Abstract
Liver cancer contributes to the increasing mortality rate in the world. Therefore, early detection may lead to a decrease in morbidity and increase the chance of survival rate. This research offers a computer-aided diagnosis system, which uses computed tomography scans to categorize hepatic tumors as benign or malignant. The 3D segmented liver from the LiTS17 dataset is passed through a Convolutional Neural Network (CNN) to detect and classify the existing tumors as benign or malignant. In this work, we propose a novel light CNN with eight layers and just one conventional layer to classify the segmented liver. This proposed model is utilized in two different tracks; the first track uses deep learning classification and achieves a 95.6% accuracy. Meanwhile, the second track uses the automatically extracted features together with a Support Vector Machine (SVM) classifier and achieves 100% accuracy. The proposed network is light, fast, reliable, and accurate. It can be exploited by an oncological specialist, which will make the diagnosis a simple task. Furthermore, the proposed network achieves high accuracy without the curation of images, which will reduce time and cost.
1. Introduction
Liver cancer (LC) is a well-known condition across the world. It is among the most frequent types of cancer that may affect humans [1]. It is a lethal disease spreading around the globe, particularly in underdeveloped nations [2]. The liver is the body’s biggest internal organ. Hepatic cancer detection is difficult given the heterogeneous nature of liver tissues. The mortality rate of primary liver cancer can be reduced if it is detected earlier. For detecting the damaged region in liver images, multiple classification algorithms have been implemented [3]. The liver is both required for living and susceptible to a variety of diseases. CT examinations may be utilized to plan and deliver radiation treatment to tumors, as well as to assist biopsies and other less invasive procedures. Manual CT image segmentation and classification is a time-consuming and inefficient method, which is unfeasible for vast amounts of data. Manual interaction is not required with fully automatic and unsupervised approaches [4]. The computer-aided diagnosis of live tumors in CT images requires automatic tumor detection and segmentation. In low-contrast images, the low-level images are too faint to identify, making it a difficult process [5]. Tumor detection and segmentation are critical pre-treatment measures in the computer-aided diagnosis of liver tumors [6,7]. In the liver, there are several different forms of tumors. The visual appearance of various tumors varies, and their visual appearance varies once the contrast medium is administered. Computer-aided diagnosis might be difficult when it comes to segmenting the liver from CT scan images accurately. Automatic liver segmentation is the initial and most important stage in the diagnosing process [8,9].
Radiologists face a difficult problem in identifying and classifying liver tumors. The liver parenchyma must be separated from the abdomen, and the liver cells with the least alteration must be classified as malignant or benign tumors. Owing to its excellent cross-sectional view, outstanding spatial resolution, quick interpretation, and strong signal-to-noise ratio (SNR), CT images remain one of the top modalities of choice. Magnetic Resonance Imaging (MRI), Positron emission tomography (PET), and Ultra Sound (US) are the other major Liver-imaging modalities. CT examinations may be performed for proper planning and managing tumor treatments, including guiding biopsies and other easily established processes. For huge amounts of data, manual segmentation and Computed Axial Tomography (CAT) image categorization are demanding and time-consuming operations. Computer-aided diagnosis (CAD) systems are a type of medical imaging that acts as a second opinion for doctors when interpreting images. Upon creating the final output, the CAD systems are interactive/semi-automated and include the results of the medical practitioner. This contrasts with a fully automated system, wherein the computer software makes all choices. CAD systems have a critical role in the early diagnosis of liver disease, lowering the fatality rate from liver cancer [10]. The utilization of CT images to identify the liver disease is prevalent. Given the various intensities, it might be challenging for even competent radiologists to remark on the type, category, and level of the tumor immediately from the CT image. Designing and developing computer-assisted imaging techniques to aid physicians/doctors in enhancing their diagnoses has become increasingly significant in recent years [11]. The diagnosis and treatment strategy are determined by classifying the lesion type and time based on CT images, which demands professional knowledge and expertise to categorize. Once the workload is severe, fatigue is common, and even competent senior specialists have trouble preventing a misdiagnosis. Deep learning may overcome the limitations of conventional machine learning, for instance, the time required to retrieve image features and conduct dimensionality reduction manually, giving high-dimensional image features. It is critical to use deep learning to aid doctors in diagnosis. The poor accuracy of tumor classification, the limited capability of feature extraction, and the sparse dataset remain challenges in the current medical image classification task [12].
In 2018, Amita Das et al. [13] developed a Watershed Gaussian Deep Learning algorithm for classifying three forms of liver cancer, including hepatocellular carcinoma, hemangioma, and metastatic carcinoma, utilizing 225 images. The watershed algorithm was utilized to segregate the liver, Gaussian Mixture Models (GMM) were utilized to detect the lesion region, and retrieved characteristics were fed into a Deep Neural Network (DNN). They were able to obtain 97.72% specificity, 100% sensitivity, and 98.38% testing accuracy. Consequently, Koichiro Yasaka et al. [14] trained a Convolutional Neural Network (CNN) to distinguish liver lesions into five categories utilizing 1068 images taken in 2013 from 460 patients and enhanced them by a factor of 52. Note that three max-pooling layers, six convolutional layers, and three fully connected layers made up the CNN. They had a median accuracy of 84% and a 92% Area Under the Curve (AUC). Moreover, Kakkar et al. [15] utilized the LiTS dataset to segment the liver, utilizing the Morphological Snake method, and predicted the liver centroid utilizing an Artificial Neural Network (ANN). They obtained a 98.11% accuracy, 88% Dice Index, and 87.71% F1-score utilizing the LiTS dataset. Furthermore, Rania Ghoniem [16] employed SegNet-UNet-BCO and LeNet5-BCO combinations to segment and categorize liver lesions in 2020, combining bio-inspired concepts with deep learning models. The models were trained to utilize the Radiopaedia and LiTS datasets, and the LiTS dataset yielded a 97.6% F1-score, 98.2% specificity, 97% Dice Index, 96.4% Jaccard Index, and 98.5% accuracy. To identify liver tumors automatically, Muhammad Suhaib Aslam et al. [17] utilized the ResUNet, a hybrid UNet and ResNet framework. Relying on the publicly accessible 3D-IRCADb01 dataset, they were able to attain a 99% accuracy and a 95% F1-score. In addition, Jiarong Zhou et al. [18] presented a multi-scale and multimodal structure in 2021, utilizing a hierarchical CNN to automatically detect and categorize focal liver lesions. Following binary class discrimination, the model produced six classes. They attained an average accuracy of 82.5% in discriminating malignant and benign tumors utilizing 3D ResNet-18 and 73.4% in solving the six classes issue. Consequently, Yasmeen Al-Saeed et al. [19] presented a comprehensive framework for separating cancerous and non-cancerous lesions in 2022. The framework is divided into three phases: liver segmentation, tumor segmentation, and lesion classification with an SVM classifier. They employed a combination of textual and statistical elements to analyze the LiTS17, MICCAI-Silver07, and 3Dircadb liver datasets. The LiTS17 dataset obtained 95.57% accuracy, 96.23% sensitivity, 95.83% specificity, and 98.2% AUC, while the 3Dircadb dataset achieved 96.88% accuracy, 97.32% sensitivity, 97.65% specificity, and 98.64% AUC. Mubasher Hussain et al. [20] introduced a revolutionary, fully automated system for liver tumor classification, which employs computer vision and machine learning. A Gabor filter was employed to denoise the images, and the Correlation-based Feature Selection (CFS) approach was employed to maximize the features. On a 17 × 17 Region of Interest (ROI), they obtained 97.48% accuracy using Random Forest and 97.08% accuracy utilizing Random Trees.
According to the literature review, the subject of medical imaging is becoming more important as the demand for a precise and efficient diagnosis in a short amount of time grows. The liver serves a variety of activities, including vascular, metabolic, secretory, and excretory. CT is a medical imaging method that doctors can use to examine pathological abnormalities in the liver. The fundamental issue with liver segmentation from CT images is the poor contrast between the intensities of the liver and adjacent organs. In addition, the liver might appear in several dimensions, making identification and segmentation even more challenging [21]. The categorization of CT images is a time-consuming and difficult operation, which is impracticable when dealing with enormous amounts of data. Manual interaction is not required with fully automatic and unsupervised approaches. Our suggested study method gives an efficient liver CT scan image classification that will be useful in medical datasets, particularly in feature selection and classification. Manually detecting liver tumors is time-consuming and tiresome; however, CAD is critical in automatically recognizing liver abnormalities. In this section, we assess and review recent breakthroughs in CT-based detection of liver tumors, with a focus on deep learning techniques that leverage the LiTS dataset. In CT images, the liver is segmented from the rest of the abdomen, utilizing a 3D technique and morphological processing. The tumor is extracted from the segmented liver area using CNN. A lot of research studies have been done to categorize liver tumor disease. Patients diagnosed with a liver tumor early on will have a better chance of being treated quickly [22]. The remainder of the article is arranged as follows: Section 2 outlines the suggested method’s technique. The experimental results and comparisons with a few selected approaches are shown in Section 3, and the study is concluded in Section 4.
2. Materials and Methods
The method that has been utilized in this paper is shown in Figure 1. The process starts from the segmented CT liver volume, which is resized to be compatible with the input layer of the proposed CNN and the existing ResNet50. The features are extracted automatically and then passed to a support vector machine classifier to discriminate between two classes of benign and malignant liver tumors.
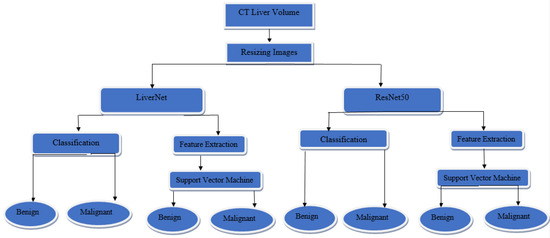
Figure 1.
The proposed method.
2.1. Dataset
The dataset was created as a consequence of liver tumor segmentation, which was held in connection with the IEEE International Symposium on Biomedical Imaging (ISBI) 2017 and the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2017. The liver images from a 3D CT image have been segmented and released. In the axial direction, pixel sizes range from 0.56 mm to 1.0 mm, and in the z-direction, they range from 0.45 mm to 6.0 mm. The number of slices per CT scan varies from 42 to 1026, and all slices were resized to 150 pixels in size. The segmented data are published in [23] and are not labeled. The data are diagnosed by the radiologist, and the following table describes the label for each case. Table 1 shows the diagnosis of the human radiologist for each volume.

Table 1.
The diagnosis of liver CT volumes by Radiologist Diagnosis.
The number of benign cases is 39, the number of malignant cases is 85, whereas 6 cases are diagnosed with no lesions, which means they are normal. The normal cases are excluded from the dataset because they are not sufficient for classification. The proposed method is just designed based on benign and malignant cases.
Figure 2 describes the malignant liver slice, the segmented liver, and the 3D view of the liver. On the other hand, Figure 3 represents the benign case of the liver and its corresponding segment with its 3D view.
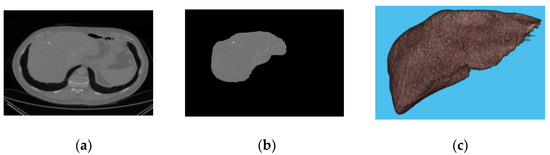
Figure 2.
(a) liver slice; (b) segmented liver; (c) 3D view of malignant liver.
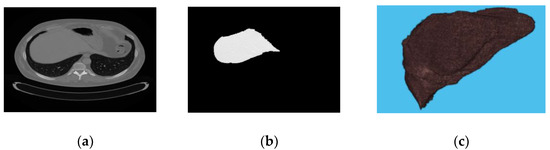
Figure 3.
(a) liver slice; (b) segmented liver; (c) 3D view of benign liver.
The benign segmented liver is augmented with a scale [0.9–1] rotated [2°–5°]. The data are also translated in the x direction with [1–2], y direction of [1–1.5], and z direction of [0.9–1.2]. The resultant beginning images after augmentation are 75 images. Table 2 shows the number of images before and after augmentation.

Table 2.
The number of images after and before augmentation.
2.2. Deep Learning
It is known that deep learning models need large data sets to train. Many scholars have used transfer learning to tune a pre-trained model to perform a certain task to overcome this issue. In this work, two pre-trained neural networks have been used, namely ResNet50 and Resnet101 [24,25,26,27,28], as shown in Figure 4, respectively. The ImageNet dataset was utilized for training these two architectures. ResNet is a deep convolutional neural network model with shortcut connections that bypass one or more layers. The number of output feature maps in this type of network is similar to the number of filters in the layer. The number of filters doubles as the size of the feature map is lowered. Down sampling is performed in a convolution layer with a stride of two, and then batch normalization and the ReLU function are applied.
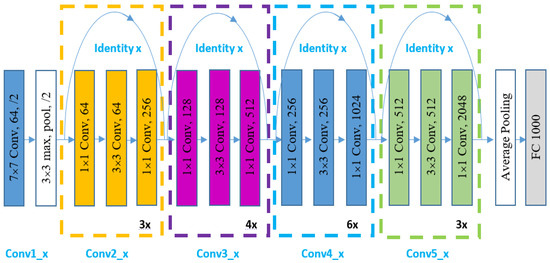
Figure 4.
ResNet50 General Structure.
Further details of both networks’ architectures are explained in Figure 4. The first convolutional layer in both networks will output a feature map of size 112 × 112 × 64 after applying 64 distinct filters of size 7 × 7 × 3 over the input of size 224 × 224. The input feature map is then processed, utilizing a max-pooling layer with a filter of 3 × 3, resulting in a feature map of 56 × 56 × 64. Furthermore, the second convolutional layer contains three building blocks, where each block contains three convolutional layers. As a result, there are nine sub-convolutional layers in the second convolutional layer. The third convolutional layer is made up of four blocks, each of which has three sub-convolutional layers. Thus, there are 12 sub-convolutional layers in the third convolutional layer. In terms of the fourth convolutional layer, ResNet50 comprises six blocks.
LiverNet
The proposed LiverNet model is light, and consists of eight layers. It consists of an input layer with size 223 × 223 × 147 × 1, a 3D convolutional layer with kernel size of 5 × 5 × 5 with six filters, and a stride by two. The output of the first layer is inserted into a 3D average pool layer of size 2 × 2 × 2, along with stride by two. This layer plays a crucial role in decreasing data variances and maintaining the most critical elements. Finally, the ReLU activation function receives the output from the previous layers, and the active output is sent into a 10-neuron fully connected layer. Afterwards, the result is sent to a fully connected layer with two neurons equivalent to the number of planned classes. The suggested network flow chart is illustrated in Figure 5.
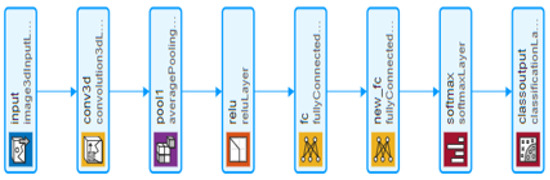
Figure 5.
The structure of the proposed network.
The fully connected layer is usually terminated with a softmax layer, which implements a softmax function to its input and whose equation corresponds to the equitation [29]:
in which denotes the input vector of size K, j = 1: K, and resembles the ith individual input. The Softmax function defines a range of values for the output, allowing it to be read as a probability. It is frequently employed in multivariant classifications. Moreover, the softmax layers are responsible for computing the probability of each class, whereas the classification layer is in charge of obtaining the classification results. Next, the proposed network is built using MATLAB® 2021b, and it is trained and tested using a PC with CPU Core i5-11 GEN processor, 8 GB RAM, and 1000 GB total storage. Table 3 shows the layer’s information for the suggested CNN architecture.

Table 3.
Layers information for the proposed LiverNet.
2.3. Classification
The classification is performed in this article by two tracks; the first one is deep learning, and the other one is a hybrid system. The deep learning approach is utilized by passing the resize images to the pretrained ResNet50 using transfer learning to discriminate between benign and malignant classes. On the other hand, the proposed network is exploited as well for classifying the available images into malignant and benign.
The hybrid approach is utilized in this paper by using the deep learning structures as feature extractors instead of applying various image processing techniques to extract the features manually. This approach is applied two times; the first one uses ResNet50, and the other one uses the proposed Liver Network. The two extracted features from the last fully connected layer in each network are passed to the Gaussian Support Vector Machine classifier independently. The results are compared between the hybrid system, which is built based on extracted features from a pretrained ResNet50 structure, and those constructed mainly on the extracted features from the proposed LiverNet. On the top of that, the corresponding results section clarified the differences between benign and malignant classification based on deep learning approaches.
3. Results and Discussion
The data are divided into 70% training data. Meanwhile, the rest resembles testing. The transfer learning strategy is employed here to be suitable for two classes. The maximum accuracy obtained using ResNet50 is 83.7%. After taking 123 min in the training stages, Figure 6 illustrates the confusion matrix for ResNet50.
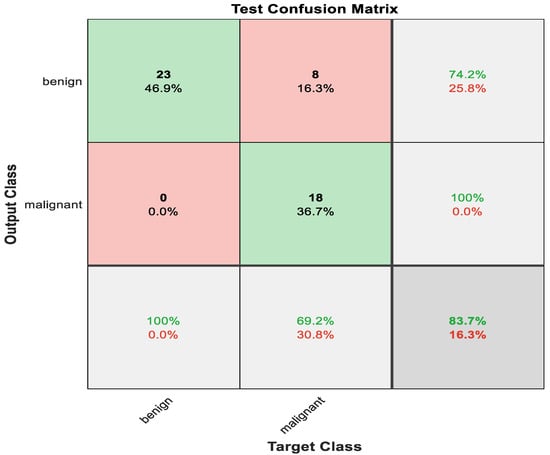
Figure 6.
Test Confusion matrix of ResNet50.
The number and percentage of correct classifications by the pre-trained network are shown in the first two diagonal cells of Figure 6. For instance, 23 occurrences were categorized as benign appropriately. This represents 46.9% of the total of 49 occurrences. In the same way, 18 occurrences were accurately labeled as malignant, in which 36.7% of all cases fell into this category.
Eight of the malignant cases were misclassified as benign, accounting for 16.3% of the total 46 cases in the study. Likewise, 0 benign biopsies were wrongly labeled as malignant, accounting for 0% of the total data.
From 31 benign predictions, 74.2% were found to be correct, meanwhile 25.8% were revealed to be wrong. Of 18 malignant predictions, 100% were correct, and 0% were wrong. Of 23 benign cases, 100% were revealed to be correctly predicted as benign, while 0% were predicted as malignant. Of 26 malignant cases, 69.2% were correctly classified as malignant, while 30.8% were categorized as benign. In total, 83.7% of the predictions were revealed to be correct, while 16.3% of them were wrong. The pertained network is very badly sensitive to malignant cases. Almost 31% of the malignant cases were diagnosed as benign, which is not acceptable in medical field applications. Figure 7 shows the ROC curve for this case.
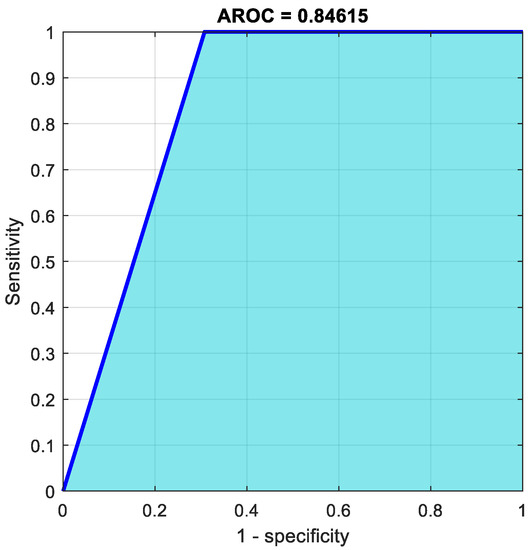
Figure 7.
AROC curve using ResNet50.
The proposed net obtained a high accuracy when compared with ResNet50, and Figure 8 shows the confusion matrix of the LiverNet. Here, the accuracy reached 95.9%.
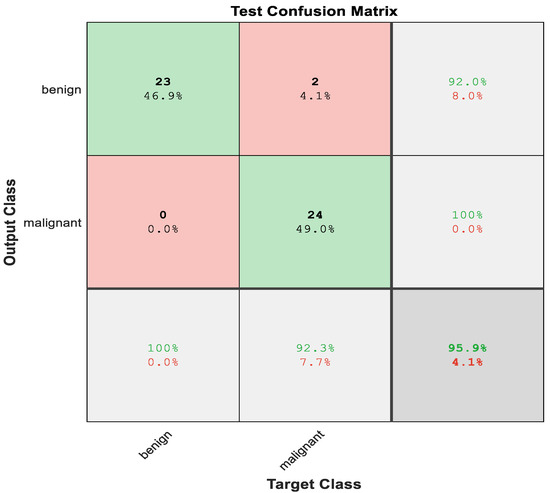
Figure 8.
Test Confusion matrix of LiverNet.
The number and percentage of correct classifications by the suggested network are shown in the first two diagonal cells of Figure 7. A total of 23 occurrences, for example, were accurately categorized as benign. This represents 46.9% of the total of 49 occurrences. In the same approach, 24 occurrences were accurately labeled as malignant. This was the case in 49% of all occurrences.
Two of the malignant instances were mistakenly categorized as benign, accounting for 4.1% of the total 49 cases in the study.
From 23 benign predictions, it was revealed that 100% were correct. Meanwhile, from 24 malignant predictions, 100% were found to be correct. Moreover, from 23 benign cases, 100% were correctly predicted as benign, meanwhile, from 24 malignant cases, 92.3% were correctly classified as malignant and were discovered to be 7.7% wrong. In total, 95.9% of the predictions were found to be correct, meanwhile 4.1% were shown to be wrong. The proposed network performance was better than the pre-trained CNN. Figure 9 illustrates the ROC curve of classification using LiverNet.
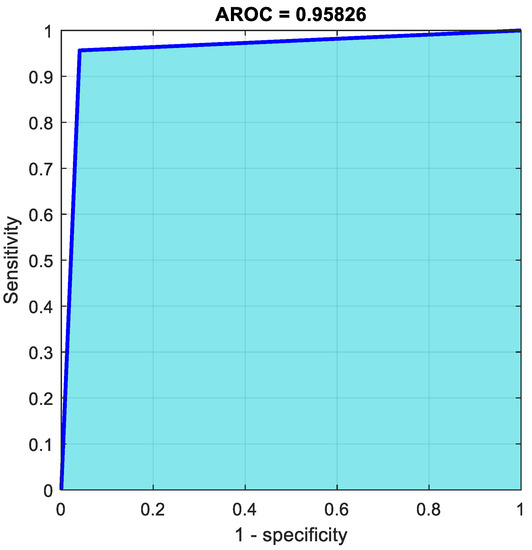
Figure 9.
AROC curve using LiverNet.
The number of convolutional layers in LiverNet is one, which makes it fast in training and testing. Table 4 shows the time required for training and test phases for both the existing CNN and the proposed one.

Table 4.
Comparison for training and testing time for both ResNet50 and LiverNet.
In the next stage ResNet50 and LiverNet are employed as feature extractors In both networks, the two features are retrieved from the final fully connected layer. Finally, the labeled data are classified using gaussian SVM.
The model is built utilizing a Gaussian SVM classifier to distinguish between malignant and benign tumors. Figure 10 describes the confusion matrix of the Gaussian SVM using 3D graphical features of ResNet50. Here, the total accuracy reached 97%.
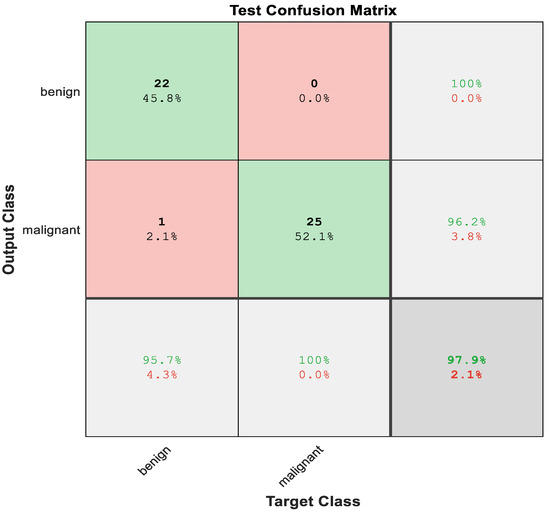
Figure 10.
Test Confusion matrix of SVM with features from ResNet50.
The first two diagonal cells in Figure 7 reflect the number and percentage of correct classifications in ResNet50 utilizing a Gaussian SVM with 3D graphical features. The benign classification for the 22 cases was valid, representing 45.8% of the total 48 cases. In the same approach, 25 cases were accurately identified as malignant, which was 52.1% of the total number of cases.
One of the benign occurrences was mistakenly labeled as malignant, accounting for 2.1% of the total 48 cases in the study. From 22 benign predictions, it was stated that 100% were correct; meanwhile, from 26 malignant predictions, 96.2% were revealed to be correct. Furthermore, from 23 benign cases, 95.7% of them were correctly predicted as benign, meanwhile, from 25 malignant cases, 100% were correctly categorized as malignant. In total, 97.9% of the predictions were revealed to be correct, while 2.1% of them were wrong. Figure 11 represents the ROC curve of the hybrid system using a pretrained CNN.
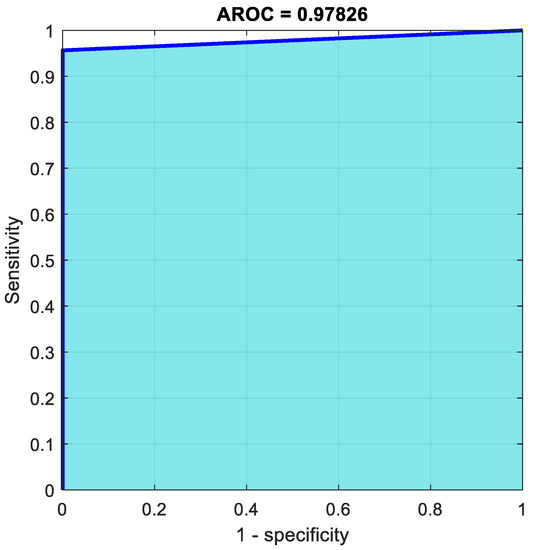
Figure 11.
AROC curve of SVM with features from ResNet50.
The first two diagonal cells in Figure 12 reflect the number and percentage of correct classifications utilizing the suggested net’s gaussian SVM with 3D graphical features. For example, the benign classification for the 23 occurrences was correct, representing 47.9% of the total 48 occurrences. In the same approach, 25 occurrences were accurately identified as malignant, which was 52.1% of the total number of cases.
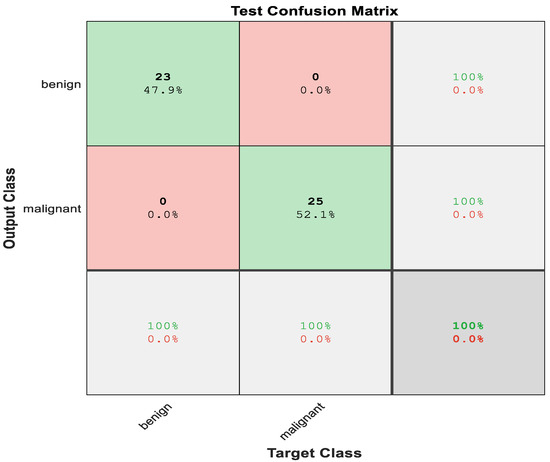
Figure 12.
Test Confusion matrix of SVM with features from LiverNet.
Of 23 benign cases, 100% of them were correctly predicted as benign, meanwhile, from 25 malignant cases, 100% were found to be correctly categorized as malignant. Overall, 100% of the predictions were correct. Figure 13 shows the ROC curve of the hybrid system using LiverNet features.
Figure 13.
AROC of hybrid system with features from LiverNet.
The evaluation criteria that have been used in this paper are clear in the corresponding equations [30]. Table 5 describes the results in the deep learning track and hybrid track for both ResNet50 and the proposed LiverNet. The following equations are used to calculate the performance of the classifier [30].
where TP is True Positive, TN is True Negative, FP is False Positive, and FN is False Negative.

Table 5.
Comparison between deep learning and the proposed hybrid model.
The high performance of the proposed net is clear as a feature extractor. Furthermore, Figure 14 below illustrates the high performance of the proposed approach in obtaining an accurate diagnosis of liver tumors.
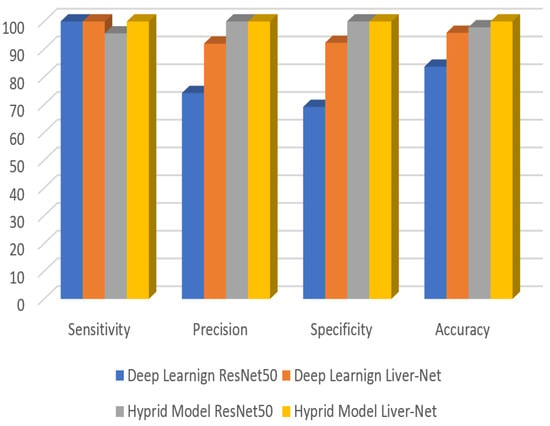
Figure 14.
Accuracy, Precision, Specificity, and Sensitivity of different approaches.
The proposed method is compared with literature that has used the LiTs17 dataset. The performance of the approach achieved the highest amongst all. Table 6 shows the comparison between this study and literature with regards to the area under the curve (AUC), specificity, sensitivity, and accuracy.

Table 6.
Comparison of the current study with the state of the art.
This paper shows the high level of confidence obtained using LiverNet as an automated feature extractor besides utilizing the benefits of machine learning to discriminate between benign and malignant liver tumors.
4. Conclusions
Patients with liver cancer have a high mortality rate attributed to the late detection of the disease. Computer-aided diagnosis systems based on a variety of medical imaging techniques can help recognize liver cancer at an early stage. With the help of both conventional machine learning and deep learning classifiers, a variety of methods have been employed to identify liver cancer. The findings of this study suggest that using CNN to automatically extract features together with SVM classifier greatly improves classification performance. Furthermore, the findings suggest that employing our suggested hybrid model can greatly reduce the processing time, which is 22 s, when contrasted to ResNet50, which takes 32 s. All performance metrics accuracy, specificity, precision, and sensitivity reached 100%. Our approach can accurately and effectively recognize tumors, even in low-contrast CT images with respect to all quantitative assessments. Lastly, we can draw the following conclusions: (1) Deep learning model performance is extremely intriguing for use in medical equipment; the experimental result demonstrates significant improvement. Moreover, the suggested technique is unaffected by discrepancies in texture and intensity across demographics, imaging devices, patients, and settings; (2) the classifier distinguishes the tumor with comparatively high precision; (3) segmentation of very small tumors is incredibly challenging, with the system being hyper-sensitive to contemplating local noise artifacts as potential tumors.
The lack of large publicly available datasets forces CAD systems to use the available small private datasets generated from hospitals and scanning facilities. This implies that additional datasets should be made available for research and classification purposes. In the future, this work can be further extended using a large clinical dataset besides applying image processing techniques to enhance the visualization of images. Using a huge dataset, a reliable and trusted system can be built and employed in clinics.
Author Contributions
Conceptualization, K.A., H.A. and M.A.; methodology, A.A., H.A., Y.A.-I. and M.A.; software, H.A., M.A. and W.A.M.; validation, K.A., H.A., Y.A.-I., W.A.M. and M.A.; formal analysis, Y.A.-I., H.A., A.B. and M.A.; writing—original draft preparation, H.A., Y.A.-I., A.A., A.B. and W.A.M.; writing—review and editing, K.A., H.A., W.A.M., M.A. and Y.A.-I.; visualization, H.A.; supervision, H.A. and W.A.M.; project administration, H.A. and K.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The dataset analyzed during the current study was derived from the LiTS (Liver Tumor Segmentation Challenge (LiTS17)) organized in conjunction with ISBI 2017 and MICCAI 2017. Available online: https://www.kaggle.com/datasets/andrewmvd/liver-tumor-segmentation (accessed on 15 January 2022).
Acknowledgments
The authors would thank the authors of the dataset for making it available online. Furthermore, they would like to thank the anonymous reviewers for their contribution towards enhancing this paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Al Sadeque, Z.; Khan, T.I.; Hossain, Q.D.; Turaba, M.Y. Automated detection and classification of liver cancer from CT Images using HOG-SVM model. In Proceedings of the 2019 5th International Conference on Advances in Electrical Engineering (ICAEE 2019), Dhaka, Bangladesh, 26–28 September 2019; pp. 21–26. [Google Scholar]
- Ba Alawi, A.E.; Saeed, A.Y.A.; Radman, B.M.N.; Alzekri, B.T. A Comparative Study on Liver Tumor Detection Using CT Images. In Lecture Notes on Data Engineering and Communications Technologies; Springer: Cham, Switzerland, 2021; Volume 72, pp. 129–137. [Google Scholar]
- Shanila, N.; Vinod Kumar, R.S.; Abin, N.A. Feature extraction and performance evaluation of classification algorithms for liver tumor diagnosis of abdominal computed tomography images. J. Adv. Res. Dyn. Control Syst. 2020, 12, 82–90. [Google Scholar] [CrossRef]
- Selvathi, D.; Malini, C.; Shanmugavalli, P. Automatic segmentation and classification of liver tumor in CT images using adaptive hybrid technique and Contourlet based ELM classifier. In Proceedings of the 2013 International Conference on Recent Trends in Information Technology (ICRTIT 2013), Dubai, United Arab Emirates, 11–12 December 2013; pp. 250–256. [Google Scholar]
- Masuda, Y.; Tateyama, T.; Xiong, W.; Zhou, J.; Wakamiya, M.; Kanasaki, S.; Furukawa, A.; Chen, Y.W. Liver tumor detection in CT images by adaptive contrast enhancement and the EM/MPM algorithm. In Proceedings of the International Conference on Image Processing (ICIP), Brussels, Belguim, 11–14 September 2011; pp. 1421–1424. [Google Scholar]
- Hasegawa, R.; Iwamoto, Y.; Han, X.; Lin, L.; Hu, H.; Cai, X.; Chen, Y.W. Automatic Detection and Segmentation of Liver Tumors in Multi- phase CT Images by Phase Attention Mask R-CNN. In Proceedings of the Digest of Technical Papers—IEEE International Conference on Consumer Electronics, Penghu, Taiwan, 15–17 September 2021. [Google Scholar]
- Salman, O.S.; Klein, R. Automatic Detection and Segmentation of Liver Tumors in Computed Tomography Images: Methods and Limitations. In Intelligent Computing; Lecture Notes in Networks and Systems; Springer: Cham, Switzerland, 2021; Volume 285, pp. 17–35. [Google Scholar]
- Das, A.; Panda, S.S.; Sabut, S. Detection of liver tumor in CT images using watershed and hidden markov random field expectation maximization algorithm. In Computational Intelligence, Communications, and Business Analytics; Springer: Singapore, 2017; Volume 776, pp. 411–419. [Google Scholar]
- Todoroki, Y.; Han, X.H.; Iwamoto, Y.; Lin, L.; Hu, H.; Chen, Y.W. Detection of liver tumor candidates from CT images using deep convolutional neural networks. In International Conference on Innovation in Medicine and Healthcare; Springer: Cham, Switzerland, 2018; Volume 71, pp. 140–145. [Google Scholar]
- Devi, R.M.; Seenivasagam, V. Automatic segmentation and classification of liver tumor from CT image using feature difference and SVM based classifier-soft computing technique. Soft Comput. 2020, 24, 18591–18598. [Google Scholar] [CrossRef]
- Krishan, A.; Mittal, D. Ensembled liver cancer detection and classification using CT images. Proc. Inst. Mech. Eng. Part H J. Eng. Med. 2021, 235, 232–244. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Song, Y.; Liu, Z. CT image classification of liver tumors based on multi-scale and deep feature extraction. J. Image Graph. 2021, 26, 1704–1715. [Google Scholar] [CrossRef]
- Das, A.; Acharya, U.R.; Panda, S.S.; Sabut, S. Deep learning based liver cancer detection using watershed transform and Gaussian mixture model techniques. Cogn. Syst. Res. 2019, 54, 165–175. [Google Scholar] [CrossRef]
- Yasaka, K.; Akai, H.; Abe, O.; Kiryu, S. Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: A preliminary study. Radiology 2018, 286, 887–896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kakkar, P.; Nagpal, S.; Nanda, N. Automatic liver segmentation in CT images using improvised techniques. In International Conference on Smart Health; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2018; Volume 10983 LNCS, pp. 41–52. [Google Scholar]
- Ghoniem, R.M. A Novel Bio-Inspired Deep Learning Approach for Liver Cancer Diagnosis. Information 2020, 11, 80. [Google Scholar] [CrossRef] [Green Version]
- Aslam, M.S.; Younas, M.; Sarwar, M.U.; Shah, M.A.; Khan, A.; Uddin, M.I.; Ahmad, S.; Firdausi, M.; Zaindin, M. Liver-Tumor detection using CNN ResUNet. Comput. Mater. Contin. 2021, 67, 1899–1914. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, W.; Lei, B.; Ge, W.; Huang, Y.; Zhang, L.; Yan, Y.; Zhou, D.; Ding, Y.; Wu, J.; et al. Automatic Detection and Classification of Focal Liver Lesions Based on Deep Convolutional Neural Networks: A Preliminary Study. Front. Oncol. 2021, 10, 581210. [Google Scholar] [CrossRef] [PubMed]
- Al-Saeed, Y.; Gab-Allah, W.A.; Soliman, H.; Abulkhair, M.F.; Shalash, W.M.; Elmogy, M. Efficient Computer Aided Diagnosis System for Hepatic Tumors Using Computed Tomography Scans. Comput. Mater. Contin. 2022, 71, 4871–4894. [Google Scholar] [CrossRef]
- Hussain, M.; Saher, N.; Qadri, S. Computer Vision Approach for Liver Tumor Classification Using CT Dataset. Appl. Artif. Intell. 2022, 1–23. [Google Scholar] [CrossRef]
- Selvathi, D.; Priyadarsini, S.; Malini, C.; Shanmugavalli, P. Performance analysis of multi resolution transforms with kernel classifiers for liver tumor detection using CT images. Int. J. Appl. Eng. Res. 2014, 9, 30935–30952. [Google Scholar]
- Krishan, A.; Mittal, D. Effective segmentation and classification of tumor on liver MRI and CT images using multi-kernel K-means clustering. Biomed. Technol. 2019, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Soler, L.; Hostettler, A.; Agnus, V.; Charnoz, A.; Fasquel, J.; Moreau, J.; Osswald, A.; Bouhadjar, M.; Marescaux, J. 3D Image Reconstruction for Comparison of Algorithm Database: A Patient Specific Anatomical and Medical Image Database; Tech Report; IRCAD: Strasbourg, France, 2010. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Poudel, S.; Kim, Y.J.; Vo, D.M.; Lee, S.W. Colorectal Disease Classification Using Efficiently Scaled Dilation in Convolutional Neural Network. IEEE Access 2020, 8, 99227–99238. [Google Scholar] [CrossRef]
- Ullah, W.; Ullah, A.; Haq, I.U.; Muhammad, K.; Sajjad, M.; Baik, S.W. CNN features with bi-directional LSTM for real-time anomaly detection in surveillance networks. Multimed. Tools Appl. 2021, 80, 16979–16995. [Google Scholar] [CrossRef]
- Wu, H.; Xin, M.; Fang, W.; Hu, H.M.; Hu, Z. Multi-Level Feature Network with Multi-Loss for Person Re-Identification. IEEE Access 2019, 7, 91052–91062. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, M.; Zhang, D.; Huang, H.; Zhang, F. Quantification of water inflow in rock tunnel faces via convolutional neural network approach. Autom. Constr. 2021, 123, 103526. [Google Scholar] [CrossRef]
- Alqudah, A.; Alqudah, A.M.; Alquran, H.; Al-zoubi, H.R.; Al-qodah, M.; Al-khassaweneh, M.A. Recognition of handwritten arabic and hindi numerals using convolutional neural networks. Appl. Sci. 2021, 11, 1573. [Google Scholar] [CrossRef]
- Alqudah, A.M.; Alquran, H.; Abu-Qasmieh, I.; Al-Badarneh, A. Employing image processing techniques and artificial intelligence for automated eye diagnosis using digital eye fundus images. J. Biomim. Biomater. Biomed. Eng. 2018, 39, 40–56. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).