Migration Groups: A Poorly Explored Point of View for Genetic Damage Assessment Using Comet Assay in Human Lymphocytes
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents Used
2.2. Obtaining Blood Samples
2.3. Genetic Damage Induced by MMS, MH, 2,4-D and NDEA
2.4. Alkaline Comet Assay
2.5. Observation and Quantification of Comets
2.6. Proposed Parameters
2.7. Statistical Analysis
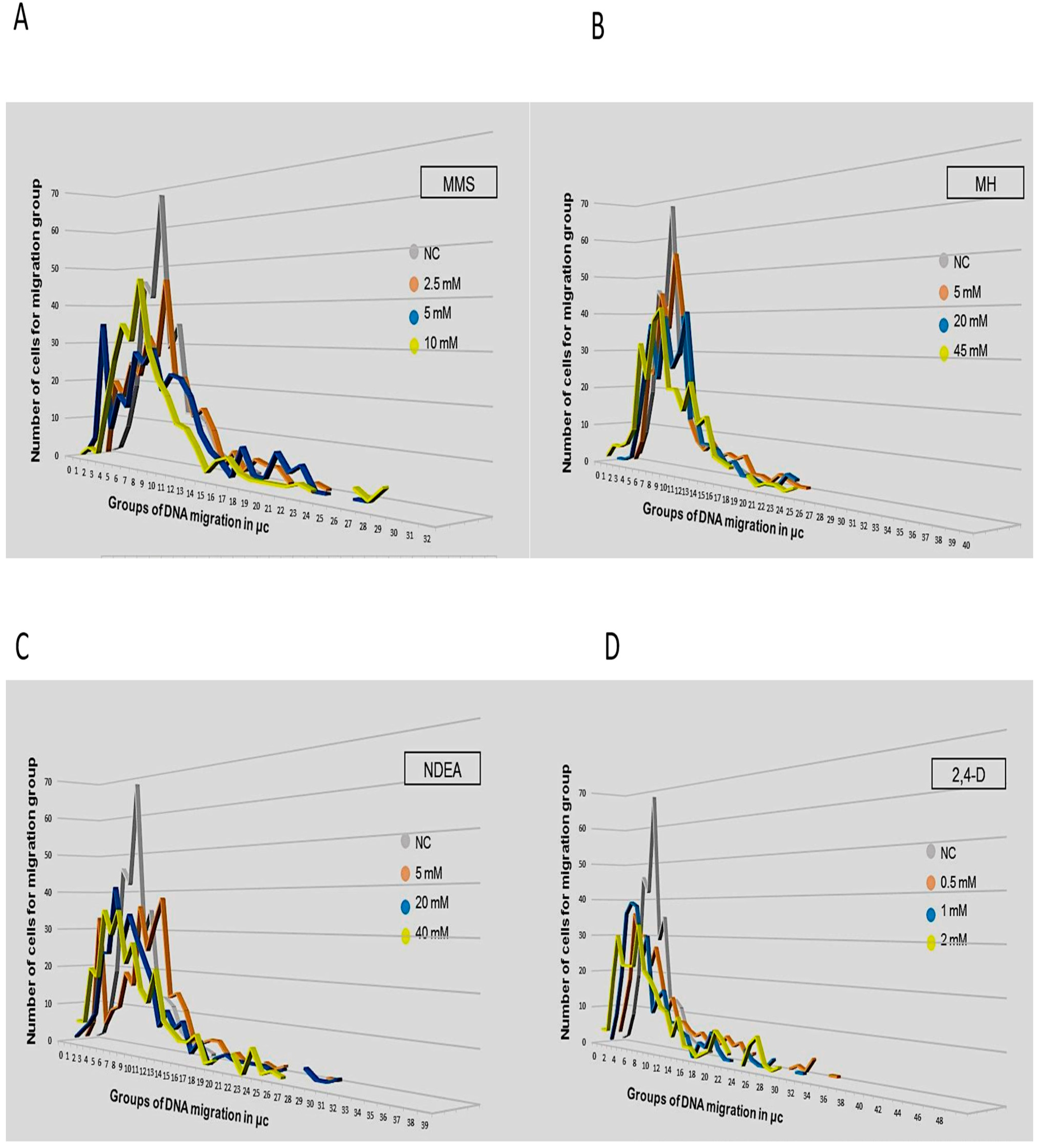
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statements
Acknowledgments
Conflicts of Interest
References
- Møller, P. The comet assay: Ready for 30 more years. Mutagenesis 2018, 33, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Cortés-Gutiérrez, E.I.; Dávila-Rodríguez, M.I.; Fernández, J.L.; López-Fernández, C.; Gosálbez, A.; Gosálvez, J. New application of the comet assay: Chromosome–comet assay. J. Histochem. Cytochem. 2011, 59, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Martus, H.J.; Hayashi, M.; Honma, M.; Kasper, P.; Gollapudi, B.; Mueller, L.; Schoeny, R.; Uno, Y.; Kirkland, D.J. Summary of major conclusions from the 6th international workshop on genotoxicity testing (IWGT), Foz do Iguaçu, Brazil. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2015, 783, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Fahim, M.; Ahmed, A.; Hussain, S. Single Cell Gel Electrophoresis and its Applications in Different Fields. J. Bioequiv. Availab. 2017, 1, 1–4. [Google Scholar]
- Rodríguez, R.A.; Noris, G.E.; Fundora, T.M.T. Principios y relevancia del ensayo cometa. Rev. Cubana Investig. Bioméd. 2016, 35, 184–194. [Google Scholar]
- Singh, N.P.; McCoy, M.T.; Tice, R.R.; Schneider, E.L. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 1988, 175, 184–191. [Google Scholar] [CrossRef]
- Alvarez-Moya, C.; Santerre-Lucas, A.; Zúñiga-González, G.; Torres-Bugarín, O.; Padilla-Camberos, E.; Feria-Velasco, A. Evaluation of genotoxic activity of maleic hydrazide, ethyl methane sulfonate, and N-nitroso diethylamine in Tradescantia. Salud Publ. Mex. 2001, 43, 563–569. [Google Scholar] [CrossRef]
- Olive, P.L.; Banáth, J.P. The comet assay: A method to measure DNA damage in individual cells. Nat. Protoc. 2006, 1, 23–29. [Google Scholar] [CrossRef]
- Gajski, G.; Žegura, B.; Ladeira, C.; Novak, M.; Sramkova, M.; Pourrut, B.; Collins, A. The comet assay in animal models: From bugs to whales—(Part 2 Vertebrates). Mutat. Res./Rev. Mutat. Res. 2019, 781, 130–164. [Google Scholar] [CrossRef]
- Koppen, G.; Toncelli, L.M.; Triest, L.; Verschaeve, L. The comet assay: A tool to study alteration of DNA integrity in developing plant leaves. Mech. Ageing Dev. 1999, 110, 13–24. [Google Scholar] [CrossRef]
- Azqueta, A.; Collins, A.R. The essential comet assay: A comprehensive guide to measuring DNA damage and repair. Arch. Toxicol. 2013, 87, 949–968. [Google Scholar] [CrossRef] [PubMed]
- Møller, P.; Loft, S.; Ersson, C.; Koppen, G.; Dusinska, M.; Collins, A. On the search for an intelligible comet assay descriptor. Front. Genet. 2014, 5, 217. [Google Scholar] [PubMed]
- Collins, A.R.; Oscoz, A.A.; Brunborg, G.; Gaivão, I.; Giovannelli, L.; Kruszewski, M.; Smith, C.C.; Štětina, R. The comet assay: Topical issues. Mutagenesis 2008, 23, 143–151. [Google Scholar] [CrossRef]
- Forchhammer, L.; Bräuner, E.V.; Folkmann, J.K.; Danielsen, P.H.; Nielsen, C.; Jensen, A.; Loft, S.; Friis, G.; Møller, P. Variation in assessment of oxidatively damaged DNA in mononuclear blood cells by the comet assay with visual scoring. Mutagenesis 2008, 23, 223–231. [Google Scholar] [CrossRef]
- Tice, R.R.; Agurell, E.; Anderson, D.; Burlinson, B.; Hartmann, A. Single cell gel/comet assay: Guidelines for in vitro and in vivo genetic toxicology testing. Environ. Mol. Mutagen. 2000, 35, 206–221. [Google Scholar] [CrossRef]
- Jha, A.N. Ecotoxicological applications and significance of the comet assay. Mutagenesis 2008, 23, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Speit, G.; Kojima, H.; Burlinson, B.; Collins, A.R.; Kasper, P. Critical issues with the in vivo comet assay: A report of the comet assay working group in the 6th International Workshop on Genotoxicity Testing (IWGT). Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2015, 783, 6–12. [Google Scholar] [CrossRef]
- Collins, A.R. The Use of Bacterial Repair Endonucleases in the Comet Assay. Methods Mol. Biol. 2011, 691, 137–147. [Google Scholar] [PubMed]
- Olive, P.L.; Banath, J.P. Detection of DNA double-strand breaks through the cell cycle after exposure to X-rays, bleomycin, etoposide and 125IdUrd. Int. J. Radiat. Biol. 1993, 64, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Moya, C.; Reynoso-Silva, M.; Canales-Aguirre, A.A.; Chavez-Chavez, J.O.; Castañeda-Vázquez, H.; Feria-Velasco, A.I. Heterogeneity of genetic damage in cervical nuclei and lymphocytes in women with different levels of dysplasia and cancer-associated risk factors. BioMed Res. Int. 2015, 2015, 293408. [Google Scholar] [CrossRef] [PubMed]
- Lovell, D.P.; Omori, T. Statistical issues in the use of the comet assay. Mutagenesis 2008, 23, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Miyaji, C.K.; Jordão, B.Q.; Ribeiro, L.R.; Eira, A.F.; Cólus, I.M. Genotoxicity and antigenotoxicity assessment of shiitake (Lentinula edodes (Berkley) Pegler) using the Comet assay. Genet. Mol. Biol. 2004, 27, 108–114. [Google Scholar] [CrossRef][Green Version]
- Nuñes, A.P.; Ferreira-Machado, S.C.; Nunes, R.M.; Dantas, F.J.; De Mattos, J.C. Analysis of genotoxic potentiality of stevioside by comet assay. Food Chem. Toxicol. 2007, 45, 662–666. [Google Scholar] [CrossRef]
- Friedberg, E.C.; Walker, G.C.; Siede, W.; Wood, R.D. DNA Repair and Mutagenesis; American Society for Microbiology Press: Whashington, DC, USA, 2005; pp. 71–98. [Google Scholar]
- Fedel-Miyasato, L.; Formagio, A.; Auharek, S.; Kassuya, C.; Navarro, S.; Cunha-Laura, A.; Monreal, A.; Vieira, M.; Oliveira, R. Antigenotoxic and antimutagenic effects of Schinus terebinthifolius Raddi in Allium cepa and Swiss mice: A comparative study. Genet. Mol. Res. 2014, 13, 3411–3425. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Oh, E.; Sul, D.; Lee, J. Use of the tail moment of the lymphocytes to evaluate DNA damage in human biomonitoring studies. Toxicol. Sci. 2004, 81, 121–132. [Google Scholar] [CrossRef]
- Møller, P.; Azqueta, A.; Boutet-Robinet, E.; Koppen, G.; Bonassi, S.; Milić, M.; Gajski, G.; Costa, S.; Teixeira, J.P.; Pereira, C.C.; et al. Minimum Information for Reporting on the Comet Assay (MIRCA): Recommendations for describing comet assay procedures and results. Nat. Protoc. 2020, 15, 3817–3826. [Google Scholar] [CrossRef]
- Mauro, M.O.; Monreal, M.T.; Silva, M.; Pesarini, J.R.; Mantovani, M.S. Evaluation of the antimutagenic and anticarcinogenic effects of inulin in vivo. Genet. Mol. Res. 2013, 12, 2281–2293. [Google Scholar] [CrossRef] [PubMed]
- Gerić, M.; Gajski, G.; Oreščanin, V.; Garaj-Vrhovac, V. Seasonal variations as predictive factors of the comet assay parameters: A retrospective study. Mutagenesis 2018, 33, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Andem, A.B.; Agbor, R.B.; Ekpo, I.A. Review on Comet Assay: A reliable tool for assessing DNA damage in animal models. J. Curr. Res. 2013, 1, 405–427. [Google Scholar]
- Dhawan, A.; Bajpayee, M.; Parmar, D. Comet assay: A reliable tool for the assessment of DNA damage in different models. Cell Biol. Toxicol. 2009, 25, 5–32. [Google Scholar] [CrossRef]
- Ateeq, B.; Farah, M.A.; Ahmad, W. Detection of DNA damage by alkaline single cell gel electrophoresis in 2, 4-dichlorophenoxyacetic-acid-and butachlor-exposed erythrocytes of Clarias batrachus. Ecotoxicol. Environ. Saf. 2015, 62, 348–354. [Google Scholar] [CrossRef]
- Alvarez-Moya, C.; Reynoso Silva, M.; Villalobos Arámbula, A.R.; Islas Sandoval, A.; Castañeda Vasquez, H.; González Montes, R.M. Evaluation of genetic damage induced by glyphosate isopropylamine salt using Tradescantia bioassays. Genet. Mol. Biol. 2011, 34, 127–130. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mihaljevic, O.; Zivancevic-Simonovic, S.; Milosevic-Djordjevic, O.; Djurdjevic, P.; Jovanovic, D.; Todorovic, Z.; Grujicic, D.; Radovic-Jakovljevic, M.; Tubic, J.; Markovic, A.; et al. Apoptosis and genome instability in children with autoimmune diseases. Mutagenesis 2018, 33, 351–357. [Google Scholar] [CrossRef] [PubMed]

| Experimental Groups | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Migration Groups in µc | NC | MMS 2.5 mM | MMS 5 mM | MMS 10 mM | MH 5 mM | MH 20 mM | MH 45 mM | 2,4-D 0.5 mM | 2,4-D 1 mM | 2,4-D 2 mM | NDEA 5 mM | NDEA 20 mM | NDEA 40 mM |
| 0 | 1 | 1 | 1 | 1 | |||||||||
| 1 | 2 | 1 | 5 | 1 | 1 | 2 | 3 | 1 | 4 | 5 | 3 | ||
| 2 | 8 | 20 | 35 | 3 | 1 | 1 | 5 | 10 | 8 | 4 | 33 | 5 | 6 |
| 3 | 19 | 16 | 8 | 2 | 6 | 2 | 5 | 18 | 19 | 18 | 5 | 8 | 6 |
| 4 | 46 | 25 | 17 | 14 | 25 | 11 | 6 | 36 | 36 | 30 | 9 | 24 | 20 |
| 5 | 42 | 22 | 14 | 26 | 24 | 24 | 10 | 30 | 39 | 22 | 10 | 24 | 18 |
| 6 | 68 | 32 | 28 | 35 | 45 | 37 | 32 | 30 | 38 | 22 | 19 | 41 | 35 |
| 7 | 29 | 27 | 25 | 31 | 36 | 23 | 23 | 22 | 24 | 22 | 16 | 27 | 29 |
| 8 | 35 | 46 | 29 | 46 | 55 | 39 | 38 | 27 | 30 | 33 | 36 | 34 | 35 |
| 9 | 13 | 22 | 19 | 29 | 39 | 26 | 41 | 19 | 10 | 21 | 25 | 28 | 22 |
| 10 | 13 | 22 | 23 | 22 | 13 | 30 | 21 | 10 | 14 | 19 | 32 | 23 | 27 |
| 11 | 11 | 13 | 22 | 18 | 8 | 40 | 21 | 14 | 16 | 16 | 38 | 18 | 16 |
| 12 | 5 | 15 | 18 | 12 | 6 | 17 | 16 | 8 | 8 | 12 | 14 | 7 | 13 |
| 13 | 3 | 10 | 10 | 11 | 8 | 8 | 23 | 8 | 8 | 11 | 15 | 11 | 21 |
| 14 | 2 | 2 | 6 | 7 | 5 | 8 | 13 | 6 | 6 | 5 | 11 | 7 | 9 |
| 15 | 2 | 6 | 4 | 2 | 6 | 7 | 15 | 5 | 5 | 10 | 4 | 9 | 7 |
| 16 | 1 | 2 | 1 | 5 | 5 | 4 | 6 | 6 | 3 | 4 | 2 | 5 | |
| 17 | 5 | 8 | 6 | 6 | 5 | 5 | 4 | 4 | 3 | 5 | 5 | 5 | |
| 18 | 4 | 2 | 3 | 2 | 2 | 4 | 7 | 3 | 1 | 5 | 7 | ||
| 19 | 4 | 2 | 2 | 2 | 2 | 4 | 7 | 2 | 2 | 2 | 1 | ||
| 20 | 1 | 8 | 2 | 1 | 1 | 3 | 6 | 3 | 3 | 3 | 3 | 2 | |
| 21 | 4 | 2 | 3 | 1 | 1 | 4 | 1 | 8 | 1 | 2 | |||
| 22 | 2 | 6 | 2 | 2 | 1 | 2 | 5 | 1 | 6 | 2 | 4 | ||
| 23 | 1 | 1 | 3 | 1 | 4 | 2 | 2 | 3 | 2 | 2 | |||
| 24 | 1 | 2 | 1 | 3 | 1 | 3 | 4 | 1 | 2 | 6 | |||
| 25 | 2 | 2 | 2 | 1 | 2 | 1 | 1 | ||||||
| 26 | 1 | 1 | 3 | 1 | 6 | 2 | 2 | ||||||
| 27 | 1 | 4 | 1 | 1 | 8 | 1 | |||||||
| 28 | 2 | 3 | 2 | 3 | |||||||||
| 29 | 1 | 5 | 1 | 1 | 1 | 1 | 1 | ||||||
| 30 | 2 | 2 | 1 | 1 | |||||||||
| 31 | 1 | 1 | 1 | 2 | |||||||||
| 32 | 4 | 1 | 2 | ||||||||||
| 33 | 2 | ||||||||||||
| 34 | 1 | 1 | 1 | ||||||||||
| 35 | 1 | 2 | |||||||||||
| 36 | 1 | ||||||||||||
| 37 | |||||||||||||
| 38 | 1 | 1 | |||||||||||
| 39 | |||||||||||||
| 40 | |||||||||||||
| 41 | 1 | 1 | |||||||||||
| 42 | |||||||||||||
| 43 | |||||||||||||
| 44 | |||||||||||||
| 45 | |||||||||||||
| 46 | 1 | ||||||||||||
| 47 | |||||||||||||
| 48 | |||||||||||||
| 49 | 1 | ||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reynoso-Silva, M.; Álvarez-Moya, C.; Ramírez-Velasco, R.; Sámano-León, A.G.; Arvizu-Hernández, E.; Castañeda-Vásquez, H.; Ruíz-Lopez, M.A. Migration Groups: A Poorly Explored Point of View for Genetic Damage Assessment Using Comet Assay in Human Lymphocytes. Appl. Sci. 2021, 11, 4094. https://doi.org/10.3390/app11094094
Reynoso-Silva M, Álvarez-Moya C, Ramírez-Velasco R, Sámano-León AG, Arvizu-Hernández E, Castañeda-Vásquez H, Ruíz-Lopez MA. Migration Groups: A Poorly Explored Point of View for Genetic Damage Assessment Using Comet Assay in Human Lymphocytes. Applied Sciences. 2021; 11(9):4094. https://doi.org/10.3390/app11094094
Chicago/Turabian StyleReynoso-Silva, Mónica, Carlos Álvarez-Moya, Rafael Ramírez-Velasco, Alexis Gerardo Sámano-León, Erandi Arvizu-Hernández, Hugo Castañeda-Vásquez, and Mario Alberto Ruíz-Lopez. 2021. "Migration Groups: A Poorly Explored Point of View for Genetic Damage Assessment Using Comet Assay in Human Lymphocytes" Applied Sciences 11, no. 9: 4094. https://doi.org/10.3390/app11094094
APA StyleReynoso-Silva, M., Álvarez-Moya, C., Ramírez-Velasco, R., Sámano-León, A. G., Arvizu-Hernández, E., Castañeda-Vásquez, H., & Ruíz-Lopez, M. A. (2021). Migration Groups: A Poorly Explored Point of View for Genetic Damage Assessment Using Comet Assay in Human Lymphocytes. Applied Sciences, 11(9), 4094. https://doi.org/10.3390/app11094094





