Pathomics and Deep Learning Classification of a Heterogeneous Fluorescence Histology Image Dataset
Abstract
:1. Introduction
Related Works
2. Materials and Methods
2.1. Dataset Description and Labeling
2.2. Data Pre-Processing
2.3. Pathomics Analysis
2.3.1. Feature Extraction
2.3.2. Feature Selection
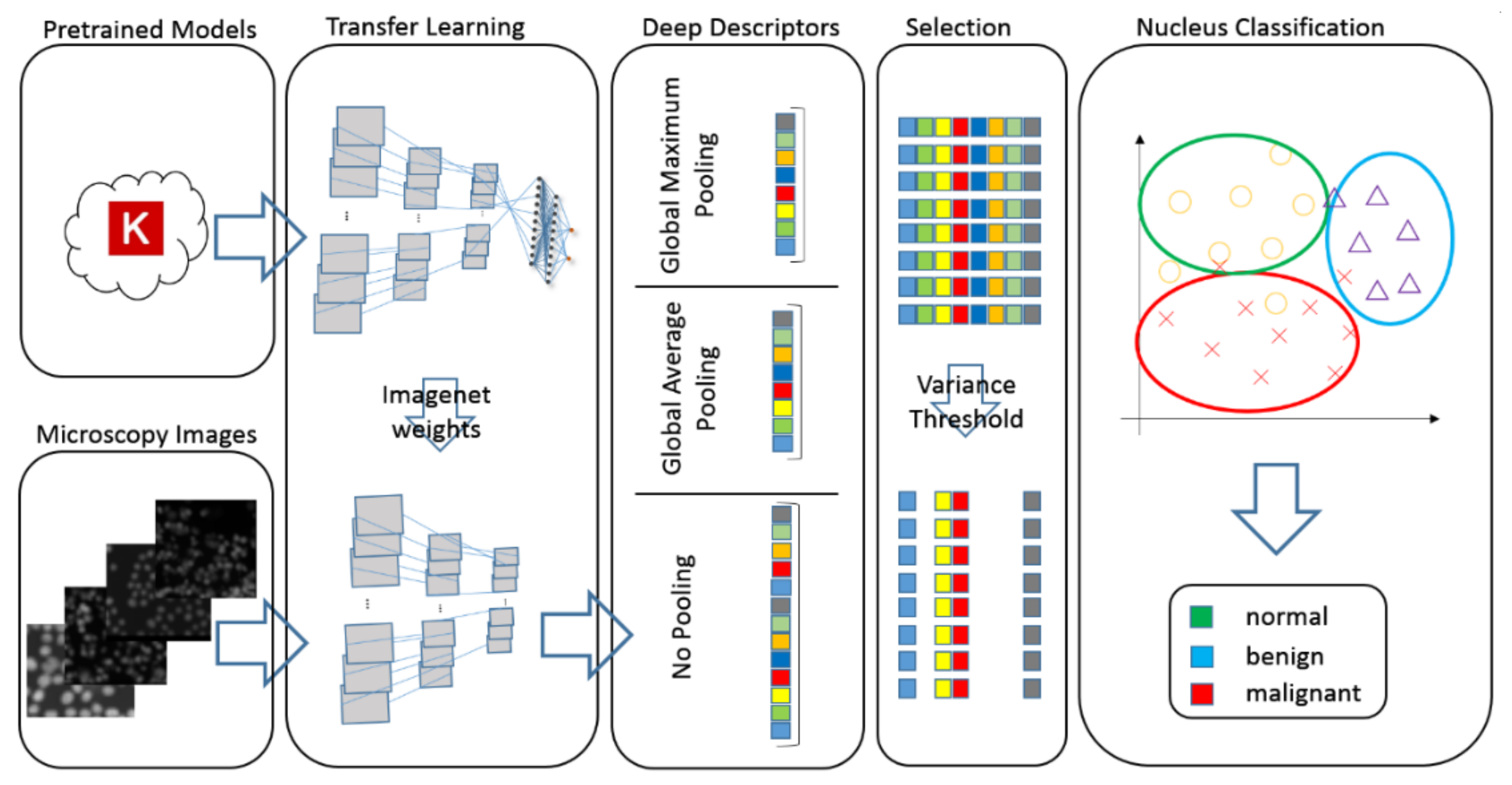
2.4. Deep–Learning Descriptors
2.4.1. Deep–Analysis Specific Image Preprocessing
2.4.2. Transfer Learning Analysis
2.5. Ternary Classification
2.6. Model Performance Evaluation Metrics
3. Results
3.1. Pathomics
3.2. Transfer Learning
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hamad, A.; Ersoy, I.; Bunyak, F. Improving Nuclei Classification Performance in H&E Stained Tissue Images Using Fully Convolutional Regression Network and Convolutional Neural Network. In Proceedings of the 2018 IEEE Applied Imagery Pattern Recognition Workshop (AIPR), Washington, DC, USA, 9–11 October 2018; pp. 1–6. [Google Scholar] [CrossRef]
- Putzu, L.; Fumera, G. An Empirical Evaluation of Nuclei Segmentation from H&E Images in a Real Application Scenario. Appl. Sci. 2020, 10, 7982. [Google Scholar] [CrossRef]
- Salvi, M.; Molinari, F. Multi-tissue and multi-scale approach for nuclei segmentation in H&E stained images. Biomed. Eng. Online 2018, 17, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Lakis, S.; Kotoula, V.; Koliou, G.-A.; Efstratiou, I.; Chrisafi, S.; Papanikolaou, A.; Zebekakis, P.; Fountzilas, G. Multisite Tumor Sampling Reveals Extensive Heterogeneity of Tumor and Host Immune Response in Ovarian Cancer. Cancer Genom. Proteom. 2020, 17, 529–541. [Google Scholar] [CrossRef] [PubMed]
- Hägele, M.; Seegerer, P.; Lapuschkin, S.; Bockmayr, M.; Samek, W.; Klauschen, F.; Müller, K.-R.; Binder, A. Resolving challenges in deep learning-based analyses of histopathological images using explanation methods. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Shapcott, M.; Hewitt, K.J.; Rajpoot, N. Deep Learning with Sampling in Colon Cancer Histology. Front. Bioeng. Biotechnol. 2019, 7, 52. [Google Scholar] [CrossRef] [Green Version]
- Dimitriou, N.; Arandjelović, O.; Caie, P.D. Deep Learning for Whole Slide Image Analysis: An Overview. Front. Med. 2019, 6, 264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurc, T.; Bakas, S.; Ren, X.; Bagari, A.; Momeni, A.; Huang, Y.; Zhang, L.; Kumar, A.; Thibault, M.; Qi, Q.; et al. Segmentation and Classification in Digital Pathology for Glioma Research: Challenges and Deep Learning Approaches. Front. Neurosci. 2020, 14, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barisoni, L.; Lafata, K.J.; Hewitt, S.M.; Madabhushi, A.; Balis, U.G.J. Digital pathology and computational image analysis in nephropathology. Nat. Rev. Nephrol. 2020, 16, 669–685. [Google Scholar] [CrossRef]
- Yetiş, S.Ç.; Çapar, A.; Ekinci, D.A.; Ayten, U.E.; Kerman, B.E.; Töreyin, B.U. Myelin detection in fluorescence microscopy images using machine learning. J. Neurosci. Methods 2020, 346, 108946. [Google Scholar] [CrossRef]
- Unger, J.; Hebisch, C.; Phipps, J.E.; Lagarto, J.L.; Kim, H.; Darrow, M.A.; Bold, R.J.; Marcu, L. Real-time diagnosis and visualization of tumor margins in excised breast specimens using fluorescence lifetime imaging and machine learning. Biomed. Opt. Express 2020, 11, 1216–1230. [Google Scholar] [CrossRef]
- Held, M.; Schmitz, M.H.A.; Fischer, B.; Walter, T.; Neumann, B.; Olma, M.H.; Peter, M.; Ellenberg, J.; Gerlich, D.W. Cell Cognition: Time-resolved phenotype annotation in high-throughput live cell imaging. Nat. Methods 2010, 7, 747–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez-Jimenez, C.; Sandino, A.A.; Prasanna, P.; Gupta, A.; Viswanath, S.E.; Romero, E. Identifying Cross-Scale Associations between Radiomic and Pathomic Signatures of Non-Small Cell Lung Cancer Subtypes: Preliminary Results. Cancers 2020, 12, 3663. [Google Scholar] [CrossRef] [PubMed]
- Rivenson, Y.; Wang, H.; Wei, Z.; de Haan, K.; Zhang, Y.; Wu, Y.; Günaydın, H.; Zuckerman, J.E.; Chong, T.; Sisk, A.E.; et al. Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning. Nat. Biomed. Eng. 2019, 3, 466–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Rivenson, Y.; Jin, Y.; Wei, Z.; Gao, R.; Günaydın, H.; Bentolila, L.A.; Kural, C.; Ozcan, A. Deep learning enables cross-modality super-resolution in fluorescence microscopy. Nat. Methods 2019, 16, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, F.; Zhang, F.; Xu, P.; Zhang, M.; Fan, M.; Li, L.; Gao, X.; Han, R. DLBI: Deep learning guided Bayesian inference for structure reconstruction of super-resolution fluorescence microscopy. Bioinformatics 2018, 34, i284–i294. [Google Scholar] [CrossRef]
- Ouyang, W.; Aristov, A.; Lelek, M.; Hao, X.; Zimmer, C. Deep learning massively accelerates super-resolution localization microscopy. Nat. Biotechnol. 2018, 36, 460–468. [Google Scholar] [CrossRef]
- Zhou, H.; Cai, R.; Quan, T.; Liu, S.; Li, S.; Huang, Q.; Ertürk, A.; Zeng, S. 3D high resolution generative deep-learning network for fluorescence microscopy imaging. Opt. Lett. 2020, 45, 1695–1698. [Google Scholar] [CrossRef]
- Oszutowska-Mazurek, D.; Parafiniuk, M.; Mazurek, P. Virtual UV Fluorescence Microscopy from Hematoxylin and Eosin Staining of Liver Images Using Deep Learning Convolutional Neural Network. Appl. Sci. 2020, 10, 7815. [Google Scholar] [CrossRef]
- Spilger, R.; Wollmann, T.; Qiang, Y.; Imle, A.; Lee, J.Y.; Müller, B.; Fackler, O.T.; Bartenschlager, R.; Rohr, K. Deep Particle Tracker: Automatic Tracking of Particles in Fluorescence Microscopy Images Using Deep Learning. Lect. Notes Comput. Sci. 2018, 128–136. [Google Scholar] [CrossRef]
- Jang, H.-J.; Song, I.H.; Lee, S.H. Generalizability of Deep Learning System for the Pathologic Diagnosis of Various Cancers. Appl. Sci. 2021, 11, 808. [Google Scholar] [CrossRef]
- Valieris, R.; Amaro, L.; Osório, C.A.B.D.T.; Bueno, A.P.; Mitrowsky, R.A.R.; Carraro, D.M.; Nunes, D.N.; Dias-Neto, E.; da Silva, I.T. Deep Learning Predicts Underlying Features on Pathology Images with Therapeutic Relevance for Breast and Gastric Cancer. Cancers 2020, 12, 3687. [Google Scholar] [CrossRef]
- Kromp, F.; Bozsaky, E.; Rifatbegovic, F.; Fischer, L.; Ambros, M.; Berneder, M.; Weiss, T.; Lazic, D.; Dörr, W.; Hanbury, A.; et al. An annotated fluorescence image dataset for training nuclear segmentation methods. Sci. Data 2020, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Shimada, H.; Ambros, I.M.; Dehner, L.P.; Hata, J.; Joshi, V.V.; Roald, B. Terminology and morphologic criteria of neuroblastic tumors: Recommendations by the International Neuroblastoma Pathology Committee. Cancer 1999, 86, 349–363. [Google Scholar] [CrossRef]
- Moch, H.; Cubilla, A.L.; Humphrey, P.A.; Reuter, V.E.; Ulbright, T.M. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs—Part A: Renal, Penile, and Testicular Tumours. Eur. Urol. 2016, 70, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Uhl, A.; Wimmer, G. A systematic evaluation of the scale invariance of texture recognition methods. Pattern Anal. Appl. 2015, 18, 945–969. [Google Scholar] [CrossRef] [Green Version]
- Coelho, L.P. Mahotas: Open source software for scriptable computer vision. J. Open Res. Softw. 2013, 1, e3. [Google Scholar] [CrossRef]
- Duron, L.; Balvay, D.; Perre, S.V.; Bouchouicha, A.; Savatovsky, J.; Sadik, J.-C.; Thomassin-Naggara, I.; Fournier, L.; Lecler, A. Gray-level discretization impacts reproducible MRI radiomics texture features. PLoS ONE 2019, 14, e0213459. [Google Scholar] [CrossRef]
- Le, N.Q.K.; Hung, T.N.K.; Do, D.T.; Lam, L.H.T.; Dang, L.H.; Huynh, T.-T. Radiomics-based machine learning model for efficiently classifying transcriptome subtypes in glioblastoma patients from MRI. Comput. Biol. Med. 2021, 132, 104320. [Google Scholar] [CrossRef] [PubMed]
- Le, N.Q.K.; Do, D.T.; Chiu, F.-Y.; Yapp, E.K.Y.; Yeh, H.-Y.; Chen, C.-Y. XGBoost Improves Classification of MGMT Promoter Methylation Status in IDH1 Wildtype Glioblastoma. J. Pers. Med. 2020, 10, 128. [Google Scholar] [CrossRef]
- Van Griethuysen, J.J.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [Green Version]
- Peng, H.; Long, F.; Ding, C. Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. Conf. Proc. 2016, 2818–2826. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, preprint. arXiv:1409.1556. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobile Nets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVRP), Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning Transferable Architectures for Scalable Image Recognition. In Proceedings of the 2018 IEEE conference on computer vision and pattern recognition (CVRP), Salt Lake City, UT, USA, 18–23 June 2018; pp. 8697–8710. [Google Scholar]
- Chollet, F. Others Keras, an Open Library for Deep Learning. Available online: http://citebay.com/how-to-cite/keras/ (accessed on 9 April 2021).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Du-Bourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Trivizakis, E.; Manikis, G.C.; Nikiforaki, K.; Drevelegas, K.; Constantinides, M.; Drevelegas, A.; Marias, K. Extending 2-D Convolutional Neural Networks to 3-D for Advancing Deep Learning Cancer Classification with Application to MRI Liver Tumor Differentiation. IEEE J. Biomed. Heal. Inform. 2018, 23, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Trivizakis, E.; Ioannidis, G.S.; Melissianos, V.D.; Papadakis, G.Z.; Tsatsakis, A.; Spandidos, D.A.; Marias, K. A novel deep learning architecture outperforming ‘off-the-shelf’ transfer learning and feature-based methods in the automated assessment of mammographic breast density. Oncol. Rep. 2019, 42, 2009–2015. [Google Scholar] [CrossRef] [Green Version]

| Logistic Regression (OVR) | SVM RBF | |||
|---|---|---|---|---|
| Selected Features | AUC | ACC | AUC | ACC |
| 3 | 0.956 ± 0.047 | 0.8 ± 0.093 | 0.965 ± 0.056 | 0.786 ± 0.064 |
| 6 | 0.968 ± 0.033 | 0.871 ± 0.103 | 0.954 ± 0.042 | 0.843 ± 0.118 |
| 10 | 0.983 ± 0.03 | 0.871 ± 0.128 | 0.981 ± 0.032 | 0.843 ± 0.159 |
| 20 | 0.986 ± 0.033 | 0.957 ± 0.105 | 0.965 ± 0.086 | 0.929 ± 0.103 |
| 30 | 0.992 ± 0.019 | 0.886 ± 0.064 | 0.978 ± 0.033 | 0.914 ± 0.035 |
| 40 | 0.996 ± 0.006 | 0.943 ± 0.049 | 0.976 ± 0.031 | 0.871 ± 0.07 |
| 50 | 0.99 ± 0.019 | 0.943 ± 0.073 | 0.986 ± 0.033 | 0.886 ± 0.064 |
| Feature Type | Model Family | SVM RBF | Logistic Regression OVR | ||||
|---|---|---|---|---|---|---|---|
| Variance Threshold | ACC | AUC | Variance Threshold | ACC | AUC | ||
| Raw | Xception | 0.0 | 0.943 ± 0.07 | 0.956 ± 0.05 | 0.0 | 0.925 ± 0.08 | 0.944 ± 0.07 |
| VGG | 0.3 | 0.646 ± 0.09 | 0.664 ± 0.08 | 0.5 | 0.905 ± 0.07 | 0.925 ± 0.06 | |
| ResNet | 0.4 | 0.876 ± 0.07 | 0.898 ± 0.07 | 0.3 | 0.924 ± 0.10 | 0.943 ± 0.07 | |
| Inception | 0.5 | 0.916 ± 0.09 | 0.929 ± 0.07 | 0.5 | 0.942 ± 0.06 | 0.960 ± 0.04 | |
| MobileNet | 0.0 | 0.905 ± 0.09 | 0.909 ± 0.07 | 0.0 | 0.924 ± 0.09 | 0.941 ± 0.07 | |
| DenseNet | 0.1 | 0.926 ± 0.13 | 0.940 ± 0.10 | 0.4 | 0.944 ± 0.06 | 0.951 ± 0.05 | |
| NasNet | 0.0 | 0.897 ± 0.10 | 0.908 ± 0.09 | 0.0 | 0.907 ± 0.07 | 0.926 ± 0.05 | |
| Global Maximum | Xception | 0.4 | 0.933 ± 0.06 | 0.943 ± 0.05 | 0.2 | 0.923 ± 0.07 | 0.944 ± 0.05 |
| VGG | 0.2 | 0.659 ± 0.06 | 0.679 ± 0.04 | 0.1 | 0.905 ± 0.06 | 0.922 ± 0.04 | |
| ResNet | 0.1 | 0.876 ± 0.10 | 0.892 ± 0.10 | 0.2 | 0.926 ± 0.11 | 0.940 ± 0.10 | |
| Inception | 0.3 | 0.918 ± 0.10 | 0.932 ± 0.10 | 0.3 | 0.945 ± 0.07 | 0.922 ± 0.04 | |
| MobileNet | 0.3 | 0.907 ± 0.08 | 0.921 ± 0.07 | 0.4 | 0.914 ± 0.07 | 0.934 ± 0.06 | |
| DenseNet | 0.5 | 0.916 ± 0.08 | 0.933 ± 0.06 | 0.3 | 0.951 ± 0.05 | 0.962 ± 0.04 | |
| NasNet | 0.0 | 0.897 ± 0.08 | 0.909 ± 0.07 | 0.2 | 0.913 ± 0.08 | 0.926 ± 0.07 | |
| Global Average | Xception | 0.2 | 0.944 ± 0.06 | 0.953 ± 0.06 | 0.2 | 0.925 ± 0.08 | 0.938 ± 0.07 |
| VGG | 0.0 | 0.763 ± 0.14 | 0.783 ± 0.13 | 0.2 | 0.905 ± 0.09 | 0.910 ± 0.08 | |
| ResNet | 0.1 | 0.875 ± 0.07 | 0.896 ± 0.07 | 0.3 | 0.942 ± 0.07 | 0.954 ± 0.06 | |
| Inception | 0.3 | 0.945 ± 0.06 | 0.959 ± 0.05 | 0.0 | 0.935 ± 0.07 | 0.951 ± 0.06 | |
| MobileNet | 0.4 | 0.913 ± 0.08 | 0.923 ± 0.07 | 0.2 | 0.924 ± 0.07 | 0.948 ± 0.06 | |
| DenseNet | 0.2 | 0.935 ± 0.06 | 0.941 ± 0.06 | 0.4 | 0.935 ± 0.07 | 0.948 ± 0.06 | |
| NasNet | 0.2 | 0.915 ± 0.09 | 0.919 ± 0.09 | 0.0 | 0.925 ± 0.07 | 0.942 ± 0.07 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ioannidis, G.S.; Trivizakis, E.; Metzakis, I.; Papagiannakis, S.; Lagoudaki, E.; Marias, K. Pathomics and Deep Learning Classification of a Heterogeneous Fluorescence Histology Image Dataset. Appl. Sci. 2021, 11, 3796. https://doi.org/10.3390/app11093796
Ioannidis GS, Trivizakis E, Metzakis I, Papagiannakis S, Lagoudaki E, Marias K. Pathomics and Deep Learning Classification of a Heterogeneous Fluorescence Histology Image Dataset. Applied Sciences. 2021; 11(9):3796. https://doi.org/10.3390/app11093796
Chicago/Turabian StyleIoannidis, Georgios S., Eleftherios Trivizakis, Ioannis Metzakis, Stilianos Papagiannakis, Eleni Lagoudaki, and Kostas Marias. 2021. "Pathomics and Deep Learning Classification of a Heterogeneous Fluorescence Histology Image Dataset" Applied Sciences 11, no. 9: 3796. https://doi.org/10.3390/app11093796
APA StyleIoannidis, G. S., Trivizakis, E., Metzakis, I., Papagiannakis, S., Lagoudaki, E., & Marias, K. (2021). Pathomics and Deep Learning Classification of a Heterogeneous Fluorescence Histology Image Dataset. Applied Sciences, 11(9), 3796. https://doi.org/10.3390/app11093796









