Pur Gene |
|---|---|---|
| F3′H-F/R | CACGGACATGCTTAGCACTTTAAT | Pur9.1 |
| GCCGTAAACATGTTCTGTAATGGA | ||
| Pur7.1-K1 | GAAGGTGACCAAGTTCATGCTCTCGTGGCTTCTCTACAATTATCG | Pur7.1 |
| GAAGGTCGGAGTCAACGGATTACTCGTCGCTTCTCTACAATTATCA | ||
| TGGTTAAGACAAAGTCACAAAACCAT | ||
| Pur7.2-K2 | GAAGGTGACCAAGTTCATGCTATCGAGTCCGAAGACGAAACGG | Pur7.1 |
| GAAGGTCGGAGTCAACGGATTCATCGAGTCAGAAGACGAAACGC | ||
| CATCACTCCCCAGTCCATAGCA |
| Parents and Lines | Growth Period (d) | Plant Height (cm) | Plant Spread (cm) | Head Length (cm) | Head Diameter (cm) | Head Shape | Stalk Diameter (cm) | Anthocyanin Content (mg/100 g FW) | Head Color | Glucoraphanin Content (µmol/g DW) | Head Weight (g) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Purple broccoli | |||||||||||
| BT126 | 90–100 | 63.2 | 82.03 | 6.36 | 13.85 | Flat | 4.13 | 63 | Purple | 8.75 | 400.0 |
| 20PBL23 | 90–100 | 62.4 * | 90 * | 8.48 * | 14.62 * | Round | 4.63 * | 46 ** | Purple | 7.11 | 523.3 |
| 20PBL62 | 90–100 | 68.5 | 78 * | 10.2 | 15.45 | Round | 5.1 | 57 ** | Purple | 5.26 | 617.3 |
| 20PBL103 | 85–95 | 66.35 | 84.55 * | 9.11 | 14.37 * | Semi-round | 4.62 * | 71 ** | Purple | 6.39 | 592.2 |
| Green broccoli | |||||||||||
| SN60 | 70–75 | 79.45 | 79.3 | 7.03 | 12.25 | Round | 4.34 | 1.5 | Light green | 3.24 | 450.2 |
| LF02 | 85–90 | 70.7 | 67.01 | 10.72 | 15.9 | Round | 5.1 | 12 | Green-purple | 7.32 | 620.0 |
| 20GBL47 | 70–80 | 70.67 | 68 | 9.56 | 14.5 * | Round | 5.14 | 1.3 ** | Green | 6.02 | 600.4 |
| 20GBL128 | 75–85 | 76.3 | 73.32 * | 9.74 | 15.35 | Round | 4.9 | 1.7 ** | Dark green | 4.65 * | 571.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Yao, X.; Li, G.; Huang, L.; Liu, C.; Xie, Z. Development of Novel Markers and Creation of Non-Anthocyanin and Anthocyanin-Rich Broccoli (Brassica oleracea var. italica) Cultivars. Appl. Sci. 2022, 12, 6267. https://doi.org/10.3390/app12126267
Liu C, Yao X, Li G, Huang L, Liu C, Xie Z. Development of Novel Markers and Creation of Non-Anthocyanin and Anthocyanin-Rich Broccoli (Brassica oleracea var. italica) Cultivars. Applied Sciences. 2022; 12(12):6267. https://doi.org/10.3390/app12126267
Chicago/Turabian StyleLiu, Chunqing, Xueqin Yao, Guangqing Li, Lei Huang, Chenghong Liu, and Zhujie Xie. 2022. "Development of Novel Markers and Creation of Non-Anthocyanin and Anthocyanin-Rich Broccoli (Brassica oleracea var. italica) Cultivars" Applied Sciences 12, no. 12: 6267. https://doi.org/10.3390/app12126267
APA StyleLiu, C., Yao, X., Li, G., Huang, L., Liu, C., & Xie, Z. (2022). Development of Novel Markers and Creation of Non-Anthocyanin and Anthocyanin-Rich Broccoli (Brassica oleracea var. italica) Cultivars. Applied Sciences, 12(12), 6267. https://doi.org/10.3390/app12126267





