CleanSeq: A Pipeline for Contamination Detection, Cleanup, and Mutation Verifications from Microbial Genome Sequencing Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Overall Flow of CleanSeq
2.2. Taxonomic Identification
- (i)
- the contamination rate (contamination reads/all reads, threshold = 10%) was over 10%,
- (ii)
- genome similarity [26] between the contaminated and target species was less than 80%.
2.3. Cleanup
2.4. Mutation Call
2.5. Mutation Verification
2.6. Report
2.7. Simulated Dataset
- (i)
- Coverage: 3× or 30×; mutation rate: 0.0001%; error rate: 0.01%.
- (ii)
- Coverage: 30×; mutation rate: 0.01% or 0.0001%; error rate: 0.01%.
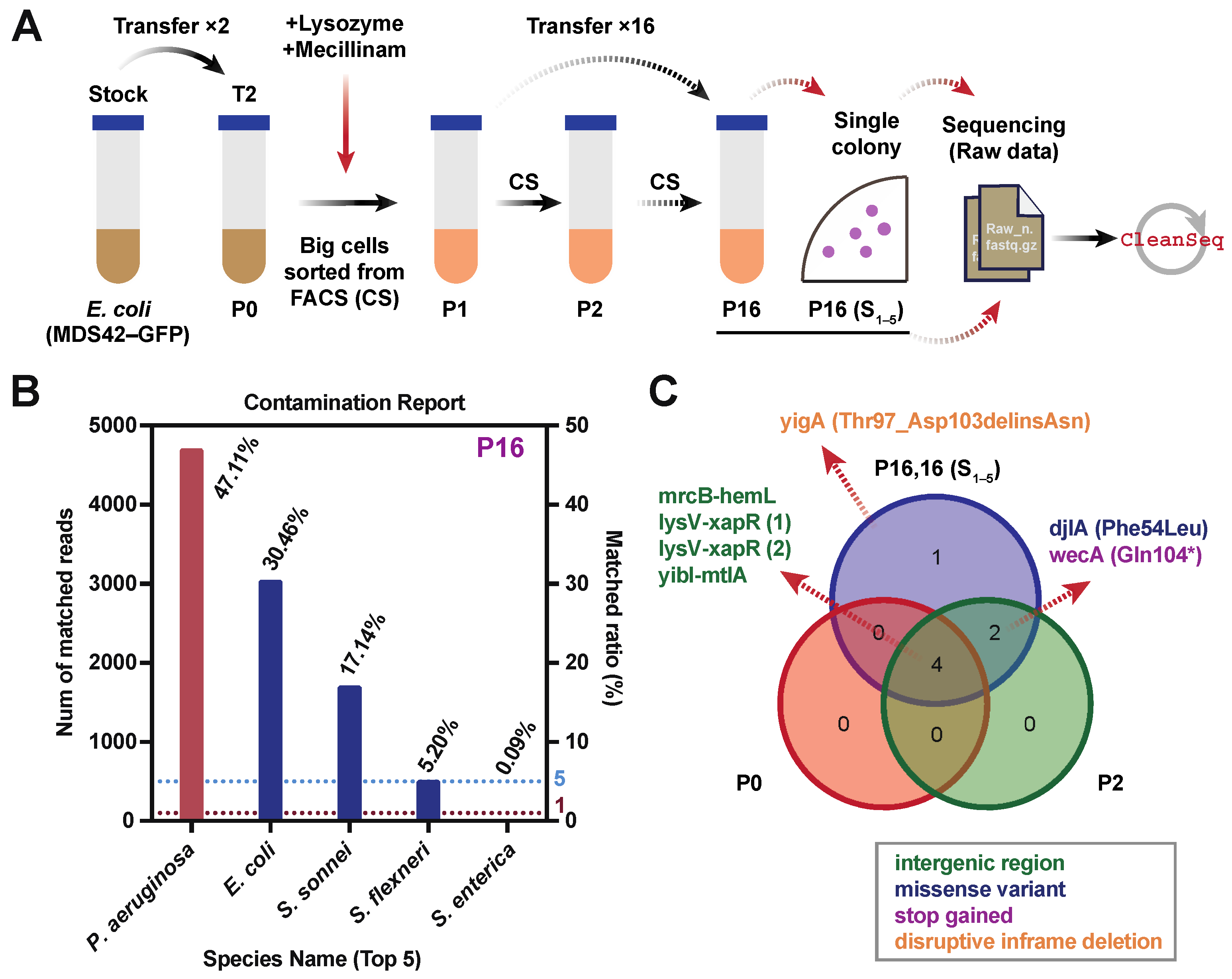
2.8. Real Dataset from Laboratory Experimental Evolution of E. coli
3. Results and Discussion
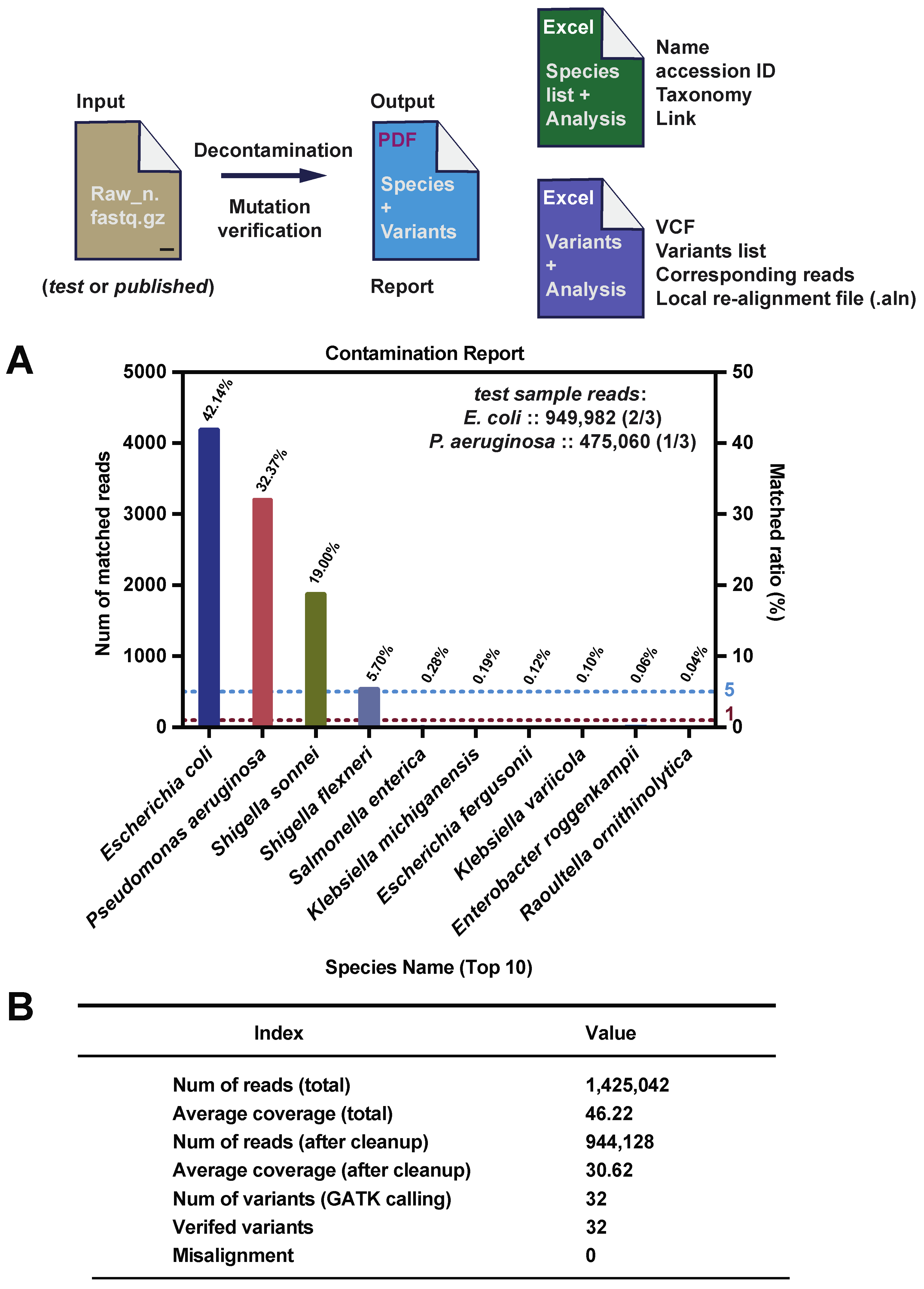
3.1. CleanSeq Processes WGS Raw Data for Contamination Detection, Decontamination, and Calling Variants
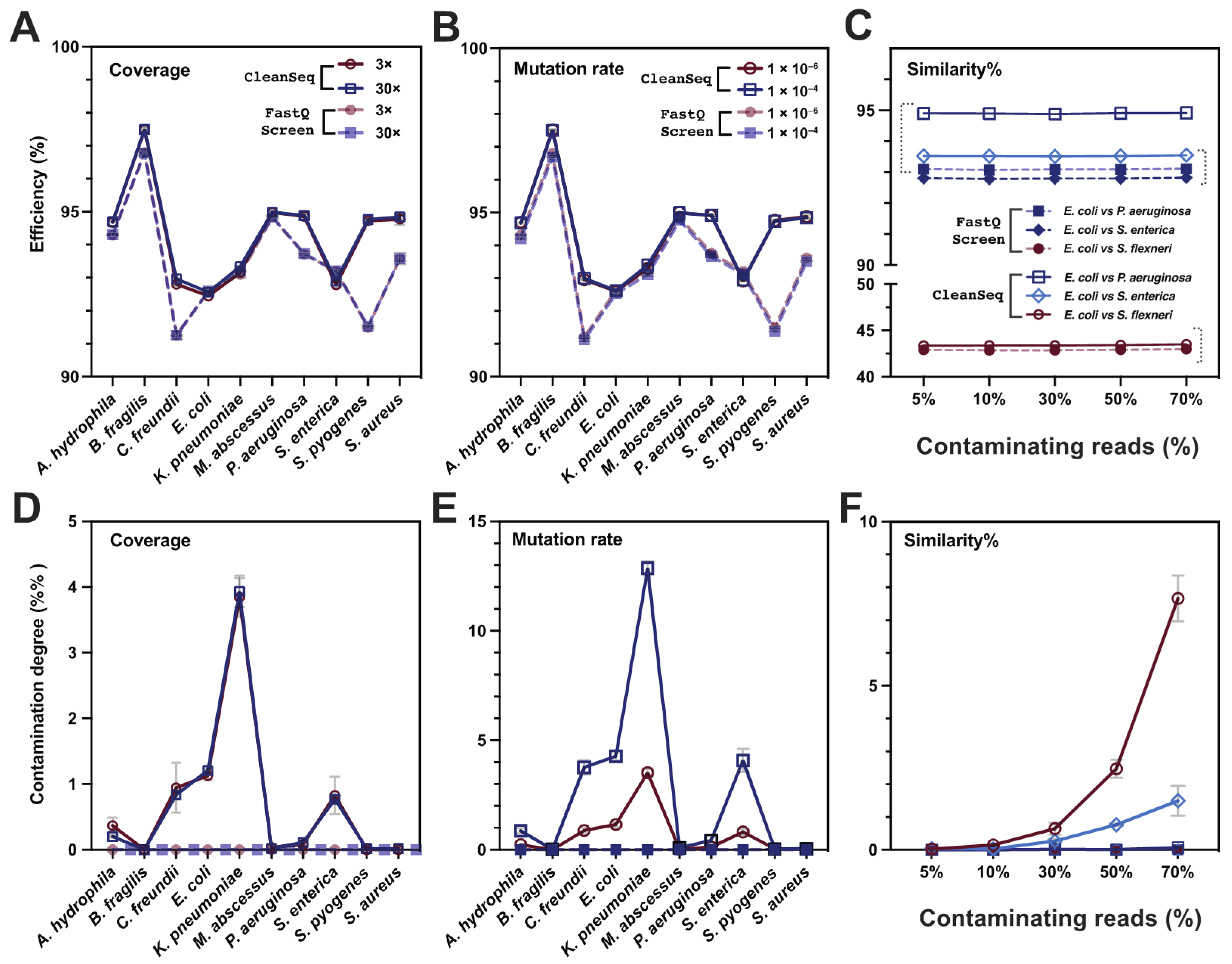
3.2. CleanSeq Detects and Eliminates Mutation Reads Efficiently as Compared with FastQ Screen
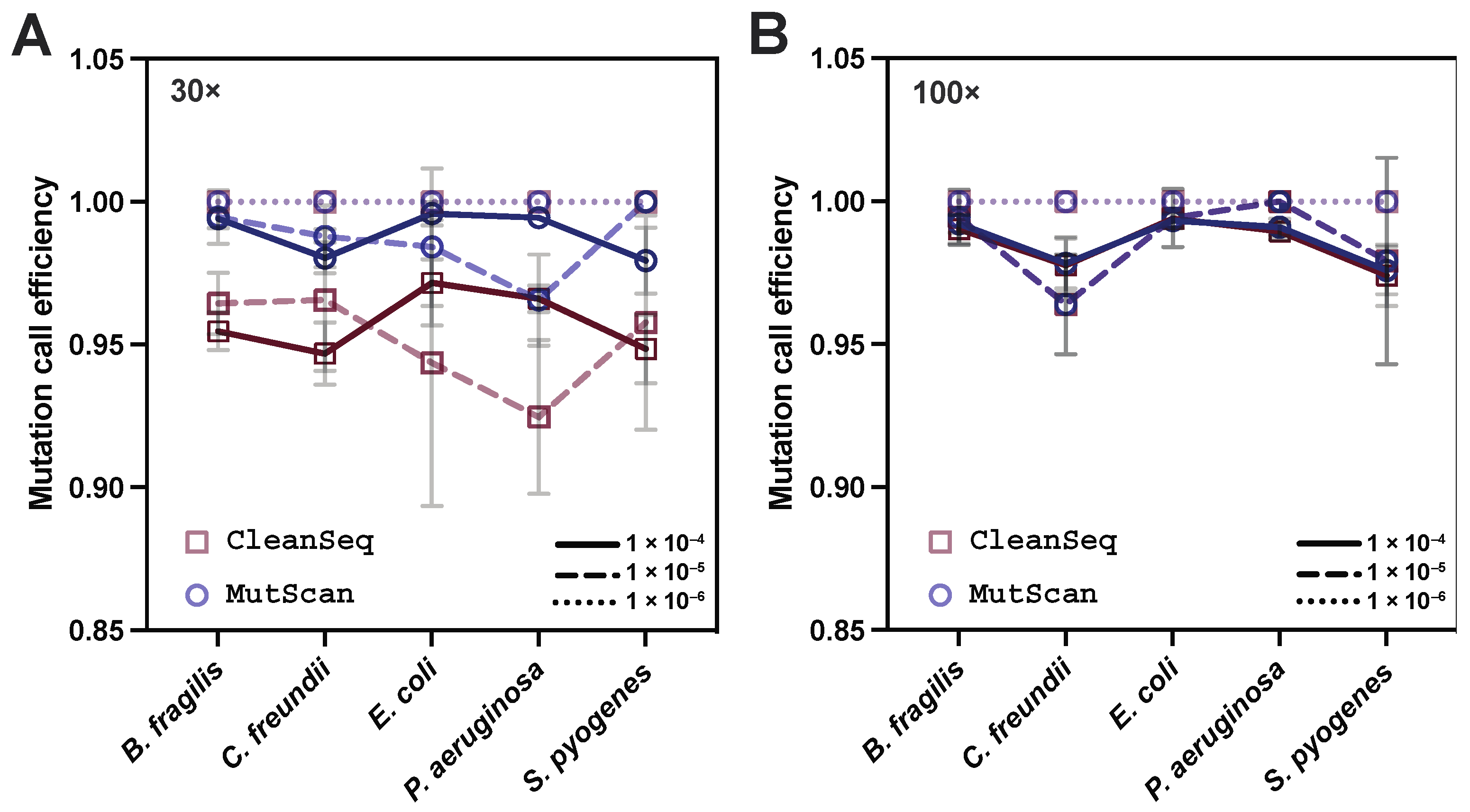
3.3. CleanSeq Calls Variants with Satisfactory Correctness as Compared with MutScan
3.4. CleanSeq Is Practical when Applied to either Simulated or Real Experimental Datasets
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hardwick, S.A.; Deveson, I.W.; Mercer, T.R. Reference standards for next-generation sequencing. Nat. Rev. Genet. 2017, 18, 473–484. [Google Scholar] [CrossRef]
- Strong, M.J.; Xu, G.; Morici, L.; Splinter Bon-Durant, S.; Baddoo, M.; Lin, Z.; Fewell, C.; Taylor, C.M.; Flemington, E.K. Microbial contamination in next generation sequencing: Implications for sequence-based analysis of clinical samples. PLoS Pathog. 2014, 10, e1004437. [Google Scholar] [CrossRef] [Green Version]
- Glassing, A.; Dowd, S.E.; Galandiuk, S.; Davis, B.; Chiodini, R.J. Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathog. 2016, 8, 24. [Google Scholar] [CrossRef] [Green Version]
- Flickinger, M.; Jun, G.; Abecasis, G.R.; Boehnke, M.; Kang, H.M. Correcting for Sample Contamination in Genotype Calling of DNA Sequence Data. Am. J. Hum. Genet. 2015, 97, 284–290. [Google Scholar] [CrossRef] [Green Version]
- Goig, G.A.; Blanco, S.; Garcia-Basteiro, A.L.; Comas, I. Contaminant DNA in bacterial sequencing experiments is a major source of false genetic variability. BMC Biol. 2020, 18, 24. [Google Scholar] [CrossRef]
- Muir, P.; Li, S.; Lou, S.; Wang, D.; Spakowicz, D.J.; Salichos, L.; Zhang, J.; Weinstock, G.M.; Isaacs, F.; Rozowsky, J.; et al. The real cost of sequencing: Scaling computation to keep pace with data generation. Genome Biol. 2016, 17, 53. [Google Scholar] [CrossRef] [Green Version]
- Gallegos, J.E.; Hayrynen, S.; Adames, N.R.; Peccoud, J. Challenges and opportunities for strain verification by whole-genome sequencing. Sci. Rep. 2020, 10, 5873. [Google Scholar] [CrossRef] [Green Version]
- Schwengers, O.; Hoek, A.; Fritzenwanker, M.; Falgenhauer, L.; Hain, T.; Chakraborty, T.; Goesmann, A. ASA3P: An automatic and scalable pipeline for the assembly, annotation and higher-level analysis of closely related bacterial isolates. PLoS Comput. Biol. 2020, 16, e1007134. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.J.C.p.i.b. From FastQ data to high-confidence variant calls: The genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef] [Green Version]
- Low, A.J.; Koziol, A.G.; Manninger, P.A.; Blais, B.; Carrillo, C.D. ConFindr: Rapid detection of intraspecies and cross-species contamination in bacterial whole-genome sequence data. PeerJ 2019, 7, e6995. [Google Scholar] [CrossRef]
- Wingett, S.W.; Andrews, S.J.F. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef]
- Chen, S.; Huang, T.; Wen, T.; Li, H.; Xu, M.; Gu, J.J.B.b. MutScan: Fast detection and visualization of target mutations by scanning FASTQ data. BMC Bioinform. 2018, 19, 16. [Google Scholar] [CrossRef] [Green Version]
- Sangiovanni, M.; Granata, I.; Thind, A.S.; Guarracino, M.R. From trash to treasure: Detecting unexpected contamination in unmapped NGS data. BMC Bioinform. 2019, 20, 168. [Google Scholar] [CrossRef]
- McKnight, D.T.; Huerlimann, R.; Bower, D.S.; Schwarzkopf, L.; Alford, R.A.; Zenger, K.R. microDecon: A highly accurate read-subtraction tool for the post-sequencing removal of contamination in metabarcoding studies. Environ. DNA 2019, 1, 14–25. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS ONE 2011, 6, e17288. [Google Scholar] [CrossRef] [Green Version]
- Caboche, S.; Even, G.; Loywick, A.; Audebert, C.; Hot, D. MICRA: An automatic pipeline for fast characterization of microbial genomes from high-throughput sequencing data. Genome Biol. 2017, 18, 233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.J.; Onizuka, S.; Seki, M.; Suzuki, Y.; Iwata, T.; Nakai, K. A systematic sequencing-based approach for microbial contaminant detection and functional inference. BMC Biol. 2019, 17, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, M.; Nayar, U.; Ludwig, L.S.; Wagle, N.; Rheinbay, E. cDNA-detector: Detection and removal of cDNA contamination in DNA sequencing libraries. BMC Bioinform. 2021, 22, 611. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B.J.B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.-A.; Lin, C.-C.; Wang, C.-D.; Wu, H.-B.; Hwang, P.-I.J.B.G. An optimized procedure greatly improves EST vector contamination removal. BMC Genom. 2007, 8, 416. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Shuaibi, A.; Bell, J.M.; Pavlichin, D.S.; Ji, H.P. Unique k-mer sequences for validating cancer-related substitution, insertion and deletion mutations. NAR Cancer 2020, 2, zcaa034. [Google Scholar] [CrossRef]
- Magoc, T.; Pabinger, S.; Canzar, S.; Liu, X.; Su, Q.; Puiu, D.; Tallon, L.J.; Salzberg, S.L.J.B. GAGE-B: An evaluation of genome assemblers for bacterial organisms. Bioinformatics 2013, 29, 1718–1725. [Google Scholar] [CrossRef]
- Pightling, A.W.; Pettengill, J.B.; Wang, Y.; Rand, H.; Strain, E. Within-species contamination of bacterial whole-genome sequence data has a greater influence on clustering analyses than between-species contamination. Genome Biol. 2019, 20, 286. [Google Scholar] [CrossRef]
- Ying, B.W.; Tsuru, S.; Seno, S.; Matsuda, H.; Yomo, T. Gene expression scaled by distance to the genome replication site. Mol. Biosyst. 2014, 10, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Aida, H.; Kurokawa, M.; Chen, F.; Xia, Y.; Xu, J.; Li, K.; Ying, B.W.; Yomo, T. Primordial mimicry induces morphological change in Escherichia coli. Commun. Biol. 2022, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Kawai, Y.; Mickiewicz, K.; Errington, J. Lysozyme counteracts β-Lactam antibiotics by promoting the emergence of L-Form bacteria. Cell 2018, 172, 1038–1049.e1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osawa, M.; Erickson, H.P. L form bacteria growth in low-osmolality medium. Microbiology 2019, 165, 842–851. [Google Scholar] [CrossRef]
- Sycuro, L.K.; Rule, C.S.; Petersen, T.W.; Wyckoff, T.J.; Sessler, T.; Nagarkar, D.B.; Khalid, F.; Pincus, Z.; Biboy, J.; Vollmer, W.; et al. Flow cytometry-based enrichment for cell shape mutants identifies multiple genes that influence Helicobacter pylori morphology. Mol. Microbiol. 2013, 90, 869–883. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, M.; Tsuru, S.; Hirata, N.; Seno, S.; Matsuda, H.; Ying, B.W.; Yomo, T. Directed evolution of cell size in Escherichia coli. BMC Evol. Biol. 2014, 14, 257. [Google Scholar] [CrossRef] [Green Version]
- Petit, R.A., 3rd; Read, T.D. Bactopia: A flexible pipeline for complete analysis of bacterial genomes. mSystems 2020, 5, e00190-20. [Google Scholar] [CrossRef]
- Quijada, N.M.; Rodriguez-Lazaro, D.; Eiros, J.M.; Hernandez, M. TORMES: An automated pipeline for whole bacterial genome analysis. Bioinformatics 2019, 35, 4207–4212. [Google Scholar] [CrossRef]
- Xavier, B.B.; Mysara, M.; Bolzan, M.; Ribeiro-Goncalves, B.; Alako, B.T.F.; Harrison, P.; Lammens, C.; Kumar-Singh, S.; Goossens, H.; Carrico, J.A.; et al. BacPipe: A rapid, user-friendly whole-genome sequencing pipeline for clinical diagnostic bacteriology. iScience 2020, 23, 100769. [Google Scholar] [CrossRef]
- Devanga Ragupathi, N.K.; Muthuirulandi Sethuvel, D.P.; Inbanathan, F.Y.; Veeraraghavan, B. Accurate differentiation of Escherichia coli and Shigella serogroups: Challenges and strategies. New Microbes New Infect. 2018, 21, 58–62. [Google Scholar] [CrossRef]
- Brenner, D.J.; Fanning, G.R.; Steigerwalt, A.G.; Orskov, I.; Orskov, F. Polynucleotide sequence relatedness among three groups of pathogenic Escherichia coli strains. Infect. Immun. 1972, 6, 308–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sims, D.; Sudbery, I.; Ilott, N.E.; Heger, A.; Ponting, C.P. Sequencing depth and coverage: Key considerations in genomic analyses. Nat. Rev. Genet. 2014, 15, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Razin, S.; Oliver, O. Morphogenesis of Mycoplasma and bacterial L-form colonies. J. Gen. Microbiol. 1961, 24, 225–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genevaux, P.; Schwager, F.; Georgopoulos, C.; Kelley, W.L. The djlA gene acts synergistically with dnaJ in promoting Escherichia coli growth. J. Bacteriol. 2001, 183, 5747–5750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genevaux, P.; Wawrzynow, A.; Zylicz, M.; Georgopoulos, C.; Kelley, W.L. DjlA is a third DnaK co-chaperone of Escherichia coli, and DjlA-mediated induction of colanic acid capsule requires DjlA-DnaK interaction. J. Biol. Chem. 2001, 276, 7906–7912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehrer, J.; Vigeant, K.A.; Tatar, L.D.; Valvano, M.A. Functional characterization and membrane topology of Escherichia coli WecA, a sugar-phosphate transferase initiating the biosynthesis of enterobacterial common antigen and O-antigen lipopolysaccharide. J. Bacteriol. 2007, 189, 2618–2628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senges, C.H.R.; Stepanek, J.J.; Wenzel, M.; Raatschen, N.; Ay, U.; Martens, Y.; Prochnow, P.; Vazquez Hernandez, M.; Yayci, A.; Schubert, B.; et al. Comparison of proteomic responses as global approach to antibiotic mechanism of action elucidation. Antimicrob. Agents Chemother. 2020, 65, e01373-20. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Xia, Y.; Liu, Y.; Kang, C.; Lu, N.; Tian, D.; Lu, H.; Han, F.; Xu, J.; Yomo, T. CleanSeq: A Pipeline for Contamination Detection, Cleanup, and Mutation Verifications from Microbial Genome Sequencing Data. Appl. Sci. 2022, 12, 6209. https://doi.org/10.3390/app12126209
Wang C, Xia Y, Liu Y, Kang C, Lu N, Tian D, Lu H, Han F, Xu J, Yomo T. CleanSeq: A Pipeline for Contamination Detection, Cleanup, and Mutation Verifications from Microbial Genome Sequencing Data. Applied Sciences. 2022; 12(12):6209. https://doi.org/10.3390/app12126209
Chicago/Turabian StyleWang, Caiyan, Yang Xia, Yunfei Liu, Chen Kang, Nan Lu, Di Tian, Hui Lu, Fuhai Han, Jian Xu, and Tetsuya Yomo. 2022. "CleanSeq: A Pipeline for Contamination Detection, Cleanup, and Mutation Verifications from Microbial Genome Sequencing Data" Applied Sciences 12, no. 12: 6209. https://doi.org/10.3390/app12126209
APA StyleWang, C., Xia, Y., Liu, Y., Kang, C., Lu, N., Tian, D., Lu, H., Han, F., Xu, J., & Yomo, T. (2022). CleanSeq: A Pipeline for Contamination Detection, Cleanup, and Mutation Verifications from Microbial Genome Sequencing Data. Applied Sciences, 12(12), 6209. https://doi.org/10.3390/app12126209






