Modeled 3D-Structures of Proteobacterial Transglycosylases from Glycoside Hydrolase Family 17 Give Insight in Ligand Interactions Explaining Differences in Transglycosylation Products
Abstract
1. Introduction
2. Materials and Methods
2.1. Modeling of the 3D Molecular Structures
2.2. Model Refinement
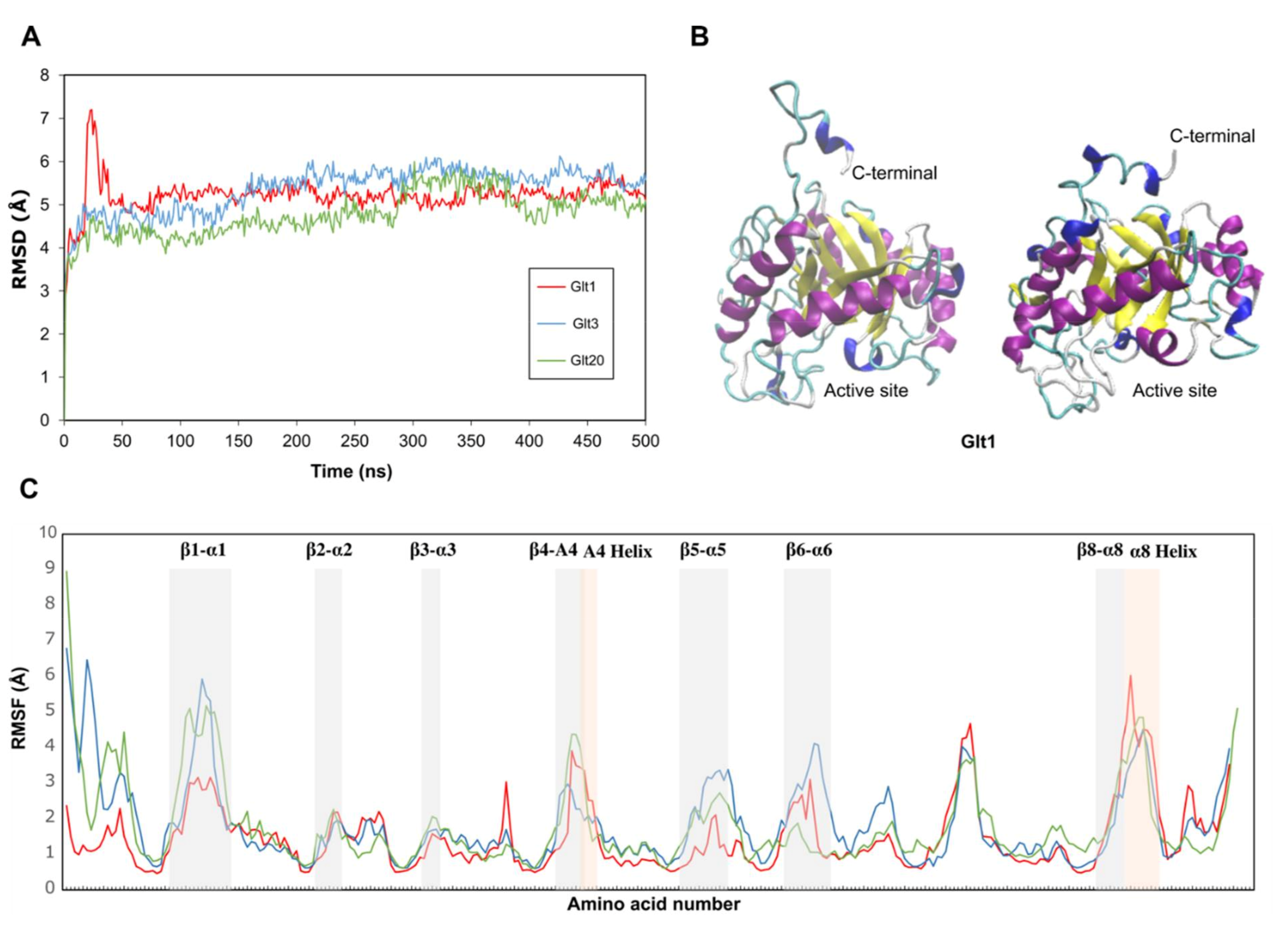
2.3. Molecular Dynamics
2.4. Comparison of Modeled Proteins with Crystallographic Structures
2.5. Mutant Construction
2.6. Expression and Purification of -Glt20 Variants
2.7. Activity Assay, and Visualization by Thin Layer Chromatography (TLC)
2.8. Ligand Modeling
2.9. Docking
3. Results and Discussion
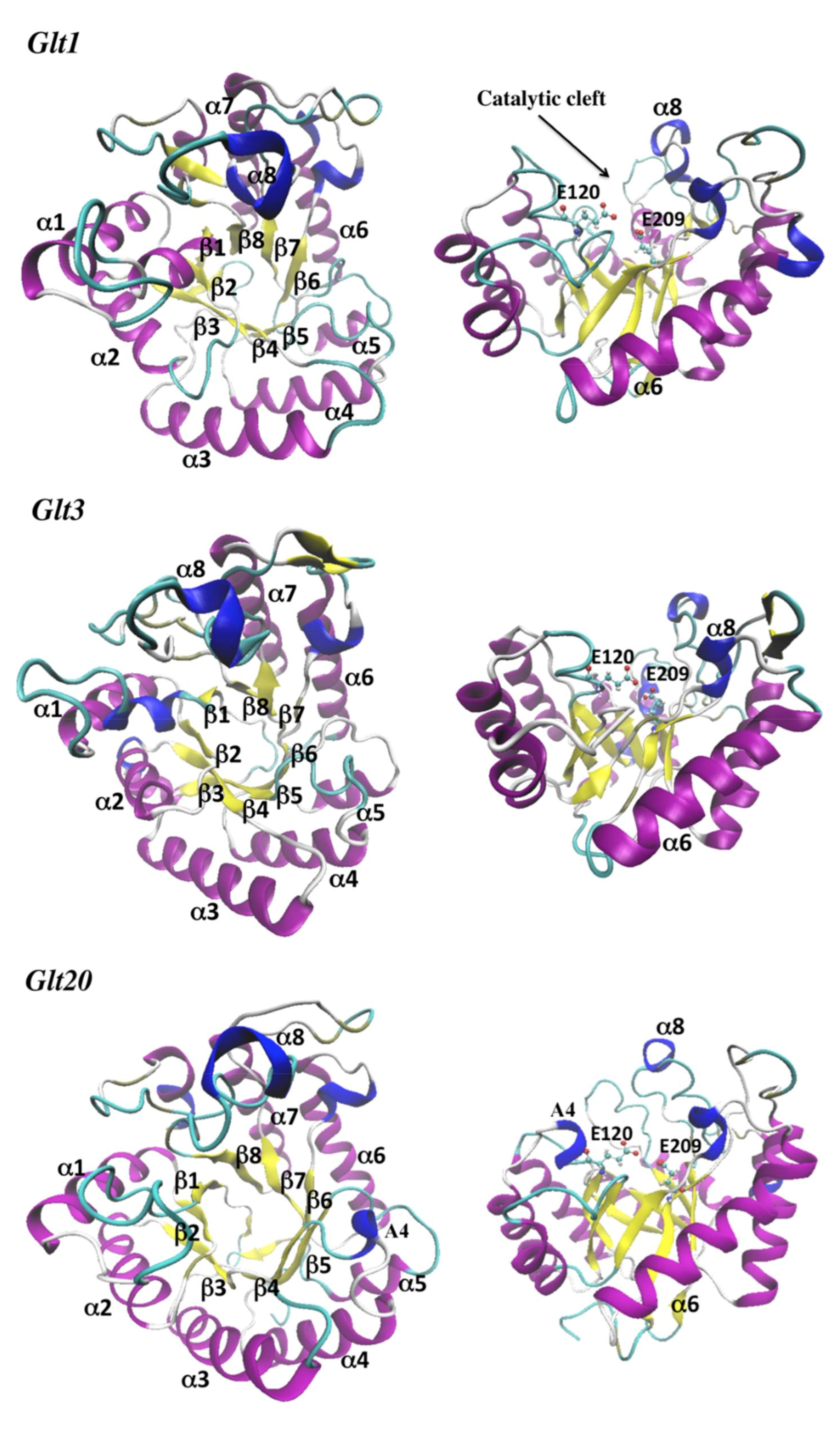
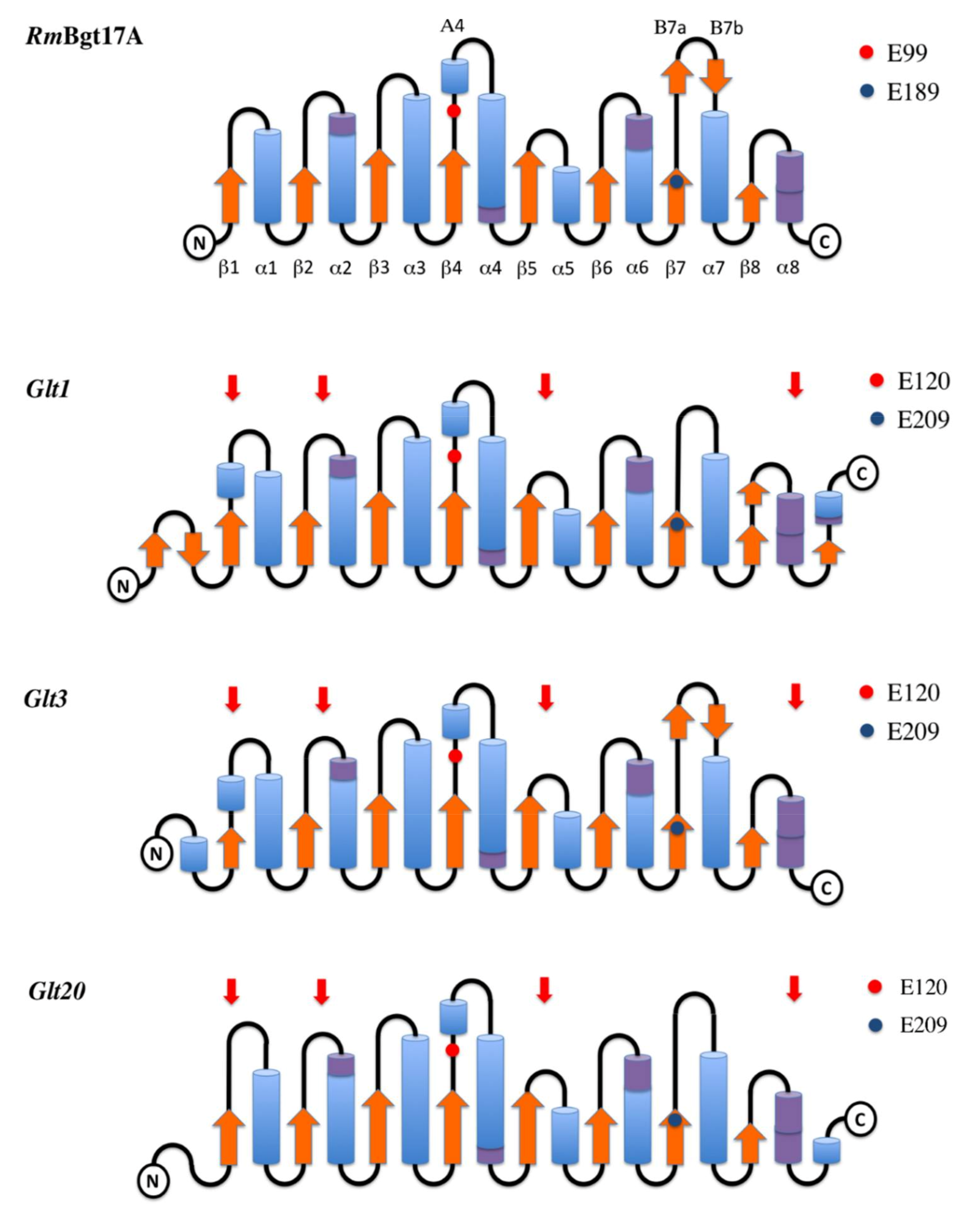
3.1. Homology Modeling and Overall Structure
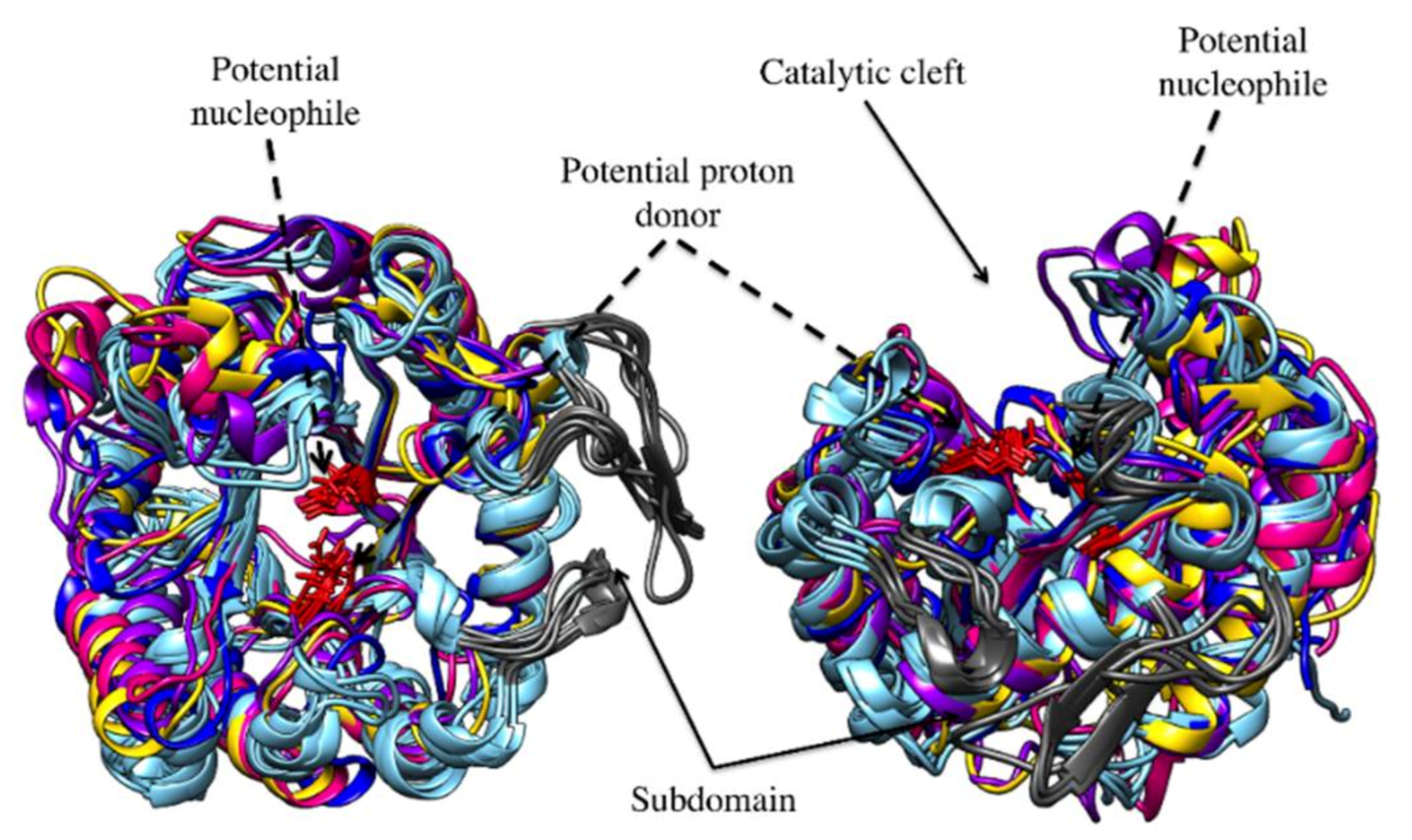
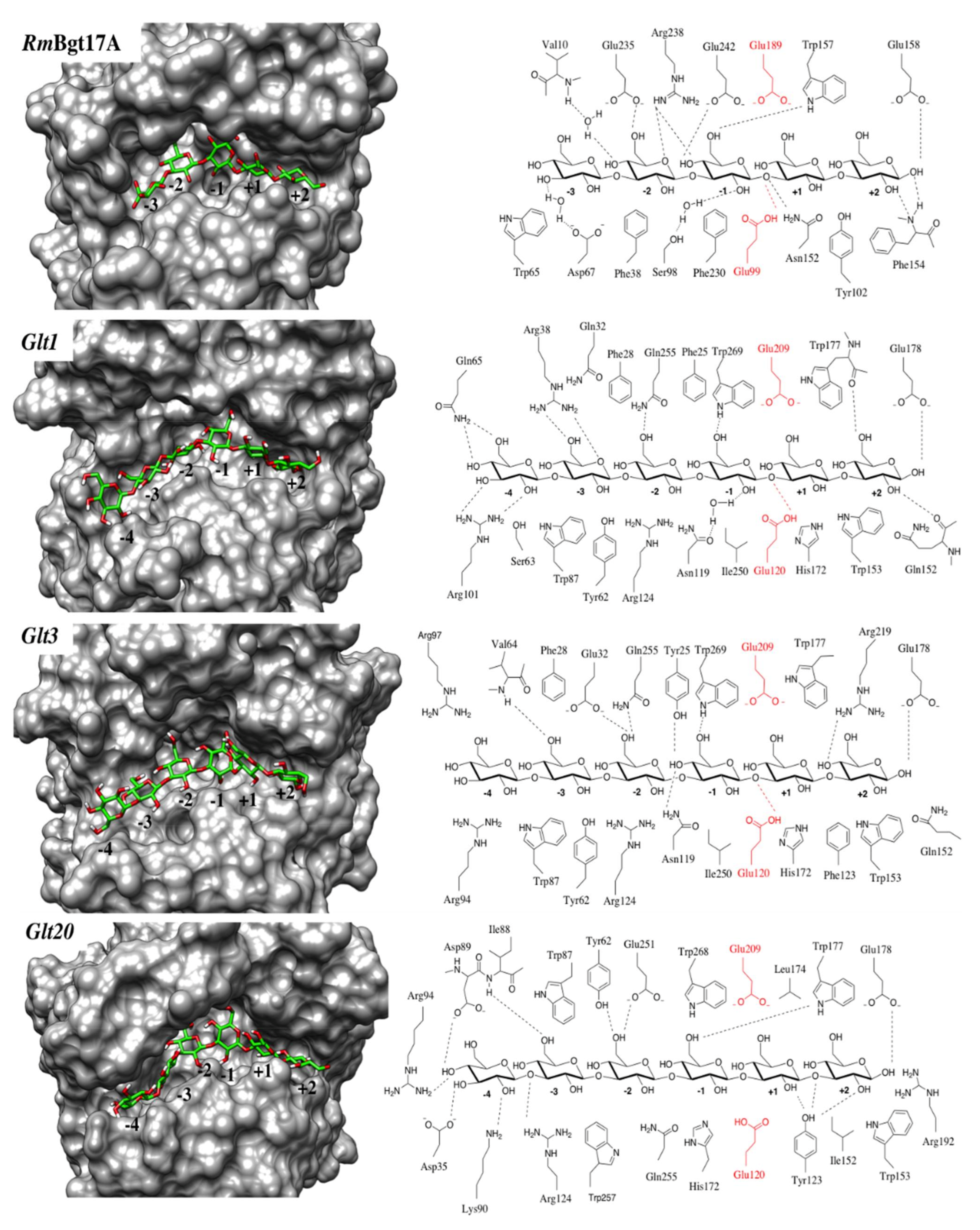
3.2. Catalytic Cleft and Sub Site Predictions
3.3. Ligand Interactions after Docking of Laminaripentaose and Laminarihexaose
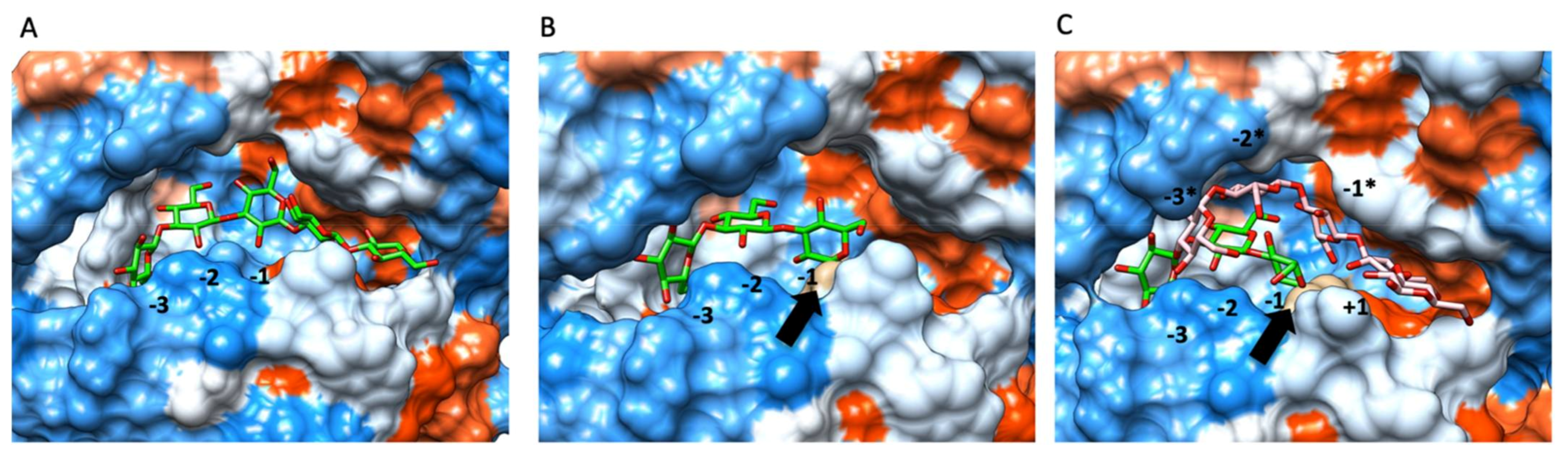
3.4. Presence of Bound Donor Allows Acceptor Binding to Branching Subsites in Glt20
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lairson, L.L.; Henrissat, B.; Davies, G.J.; Withers, S.G. Glycosyltransferases: Structures, functions, and mechanisms. Annu. Rev. Biochem. 2008, 77, 521–555. [Google Scholar] [CrossRef] [PubMed]
- Bissaro, B.; Monsan, P.; Faure, R.; O’Donohue, M.J. Glycosynthesis in a waterworld: New insight into the molecular basis of transglycosylation in retaining glycoside hydrolases. Biochem. J. 2015, 467, 17–35. [Google Scholar] [CrossRef]
- Lundemo, P.; Karlsson, E.N.; Adlercreutz, P. Eliminating hydrolytic activity without affecting the transglycosylation of a GH1 β-glucosidase. Appl. Microbiol. Biotechnol. 2017, 101, 1121–1131. [Google Scholar] [CrossRef]
- Moulis, C.; Guieysse, D.; Morel, S.; Séverac, E.; Remaud-Siméon, M. Natural and engineered transglycosylases: Green tools for the enzyme-based synthesis of glycoproducts. Curr. Opin. Chem. Biol. 2021, 61, 96–106. [Google Scholar] [CrossRef]
- Seibel, J.; Jördening, H.-J.; Buchholz, K. Glycosylation with activated sugars using glycosyltransferases and transglycosidases. Biocatal. Biotransform. 2006, 24, 311–342. [Google Scholar] [CrossRef]
- Desmet, T.; Soetaert, W. Enzymatic glycosyl transfer: Mechanisms and applications. Biocatal. Biotransform. 2011, 29, 1–18. [Google Scholar] [CrossRef]
- Ara, K.Z.G.; Linares-Pastén, J.A.; Jönsson, J.; Viloria-Cols, M.; Ulvenlund, S.; Adlercreutz, P.; Nordberg Karlsson, E. Engineering CGTase to improve synthesis of alkyl glycosides. Glycobiology 2020, 1–10. [Google Scholar] [CrossRef]
- Ngo, N.T.; Linares-Pastén, J.A.; Grey, C.; Adlercreutz, P. Synthesis of novel oligomeric anionic alkyl glycosides using laccase/TEMPO oxidation and cyclodextrin glucanotransferase (CGTase)-catalysed transglycosylation. Biotechnol. Bioeng. 2021. [CrossRef]
- Cho, E.; Jeong, D.; Choi, Y.; Jung, S. Properties and current applications of bacterial cyclic β-glucans and their derivatives. J. Incl. Phenom. Macrocycl. Chem. 2016, 85, 175–185. [Google Scholar] [CrossRef]
- Mah, T.-F.; Pitts, B.; Pellock, B.; Walker, G.C.; Stewart, P.S.; O’Toole, G.A. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 2003, 426, 306–310. [Google Scholar] [CrossRef]
- Bhagwat, A.A.; Gross, K.C.; Tully, R.E.; Keister, D.L. Beta-glucan synthesis in Bradyrhizobium japonicum: Characterization of a new locus (ndvC) influencing beta-(1→6) linkages. J. Bacteriol. 1996, 178, 4635–4642. [Google Scholar] [CrossRef]
- Hreggvidsson, G.O.; Dobruchowska, J.M.; Fridjonsson, O.H.; Jonsson, J.O.; Gerwig, G.J.; Aevarsson, A.; Kristjansson, J.K.; Curti, D.; Redgwell, R.R.; Hansen, C.-E.E.; et al. Exploring novel non-Leloir β-glucosyltransferases from proteobacteria for modifying linear (β1->3)-linked gluco-oligosaccharide chains. Glycobiology 2011, 21, 304–328. [Google Scholar] [CrossRef]
- Jonsson Wheat, J.O.; Hreggvidsson, G.O.; Fridjonsson, O.H.; Dobruchowska, J.M.; Kamerling, J.P. Glucan Branching Enzymes and Their Methods of Use. U.S. Patent 14/780,106, 15 September 2016. [Google Scholar]
- Qin, Z.; Yan, Q.; Lei, J.; Yang, S.; Jiang, Z.; Wu, S. The first crystal structure of a glycoside hydrolase family 17 beta-1,3-glucanosyltransferase displays a unique catalytic cleft. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 1714–1724. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. YASARA View—Molecular graphics for all devices—From smartphones to workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Darden, T.; Nabuurs, S.B.; Finkelstein, A.; Vriend, G. Making optimal use of empirical energy functions: Force-field parameterization in crystal space. Proteins 2004, 57, 678–683. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Berendsen, H.J.; van der Spoel, D.; van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef]
- Aronsson, A.; Güler, F.; Petoukhov, M.V.; Crennell, S.J.; Svergun, D.I.; Linares-Pastén, J.A.; Nordberg Karlsson, E. Structural insights of RmXyn10A–A prebiotic-producing GH10 xylanase with a non-conserved aglycone binding region. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 292–306. [Google Scholar] [CrossRef]
- Bustos, A.-S.; Håkansson, A.; Linares-Pastén, J.A.; Nilsson, L. Interaction between myricetin aggregates and lipase under simplified intestinal conditions. Foods 2020, 9, 777. [Google Scholar] [CrossRef] [PubMed]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed]
- Nosé, S.; Klein, M. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983, 50, 1055–1076. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera— A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Zheng, L.; Baumann, U.; Reymond, J.L. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004, 32, e115. [Google Scholar] [CrossRef]
- Motejadded, H.; Altenbuchner, J. Construction of a dual-tag system for gene expression, protein affinity purification and fusion protein processing. Biotechnol. Lett. 2009, 31, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Wojtkowiak, A.; Witek, K.; Hennig, J.; Jaskolski, M. Structures of an active-site mutant of a plant 1,3-beta-glucanase in complex with oligosaccharide products of hydrolysis. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Yan, Q.; Yang, S.; Jiang, Z. Modulating the function of a β-1, 3-glucanosyltransferase to that of an endo-β-1, 3-glucanase by structure-based protein engineering. Appl. Microbiol. Biotechnol. 2016, 100, 1765–1776. [Google Scholar] [CrossRef] [PubMed]
- Dobruchowska, J.M.; Jonsson, J.O.; Fridjonsson, O.H.; Aevarsson, A.; Kristjansson, J.K.; Altenbuchner, J.; Watzlawick, H.; Gerwig, G.J.; Dijkhuizen, L.; Kamerling, J.P. Modification of linear (β1→3)-linked gluco-oligosaccharides with a novel recombinant β-glucosyltransferase (trans-β-glucosidase) enzyme from Bradyrhizobium diazoefficiens. Glycobiology 2016, 26, 1157–1170. [Google Scholar] [CrossRef] [PubMed]

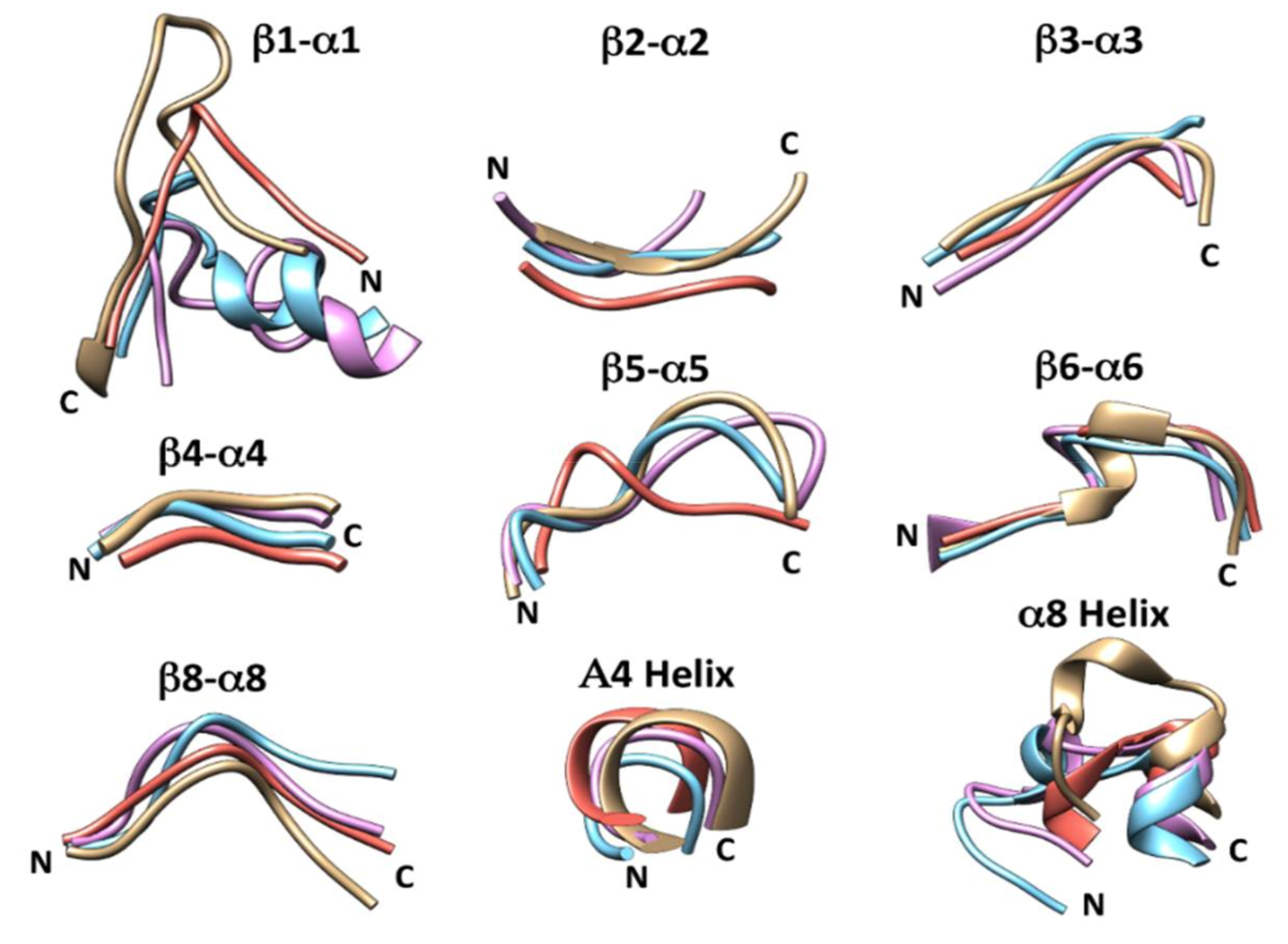
| Protein | Preferred Regions | Allowed Regions | Outliers |
|---|---|---|---|
| Glt1 | 268 (95.04%) | 11 (3.90%) | 3 (1.06%) Gln65, Arg217, Ala252 |
| Glt3 | 268 (95.01%) | 11 (3.90%) | 3 (1.06%) Ala15, Ala252, Arg217 |
| Glt20 | 272 (95.77%) | 11 (3.87%) | 1 (0.35%) Asp15 |
| Protein | AA | Z-Score |
|---|---|---|
| RmBgt17A (4WTS) | 264 | −8.92 |
| Glt1 | 284 | −8.22 |
| Glt3 | 284 | −8.48 |
| Glt20 | 286 | −8.38 |
| Enzyme | Source | PDB Code | Resolution (Å) | Glt1 | Glt3 | Glt20 | |||
|---|---|---|---|---|---|---|---|---|---|
| RMSD (Å) | Atom Pairs | RMSD (Å) | Atom Pairs | RMSD (Å) | Atom Pairs | ||||
| β-1,3-glucanosyl transferase (Bgt17A) | Rhizomucor miehei | 4WTP | 1.3 | 1.143 | 176 | 1.059 | 140 | 1.214 | 152 |
| endo-β-1,3-glucanase/allergen (Mus a 5) | Musa acuminata | 2CYG | 1.45 | 1.273 | 89 | 1.198 | 80 | 1.248 | 115 |
| endo-β-1,3-glucanase (Bgl32) | Hordeum vulgare | 1GHS | 2.30 | 1.148 | 87 | 1.151 | 81 | 1.311 | 98 |
| endo-β-1,3-1,4-glucanase II (EII)/lichenase II β-glucosidase II (Glb2) | Hordeum vulgare | 1AQ0 | 2.00 | 1.141 | 88 | 1.100 | 81 | 1.194 | 100 |
| endo-β-1,3-glucanase (GluB20-2, GluB20-2) | Solanum tuberosum | 3UR8 | 1.26 | 1.153 | 96 | 1.222 | 72 | 1.296 | 103 |
| β-1,3-glucanase (Glu1, Glu2, Hev b 2) | Hevea brasiliensis | 4HPG | 2.54 | 1.183 | 94 | 1.207 | 93 | 1.243 | 82 |
| Loop | β1-α1 | β2-α2 | β3-α3 | β4-A4 | β5-α5 | β6-α6 | β8-α8 | A4 Helix | α8 Helix |
|---|---|---|---|---|---|---|---|---|---|
| RmBgt17A | 9-17 GVNE-NSCPT | 38-41 FALS | 65-68 WIDR- | 97-99 GSE | 134-139 VYYKFP- | 153-161 AFPYWEGVT | 230-235 | 100-103 VLYR | 236-243 PYRG-GVEA |
| Glt1 | 26-41 (26-31) * SPFRLNQNPQSGRYPS | 62-65 YSVQ | 87-91 WLGPD | 118-120 GNE | 152-158 QWHVYRE | 173-181 VLPYWEATP | 250-255 IEAFDQ | 121-124 ALFR | 256-267 PWKVSAEGSVGA |
| Glt3 | 26-41 (26-32) * SPFRLGESPQKGQYPT | 62-65 YTVE | 87-91 WISPD | 118-120 GNE | 152-158 QWHIWKE | 173-181 LPYWEFVP | 250-255 IEAYDQ | 121-124 ALFR | 256-267 PWKASDEGSVGA |
| Glt20 | 26-41 APFEGSAHPDIDNIPT | 61-64 YSST | 87-91 WIDKD | 118-120 GNE | 152-158 IWNIWRD | 173-181 VLPYWENFR | 250-255 VEAIDQ | 121-124 IYR | 256-266 PWKF-FEGGVGP |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Linares-Pastén, J.A.; Jonsdottir, L.B.; Hreggvidsson, G.O.; Fridjonsson, O.H.; Watzlawick, H.; Karlsson, E.N. Modeled 3D-Structures of Proteobacterial Transglycosylases from Glycoside Hydrolase Family 17 Give Insight in Ligand Interactions Explaining Differences in Transglycosylation Products. Appl. Sci. 2021, 11, 4048. https://doi.org/10.3390/app11094048
Linares-Pastén JA, Jonsdottir LB, Hreggvidsson GO, Fridjonsson OH, Watzlawick H, Karlsson EN. Modeled 3D-Structures of Proteobacterial Transglycosylases from Glycoside Hydrolase Family 17 Give Insight in Ligand Interactions Explaining Differences in Transglycosylation Products. Applied Sciences. 2021; 11(9):4048. https://doi.org/10.3390/app11094048
Chicago/Turabian StyleLinares-Pastén, Javier A., Lilja Björk Jonsdottir, Gudmundur O. Hreggvidsson, Olafur H. Fridjonsson, Hildegard Watzlawick, and Eva Nordberg Karlsson. 2021. "Modeled 3D-Structures of Proteobacterial Transglycosylases from Glycoside Hydrolase Family 17 Give Insight in Ligand Interactions Explaining Differences in Transglycosylation Products" Applied Sciences 11, no. 9: 4048. https://doi.org/10.3390/app11094048
APA StyleLinares-Pastén, J. A., Jonsdottir, L. B., Hreggvidsson, G. O., Fridjonsson, O. H., Watzlawick, H., & Karlsson, E. N. (2021). Modeled 3D-Structures of Proteobacterial Transglycosylases from Glycoside Hydrolase Family 17 Give Insight in Ligand Interactions Explaining Differences in Transglycosylation Products. Applied Sciences, 11(9), 4048. https://doi.org/10.3390/app11094048







