Polymorphism of 16s rRNA Gene: Any Effect on the Biomolecular Quantitation of the Honey Bee (Apis mellifera L., 1758) Pathogen Nosema ceranae?
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. DNA Extraction and qPCR
2.3. Statistical Analysis
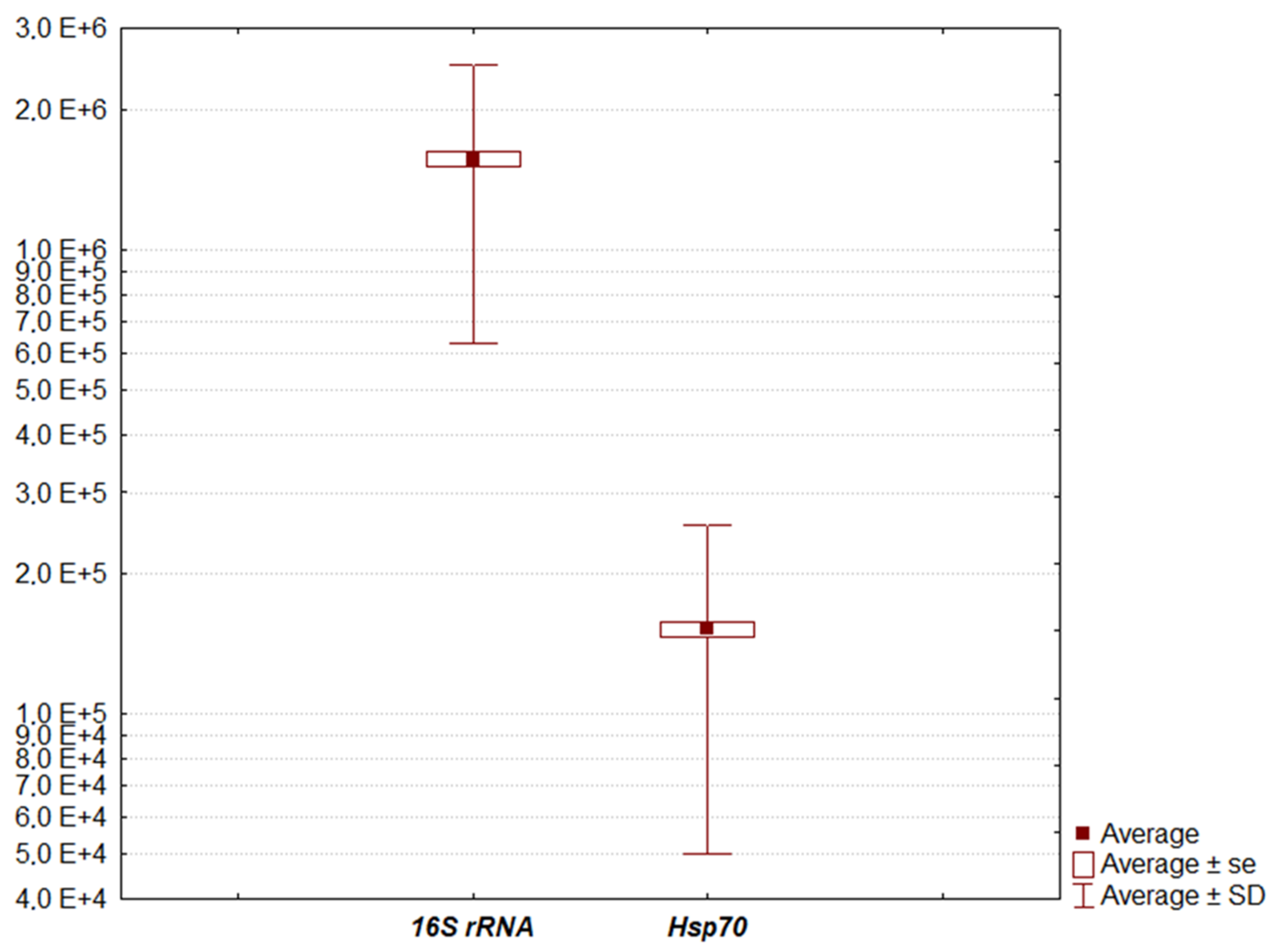
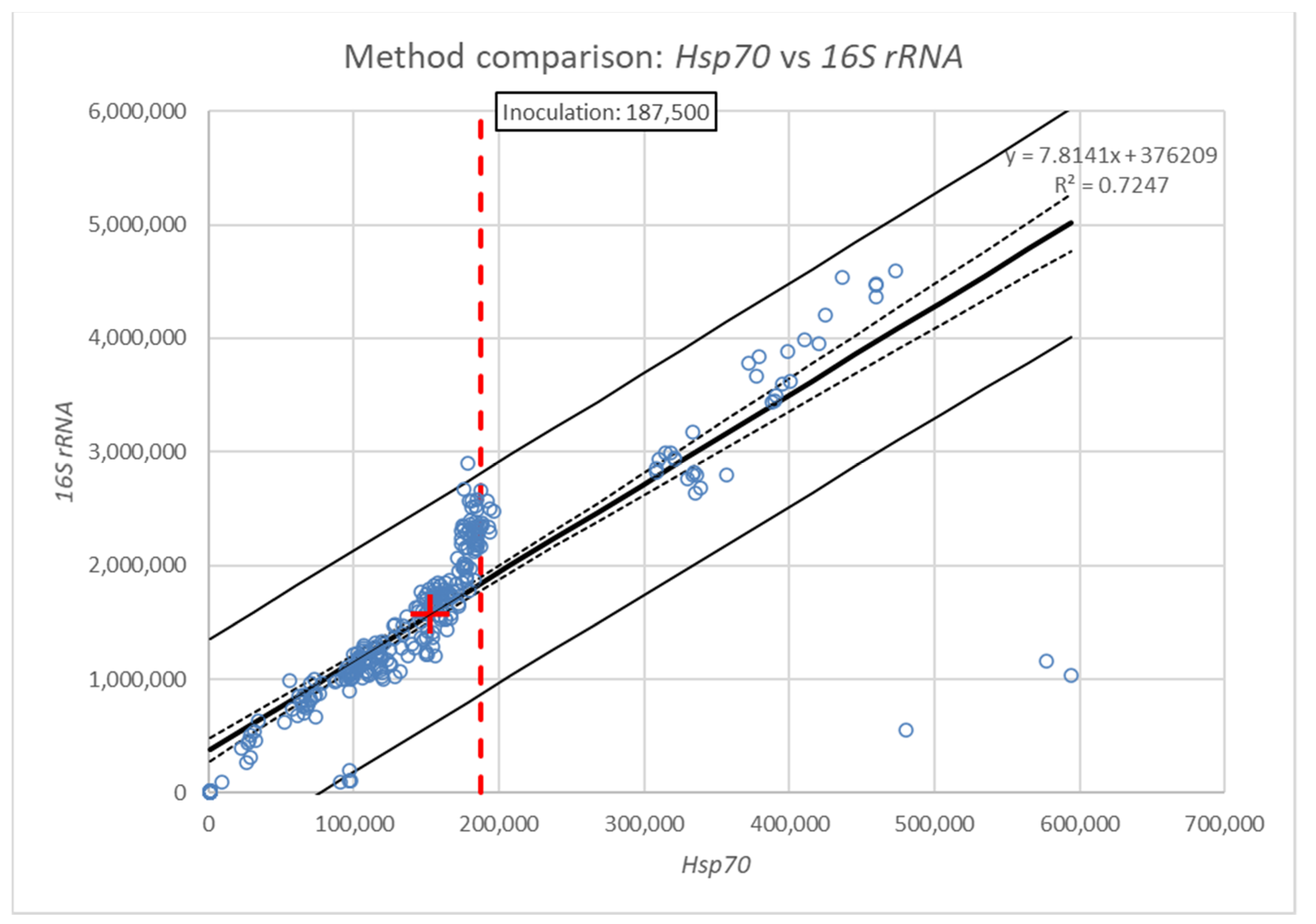
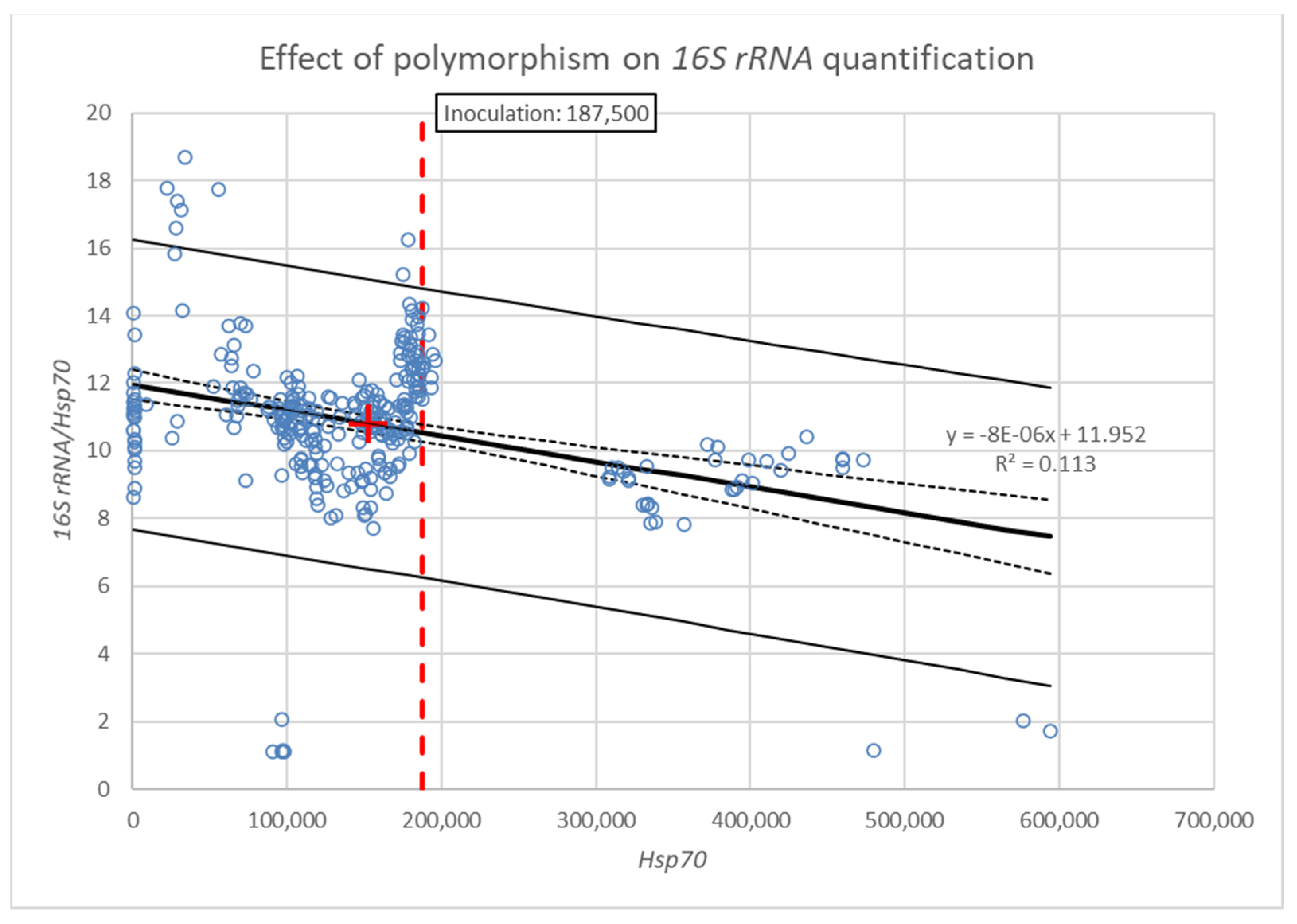
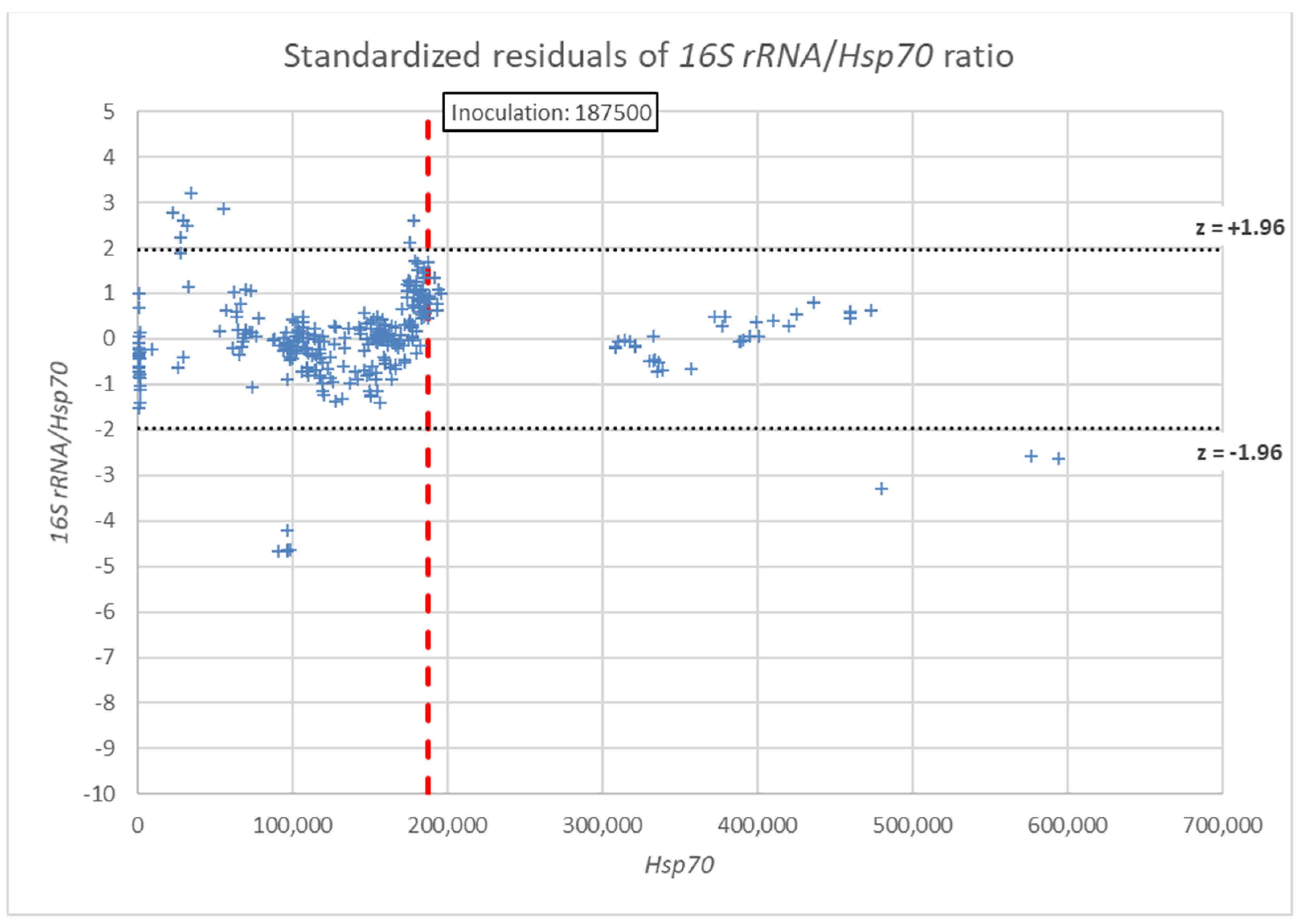
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fries, I. Nosema ceranae in European honey bees (Apis mellifera). J. Invertebr. Pathol. 2010, 103, S73–S79. [Google Scholar] [CrossRef]
- Botías, C.; Martín-hernández, R.; Barrios, L.; Meana, A.; Higes, M. Nosema spp. infection and its negative effects on honey bees (Apis mellifera iberiensis ) at the colony level. Vet. Res. 2013, 44, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Fries, I.; Feng, F.; Da Silva, A.; Slemenda, S.B.; Pieniazek, N.J. Nosema ceranae n. sp. (Microspora, Nosematidae), morphological and molecular characterization of a microsporidian parasite of the Asian honey bee Apis cerana (Hymenoptera, Apidae). Eur. J. Protistol. 1996, 32, 356–365. [Google Scholar] [CrossRef]
- Klee, J.; Besana, A.M.; Genersch, E.; Gisder, S.; Nanetti, A.; Tam, D.Q.; Chinh, T.X.; Puerta, F.; Ruz, J.M.; Kryger, P.; et al. Widespread dispersal of the microsporidian Nosema ceranae, an emergent pathogen of the western honey bee, Apis mellifera. J. Invertebr. Pathol. 2007, 96, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Higes, M.; Martín, R.; Meana, A. Nosema ceranae, a new microsporidian parasite in honeybees in Europe. J. Invertebr. Pathol. 2006, 92, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Roudel, M.; Aufauvre, J.; Corbara, B.; Delbac, F.; Blot, N. New insights on the genetic diversity of the honeybee parasite Nosema ceranae based on multilocus sequence analysis. Parasitology 2013, 140, 1346–1356. [Google Scholar] [CrossRef]
- Van der Zee, R.; Gómez-Moracho, T.; Pisa, L.; Sagastume, S.; García-Palencia, P.; Maside, X.; Bartolomé, C.; Martín-Hernández, R.; Higes, M. Virulence and polar tube protein genetic diversity of Nosema ceranae (Microsporidia) field isolates from Northern and Southern Europe in honeybees (Apis mellifera iberiensis). Environ. Microbiol. Rep. 2014, 6, 401–413. [Google Scholar] [CrossRef] [PubMed]
- Cilia, G.; Sagona, S.; Giusti, M.; Jarmela dos Santos, P.E.; Nanetti, A.; Felicioli, A. Nosema ceranae infection in honeybee samples from Tuscanian Archipelago (Central Italy) investigated by two qPCR methods. Saudi J. Biol. Sci. 2019, 26, 1553–1556. [Google Scholar] [CrossRef]
- Higes, M.; García-Palencia, P.; Martín-Hernández, R.; Meana, A. Experimental infection of Apis mellifera honeybees with Nosema ceranae (Microsporidia). J. Invertebr. Pathol. 2007, 94, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Higes, M.; Martín-Hernández, R.; Botías, C.; Bailón, E.G.; González-Porto, A.V.; Barrios, L.; Del Nozal, M.J.; Bernal, J.L.; Jiménez, J.J.; Palencia, P.G.; et al. How natural infection by Nosema ceranae causes honeybee colony collapse. Environ. Microbiol. 2008, 10, 2659–2669. [Google Scholar] [CrossRef]
- Higes, M.; García-Palencia, P.; Urbieta, A.; Nanetti, A.; Martín-Hernández, R. Nosema apis and Nosema ceranae Tissue Tropism in Worker Honey Bees (Apis mellifera). Vet. Pathol. 2020, 57, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Giersch, T.; Berg, T.; Galea, F.; Hornitzky, M. Nosema ceranae infects honey bees ( Apis mellifera ) and contaminates honey in Australia. Apidologie 2009, 40, 117–123. [Google Scholar] [CrossRef]
- Eiri, D.M.; Suwannapong, G.; Endler, M.; Nieh, J.C. Nosema ceranae Can Infect Honey Bee Larvae and Reduces Subsequent Adult Longevity. PLoS ONE 2015, 10, e0126330. [Google Scholar] [CrossRef] [PubMed]
- Higes, M.; Martín-Hernández, R.; Garrido-Bailón, E.; González-Porto, A.V.; García-Palencia, P.; Meana, A.; del Nozal, M.J.; Mayo, R.; Bernal, J.L.; Kamer, Y.; et al. Honeybee colony collapse due to Nosema ceranae in professional apiaries. Environ. Microbiol. Rep. 2009, 1, 110–113. [Google Scholar] [CrossRef]
- Nanetti, A.; Bortolotti, L.; Cilia, G. Pathogens Spillover from Honey Bees to Other Arthropods. Pathogen 2021, 10, 1044. [Google Scholar] [CrossRef]
- Graystock, P.; Yates, K.; Darvill, B.; Goulson, D.; Hughes, W.O.H. Emerging dangers: Deadly effects of an emergent parasite in a new pollinator host. J. Invertebr. Pathol. 2013, 114, 114–119. [Google Scholar] [CrossRef]
- Purkiss, T.; Lach, L. Pathogen spillover from Apis mellifera to a stingless bee. Proc. R. Soc. B Biol. Sci. 2019, 286, 20191071. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.J.F. Microsporidia: An Emerging Threat to Bumblebees? Trends Parasitol. 2017, 33, 754–762. [Google Scholar] [CrossRef]
- Porrini, M.P.; Porrini, L.P.; Garrido, P.M.; De Melo, C.; Neto, S.; Porrini, D.P.; Muller, F.; Nuñez, L.A.; Alvarez, L.; Fernandez Iriarte, P.; et al. Nosema ceranae in South American Native Stingless Bees and Social Wasp. Microb. Ecol. 2017, 74, 761–764. [Google Scholar] [CrossRef]
- Ravoet, J.; De Smet, L.; Meeus, I.; Smagghe, G.; Wenseleers, T.; de Graaf, D.C. Widespread occurrence of honey bee pathogens in solitary bees. J. Invertebr. Pathol. 2014, 122, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Ilyasov, R.A.; Gaifullina, L.R.; Saltykova, E.S.; Poskryakov, A.V.; Nikolenko, A.G. Review of the expression of antimicrobial peptide defensin in honey bees Apis mellifera L. J. Apic. Sci. 2012, 56, 115–124. [Google Scholar] [CrossRef]
- Higes, M.; Martín-Hernández, R.; Garrido-Bailón, E.; Botías, C.; García-Palencia, P.; Meana, A. Regurgitated pellets of Merops apiaster as fomites of infective Nosema ceranae (Microsporidia) spores. Environ. Microbiol. 2008, 10, 1374–1379. [Google Scholar] [CrossRef] [PubMed]
- Valera, F.; Martín-Hernández, R.; Higes, M. Evaluation of large-scale dissemination of Nosema ceranae spores by European bee-eaters Merops apiaster. Environ. Microbiol. Rep. 2011, 3, 47–53. [Google Scholar] [CrossRef]
- Cilia, G.; Cardaio, I.; dos Santos, P.E.J.; Ellis, J.D.; Nanetti, A. The first detection of Nosema ceranae (Microsporidia) in the small hive beetle, Aethina tumida Murray (Coleoptera: Nitidulidae). Apidologie 2018, 49, 619–624. [Google Scholar] [CrossRef]
- Wadi, L.; Reinke, A.W. Evolution of microsporidia: An extremely successful group of eukaryotic intracellular parasites. PLoS Pathog. 2020, 16, e1008276. [Google Scholar] [CrossRef]
- Cornman, R.S.; Chen, Y.; Schatz, M.C.; Street, C.; Zhao, Y.; Desany, B.; Egholm, M.; Hutchison, S.; Pettis, J.S.; Lipkin, W.I.; et al. Genomic analyses of the microsporidian Nosema ceranae, an emergent pathogen of honey bees. PLoS Pathog. 2009, 5, 1000466. [Google Scholar] [CrossRef]
- Chen, Y.; Pettis, J.S.; Zhao, Y.; Liu, X.; Tallon, L.J.; Sadzewicz, L.D.; Li, R.; Zheng, H.; Huang, S.; Zhang, X.; et al. Genome sequencing and comparative genomics of honey bee microsporidia, Nosema apis reveal novel insights into host-parasite interactions. BMC Genom. 2013, 14, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Gregory, R.T. Genome Size Evolution in Animals; Elsevier Inc.: Amsterdam, The Netherlands, 2005; ISBN 9780123014634. [Google Scholar]
- Ironside, J.E. Diversity and Recombination of Dispersed Ribosomal DNA and Protein Coding Genes in Microsporidia. PLoS ONE 2013, 8, 55878. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.F.; Jiang, J.H.; Chen, Y.W.; Wang, C.H. A Nosema ceranae isolate from the honeybee Apis mellifera. Apidologie 2007, 38, 30–37. [Google Scholar] [CrossRef]
- Chen, Y.; Evans, J.D.; Zhou, L.; Boncristiani, H.; Kimura, K.; Xiao, T.; Litkowski, A.M.; Pettis, J.S. Asymmetrical coexistence of Nosema ceranae and Nosema apis in honey bees. J. Invertebr. Pathol. 2009, 101, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Martín-Hernández, R.; Meana, A.; Prieto, L.; Salvador, A.M.; Garrido-Bailón, E.; Higes, M. Outcome of colonization of Apis mellifera by Nosema ceranae. Appl. Environ. Microbiol. 2007, 73, 6331–6338. [Google Scholar] [CrossRef]
- Williams, G.R.; Shafer, A.B.A.; Rogers, R.E.L.; Shutler, D.; Stewart, D.T. First detection of Nosema ceranae, a microsporidian parasite of European honey bees (Apis mellifera), in Canada and central USA. J. Invertebr. Pathol. 2008, 97, 189–192. [Google Scholar] [CrossRef] [PubMed]
- Sagastume, S.; del Águila, C.; Martín-Hernández, R.; Higes, M.; Henriques-Gil, N. Polymorphism and recombination for rDNA in the putatively asexual microsporidian Nosema ceranae, a pathogen of honeybees. Environ. Microbiol. 2011, 13, 84–95. [Google Scholar] [CrossRef]
- Huang, W.F.; Bocquet, M.; Lee, K.C.; Sung, I.H.; Jiang, J.H.; Chen, Y.W.; Wang, C.H. The comparison of rDNA spacer regions of Nosema ceranae isolates from different hosts and locations. J. Invertebr. Pathol. 2008, 97, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Fries, I.; Chauzat, M.-P.; Chen, Y.-P.; Doublet, V.; Genersch, E.; Gisder, S.; Higes, M.; McMahon, D.P.; Martín-Hernández, R.; Natsopoulou, M.; et al. Standard methods for Nosema research. J. Apic. Res. 2013, 52, 1–28. [Google Scholar] [CrossRef]
- Sagastume, S.; Martín-Hernández, R.; Higes, M.; Henriques-Gil, N. Ribosomal gene polymorphism in small genomes: Analysis of different 16s rRNA sequences expressed in the honeybee parasite Nosema ceranae (Microsporidia). J. Eukaryot. Microbiol. 2014, 61, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Katinka, M.D.; Duprat, S.; Cornillott, E.; Méténler, G.; Thomarat, F.; Prensier, G.; Barbe, V.; Peyretaillade, E.; Brottier, P.; Wincker, P.; et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 2001, 414, 450–453. [Google Scholar] [CrossRef]
- Liao, D. Concerted evolution: Molecular mechanism and biological implications. Am. J. Hum. Genet. 1999, 64, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Ganley, A.R.D. Concerted Evolution. In Reference Module in Life Sciences; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Rooney, A.P. Mechanisms underlying the evolution and maintenance of functionally heterogeneous 18S rRNA genes in apicomplexans. Mol. Biol. Evol. 2004, 21, 1704–1711. [Google Scholar] [CrossRef] [PubMed]
- Nishimoto, Y.; Arisue, N.; Kawai, S.; Escalante, A.A.; Horii, T.; Tanabe, K.; Hashimoto, T. Evolution and phylogeny of the heterogeneous cytosolic SSU rRNA genes in the genus Plasmodium. Mol. Phylogenet. Evol. 2008, 47, 45–53. [Google Scholar] [CrossRef]
- Ueno, R.; Huss, V.A.R.; Urano, N.; Watabe, S. Direct evidence for redundant segmental replacement between multiple 18S rRNA genes in a single Prototheca strain. Microbiology 2007, 153, 3879–3893. [Google Scholar] [CrossRef][Green Version]
- Keller, I.; Chintauan-Marquier, I.C.; Veltsos, P.; Nichols, R.A. Ribosomal DNA in the grasshopper Podisma pedestris: Escape from concerted evolution. Genetics 2006, 174, 863–874. [Google Scholar] [CrossRef] [PubMed]
- Carranza, S.; Baguñà, J.; Riutort, M. Origin and evolution of paralogous rRNA gene clusters within the flatworm family Dugesiidae (Platyhelminthes, Tricladida). J. Mol. Evol. 1999, 49, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Daugaard, M.; Rohde, M.; Jäättelä, M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007, 581, 3702–3710. [Google Scholar] [CrossRef] [PubMed]
- Schwerin, M.; Maak, S.; Kalbe, C.; Fuerbass, R. Functional promoter variants of highly conserved inducible hsp70 genes significantly affect stress response. Biochim. Biophys. Acta 2001, 1522, 108–111. [Google Scholar] [CrossRef]
- Gupta, R.S.; Singh, B. Phylogenetic analysis of 70 kD heat shock protein sequences suggests a chimeric origin for the eukaryotic cell nucleus. Curr. Biol. 1994, 4, 1104–1114. [Google Scholar] [CrossRef]
- Gómez-Moracho, T.; Maside, X.; Martín-Hernández, R.; Higes, M.; Bartolomé, C. High levels of genetic diversity in Nosema ceranae within Apis mellifera colonies. Parasitology 2014, 141, 475–481. [Google Scholar] [CrossRef]
- Cilia, G.; Cabbri, R.; Maiorana, G.; Cardaio, I.; Dall’Olio, R.; Nanetti, A. A novel TaqMan® assay for Nosema ceranae quantification in honey bee, based on the protein coding gene Hsp70. Eur. J. Protistol. 2018, 63, 44–50. [Google Scholar] [CrossRef]
- Dong, Z.; Long, J.; Huang, L.; Hu, Z.; Chen, P.; Hu, N.; Zheng, N.; Huang, X.; Lu, C.; Pan, M. Construction and application of an HSP70 promoter-inducible genome editing system in transgenic silkworm to induce resistance to Nosema bombycis. Appl. Microbiol. Biotechnol. 2019, 103, 9583–9592. [Google Scholar] [CrossRef]
- Wang, Y.; Ru, Y.; Liu, W.; Wang, D.; Zhou, J.; Jiang, Y.; Shi, S.; Qin, L. Morphological characterization and HSP70-, IGS-based phylogenetic analysis of two microsporidian parasites isolated from Antheraea pernyi. Parasitol. Res. 2017, 116, 971–977. [Google Scholar] [CrossRef] [PubMed]
- Gobet, P.; Toze, S. Sensitive genotyping of Cryptosporidium parvum by PCR-RFLP analysis of the 70-kilodalton heat shock protein (HSP70) gene. FEMS Microbiol. Lett. 2001, 200, 37–41. [Google Scholar] [CrossRef]
- Garcés-Sanchez, G.; Wilderer, P.A.; Munch, J.C.; Horn, H.; Lebuhn, M. Evaluation of two methods for quantification of hsp70 mRNA from the waterborne pathogen Cryptosporidium parvum by reverse transcription real-time PCR in environmental samples. Water Res. 2009, 43, 2669–2678. [Google Scholar] [CrossRef]
- Germot, A.; Philippe, H.; Le Guyader, H. Evidence for loss of mitochondria in Microsporidia from a mitochondrial-type HSP70 in Nosema locustae. Mol. Biochem. Parasitol. 1997, 87, 159–168. [Google Scholar] [CrossRef]
- Zampieri, R.A.; Laranjeira-Silva, M.F.; Muxel, S.M.; Stocco de Lima, A.C.; Shaw, J.J.; Floeter-Winter, L.M. High Resolution Melting Analysis Targeting hsp70 as a Fast and Efficient Method for the Discrimination of Leishmania Species. PLoS Negl. Trop. Dis. 2016, 10, e0004485. [Google Scholar] [CrossRef]
- Deane, E.E.; Woo, N.Y.S. Impact of heavy metals and organochlorines on hsp70 and hsc70 gene expression in black sea bream fibroblasts. Aquat. Toxicol. 2006, 79, 9–15. [Google Scholar] [CrossRef]
- Lee, G.C.; Nam, S.H.; Chae, J.C.; Lee, C.H. Giardia duodenalis: Improved detection of viable cysts by reverse transcription-PCR of heat shock-inducible hsp70 gene. Exp. Parasitol. 2009, 123, 377–380. [Google Scholar] [CrossRef]
- Liang, Z.; Keeley, A. Detection of viable Cryptosporidium parvum in soil by reverse transcription-real-time PCR targeting hsp70 mRNA. Appl. Environ. Microbiol. 2011, 77, 6476–6485. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Piao, L.X.; Aosai, F.; Chen, M.; Fang, H.; Mun, H.S.; Norose, K.; Yano, A. A quantitative assay method of Toxoplasma gondii HSP70 mRNA by quantitative competitive-reverse transcriptase-PCR. Parasitol. Int. 2004, 53, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Moracho, T.; Bartolomé, C.; Martín-Hernández, R.; Higes, M.; Maside, X. Evidence for weak genetic recombination at the PTP2 locus of Nosema ceranae. Environ. Microbiol. 2015, 17, 1300–1309. [Google Scholar] [CrossRef]
- Maside, X.; Gómez-Moracho, T.; Jara, L.; Martín-Hernández, R.; De La Rúa, P.; Higes, M.; Bartolomé, C. Population genetics of Nosema apis and Nosema ceranae: One host (apis mellifera) and two different histories. PLoS ONE 2015, 10, e0145609. [Google Scholar] [CrossRef]
- Gómez-Moracho, T.; Bartolomé, C.; Bello, X.; Martín-Hernández, R.; Higes, M.; Maside, X. Recent worldwide expansion of Nosema ceranae (Microsporidia) in Apis mellifera populations inferred from multilocus patterns of genetic variation. Infect. Genet. Evol. 2015, 31, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Sagastume, S.; Martín-Hernández, R.; Higes, M.; Henriques-Gil, N. Genotype diversity in the honey bee parasite Nosema ceranae: Multi-strain isolates, cryptic sex or both? BMC Evol. Biol. 2016, 16, 216. [Google Scholar] [CrossRef]
- Krebes, L.; Zeidler, L.; Frankowski, J.; Bastrop, R. (Cryptic) sex in the microsporidian Nosema granulosis – Evidence from parasite rDNA and host mitochondrial DNA. Infect. Genet. Evol. 2014, 21, 259–268. [Google Scholar] [CrossRef]
- Ironside, J.E. Multiple losses of sex within a single genus of Microsporidia. BMC Evol. Biol. 2007 71 2007, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Pelin, A.; Selman, M.; Aris-Brosou, S.; Farinelli, L.; Corradi, N. Genome analyses suggest the presence of polyploidy and recent human-driven expansions in eight global populations of the honeybee pathogen Nosema ceranae. Environ. Microbiol. 2015, 17, 4443–4458. [Google Scholar] [CrossRef]
- Nanetti, A.; Ugolini, L.; Cilia, G.; Pagnotta, E.; Malaguti, L.; Cardaio, I.; Matteo, R.; Lazzeri, L. Seed Meals from Brassica nigra and Eruca sativa Control Artificial Nosema ceranae Infections in Apis mellifera. Microorganisms 2021, 9, 949. [Google Scholar] [CrossRef]
- Cilia, G.; Garrido, C.; Bonetto, M.; Tesoriero, D.; Nanetti, A. Effect of Api-Bioxal ® and ApiHerb ® Treatments against Nosema ceranae Infection in Apis mellifera Investigated by Two qPCR Methods. Vet. Sci. 2020, 7, 125. [Google Scholar] [CrossRef]
- Bourgeois, A.L.; Rinderer, T.E.; Beaman, L.D.; Danka, R.G. Genetic detection and quantification of Nosema apis and N. ceranae in the honey bee. J. Invertebr. Pathol. 2010, 103, 53–58. [Google Scholar] [CrossRef]
- Paxton, R.J.; Klee, J.; Korpela, S.; Fries, I. Nosema ceranae has infected Apis mellifera in Europe since at least 1998 and may be more virulent than Nosema apis. Apidologie 2007, 38, 558–565. [Google Scholar] [CrossRef]
- Wilson, D.M.; Thompson, L.H. Molecular mechanisms of sister-chromatid exchange. Mutat. Res. -Fundam. Mol. Mech. Mutagen. 2007, 616, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Alexander, W.G.; Wisecaver, J.H.; Rokas, A.; Hittinger, C.T. Horizontally acquired genes in early-diverging pathogenic fungi enable the use of host nucleosides and nucleotides. Proc. Natl. Acad. Sci. USA 2016, 113, 4116–4121. [Google Scholar] [CrossRef] [PubMed]
- Husnik, F.; McCutcheon, J.P. Functional horizontal gene transfer from bacteria to eukaryotes. Nat. Publ. Gr. 2017, 16, 67–79. [Google Scholar] [CrossRef]
- Mortier, J.; Gayán, E.; Van Eyken, R.; Torres Montaguth, O.E.; Khodaparast, L.; Khodaparast, L.; Houben, B.; Carpentier, S.; Rousseau, F.; Schymkowitz, J.; et al. Gene Erosion Can Lead to Gain-of-Function Alleles That Contribute to Bacterial Fitness. MBio 2021, 12. [Google Scholar] [CrossRef]
- Brown, B.P.; Wernegreen, J.J. Genomic erosion and extensive horizontal gene transfer in gut-associated Acetobacteraceae. BMC Genomics 2019, 20, 1–15. [Google Scholar] [CrossRef]
- Kwong, W.K.; Moran, N.A. Gut microbial communities of social bees. Nat. Rev. Microbiol. 2016, 14, 374–384. [Google Scholar] [CrossRef]
- Gisder, S.; Genersch, E. Molecular differentiation of Nosema apis and Nosema ceranae based on species–specific sequence differences in a protein coding gene. J. Invertebr. Pathol. 2013, 113, 1–6. [Google Scholar] [CrossRef]
- Stevanovic, J.; Stanimirovic, Z.; Genersch, E.; Kovacevic, S.R.; Ljubenkovic, J.; Radakovic, M.; Aleksic, N. Dominance of Nosema ceranae in honey bees in the Balkan countries in the absence of symptoms of colony collapse disorder. Apidologie 2011, 42, 49–58. [Google Scholar] [CrossRef]
- Erler, S.; Lommatzsch, S.; Lattorff, H.M.G. Comparative analysis of detection limits and specificity of molecular diagnostic markers for three pathogens (Microsporidia, Nosema spp.) in the key pollinators Apis mellifera and Bombus terrestris. Parasitol. Res. 2012, 110, 1403–1410. [Google Scholar] [CrossRef] [PubMed]
| Gene | Primer | Sequence (5′-3′) | Reference |
|---|---|---|---|
| 16S rRNA | Forward | AAGAGTGAGACCTATCAGCTAGTTG | |
| Reverse | CCGTCTCTCAGGCTCCTTCTC | [70] | |
| Taqman Probe | ACCGTTACCCGTCACAGCCTTGTT | ||
| Hsp70 | Forward | GGGATTACAAGTGCTTAGAGTGATT | |
| Reverse | TGTCAAGCCCATAAGCAAGTG | [50] | |
| Taqman Probe | TGAGCCTACTGCGGC |
| Model | F (1.304) = 800.403, p = 0.000, Adj. R2 = 0.724 |
| Intercept | 376,209.4 ± 50,726.0 s.e. (95% CI = 276,390.9, 476,028.0), t (304) = 7.417, p = 0.000 |
| Slope | 7.8 ± 0.3 s.e. (95% CI = 7.3, 8.4), t (304) = 28.291, p = 0.000 |
| Model | F (1.304) = 38.740, p = 0.000, Adj. R2 = 0.110 |
| Intercept | 11.9 ± 0.2 s.e. (95% CI = 11.5, 12.4), t (304) = 53.551, p = 0.000 |
| Slope | −7.6 × 10−6 ± 1.2 × 10−6 s.e. (95% CI = −10 × 10−6, −5.2 × 10−6), t (304) = −6.224, p = 0.000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cilia, G.; Luchetti, G.; Nanetti, A. Polymorphism of 16s rRNA Gene: Any Effect on the Biomolecular Quantitation of the Honey Bee (Apis mellifera L., 1758) Pathogen Nosema ceranae? Appl. Sci. 2022, 12, 422. https://doi.org/10.3390/app12010422
Cilia G, Luchetti G, Nanetti A. Polymorphism of 16s rRNA Gene: Any Effect on the Biomolecular Quantitation of the Honey Bee (Apis mellifera L., 1758) Pathogen Nosema ceranae? Applied Sciences. 2022; 12(1):422. https://doi.org/10.3390/app12010422
Chicago/Turabian StyleCilia, Giovanni, Giacomo Luchetti, and Antonio Nanetti. 2022. "Polymorphism of 16s rRNA Gene: Any Effect on the Biomolecular Quantitation of the Honey Bee (Apis mellifera L., 1758) Pathogen Nosema ceranae?" Applied Sciences 12, no. 1: 422. https://doi.org/10.3390/app12010422
APA StyleCilia, G., Luchetti, G., & Nanetti, A. (2022). Polymorphism of 16s rRNA Gene: Any Effect on the Biomolecular Quantitation of the Honey Bee (Apis mellifera L., 1758) Pathogen Nosema ceranae? Applied Sciences, 12(1), 422. https://doi.org/10.3390/app12010422







