Fake It Till You Make It: Guidelines for Effective Synthetic Data Generation
Abstract
1. Introduction
1.1. Background
1.2. Contributions
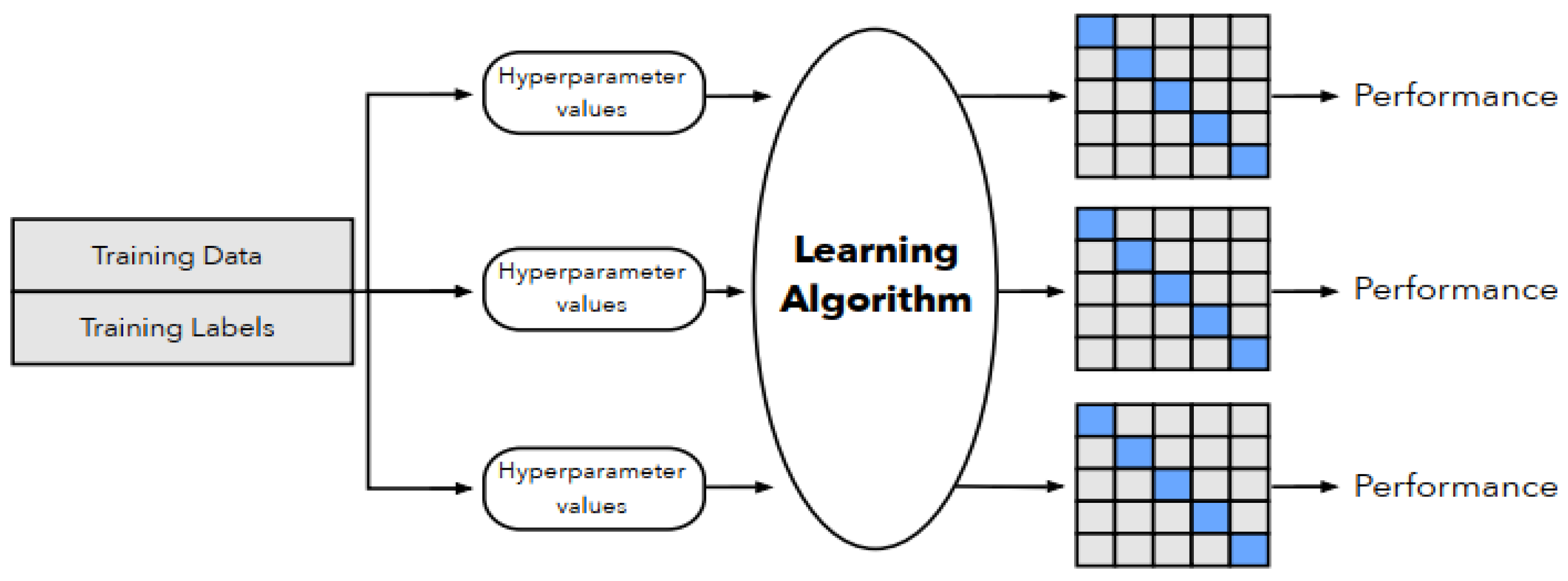
2. Methods: Synthetic Data Generation and Utility
2.1. Synthetic Data Generators
2.2. Utility Measures
3. Materials and Methods
- Q 1.
- Does preprocessing real data prior to the generation of synthetic data improve the utility of the generated synthetic data?
- Q 2.
- When calculating prediction accuracy, does importing the real data settings (tuning-settings) to the synthetic data lead to improved accuracy results?
- Q 3.
- If real-data settings are not imported, should we apply tuning on the synthetic datasets at all? Or would non-tuning lead to a better accuracy?
- Q 4.
- As propensity is widely regarded as the most practical measure of synthetic data utility, does a better propensity score lead to better prediction accuracy? in other words, is propensity score a good indicator for Prediction accuracy?
3.1. Paths Tested
3.1.1. Data Generation Paths
- Path 1
- Preprocess real data prior to synthesizing it versus
- Path 2
- Generate synthetic data without applying any preprocessing on the real data (note that this path is the one followed across the literature). In this path, pre-processing is done independently for real and synthetic datasets when applying machine learning algorithms.
3.1.2. Data Training Paths
- Case 1
- Perform feature selection and hyper-parameter tuning on the real data and apply the obtained results on the generated synthetic data.
- Case 2
- Perform tuning independently on real and on synthetic datasets, and
- Case 3
- Apply no tuning, instead use the default parameter values and include all features in the model (note that, this path is the one followed across all literature).
3.1.3. Propensity Versus Accuracy
3.2. Synthetic Data Generation Process
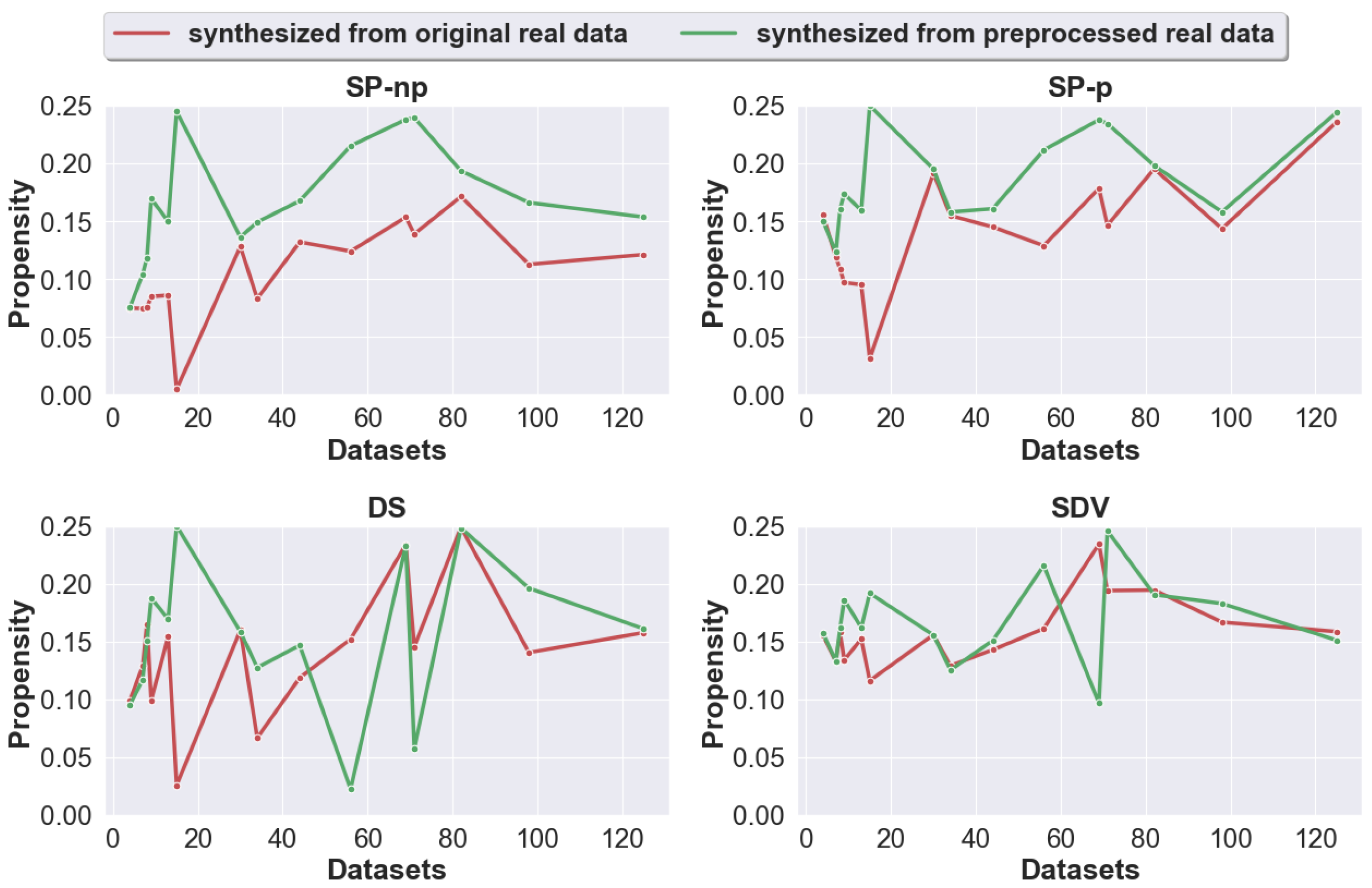
3.2.1. Datasets and Generation Process
- We performed a repeated holdout method, where we randomly generate 4 splits of each real dataset into 70% training and 30% testing. For each split, we repeatedly apply the 4 synthetic data generators 5 times with the real training data as the input. The generated synthetic data is of equal length as the real training data. Thus, the total number of synthetic data generated for each [path, generator, dataset] is 4 × 5 = 20, for each [generator, dataset] is 2 × 20 = 40, and for each dataset is 40 × 4 = 160.
- The propensity score is calculated for each of the synthetic datasets generated. The final propensity score for each [path, dataset, generator], is the average across the 20 corresponding synthetic datasets.
- Similarly, Prediction accuracy is calculated for each synthetic dataset generated in Path 2, for each of the three training cases, and each of the four machine learning algorithms. The accuracy is evaluated on the (corresponding) real testing dataset. The final accuracy for each [Case, dataset, generator, algorithm] is obtained by averaging across the 20 corresponding synthetic datasets.
- (i)
- Each real dataset, , is randomly split 4 times into 70% training and 30% testing, where are the training sets and , …, their corresponding testing sets.
- (ii)
- For each [synthesizer, training dataset] pairs: we generate 5 synthetic datasets: .
- (iii)
- The propensity score is then calculated for each generated synthetic dataset
- (iv)
- The final score for each [synthesizer, dataset] pair: is the average across the 20 generated datasets: .
- (v)
- For Case , dataset , and algorithm , the prediction accuracy is calculated for each of the synthetic dataset generated from
- (vi)
- The final accuracy (in case ) for each [synthesizer, dataset, algorithm]: is the average across the 20 generated datasets: , measured from the corresponding real testing sets: .
3.2.2. Generation Set-Up
4. Results and Discussion
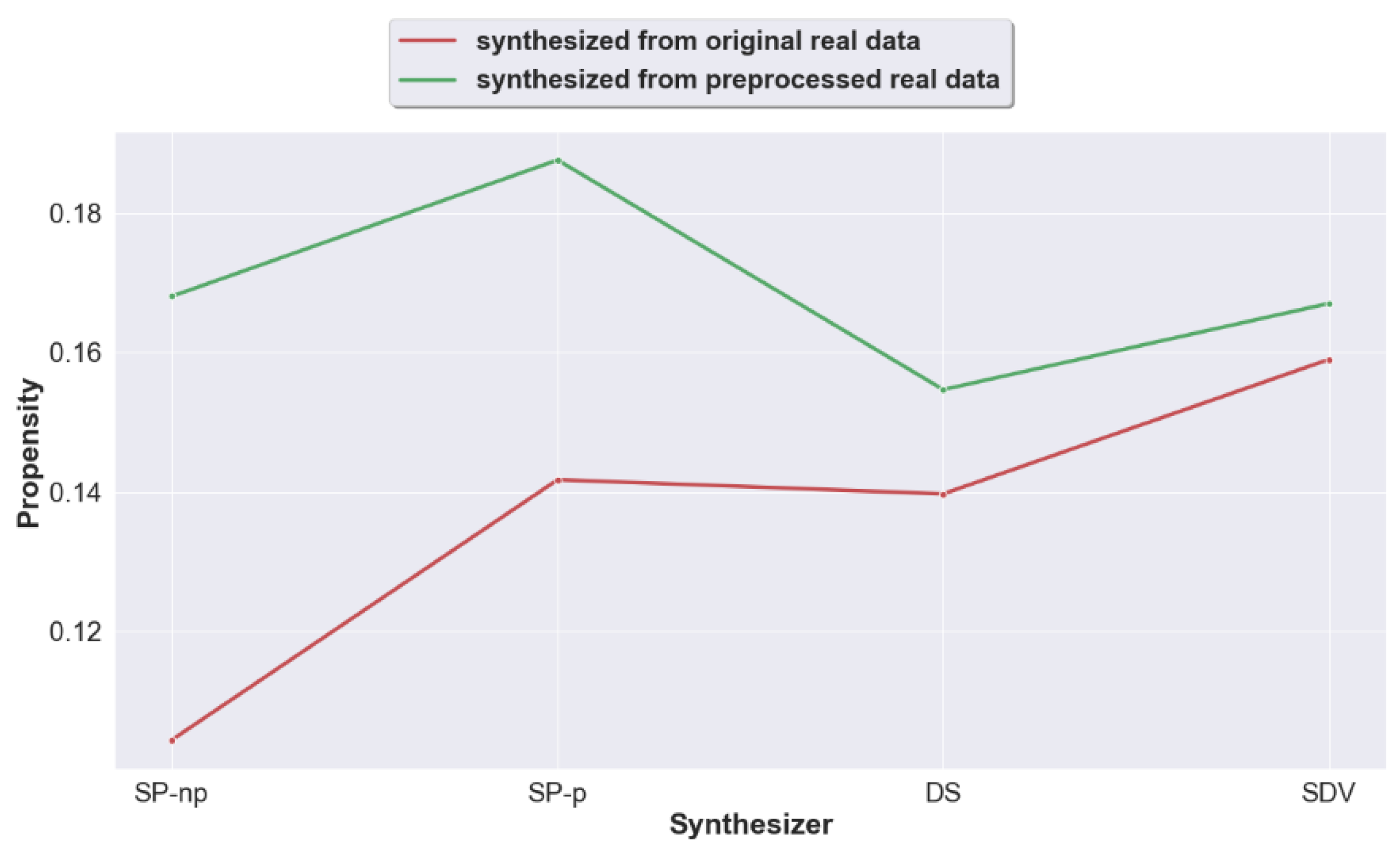
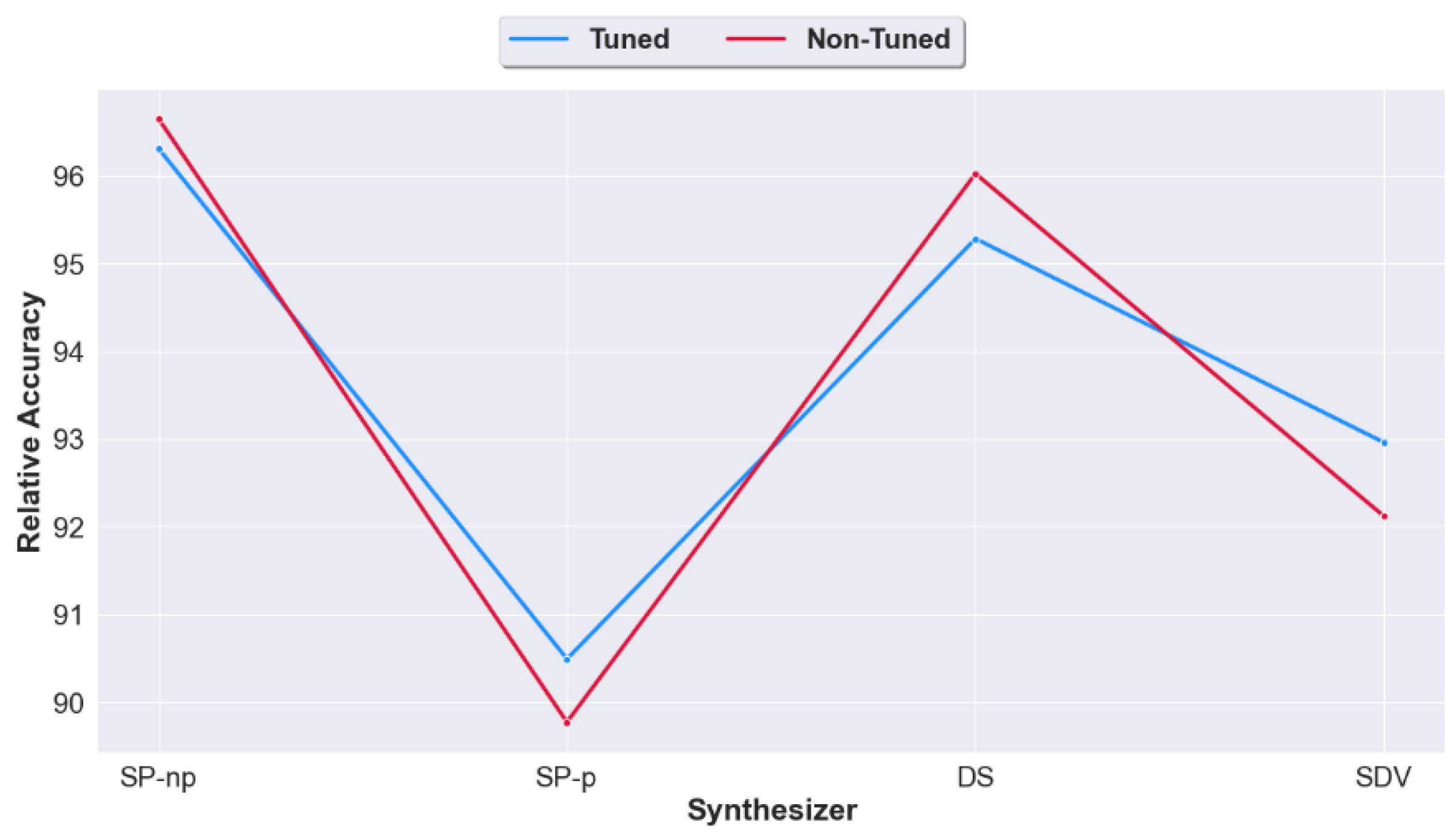
4.1. Question 1: Does Pre-Processing Real Data Prior to the Generation of Synthetic Data Improve the Utility of the Generated Synthetic Data?
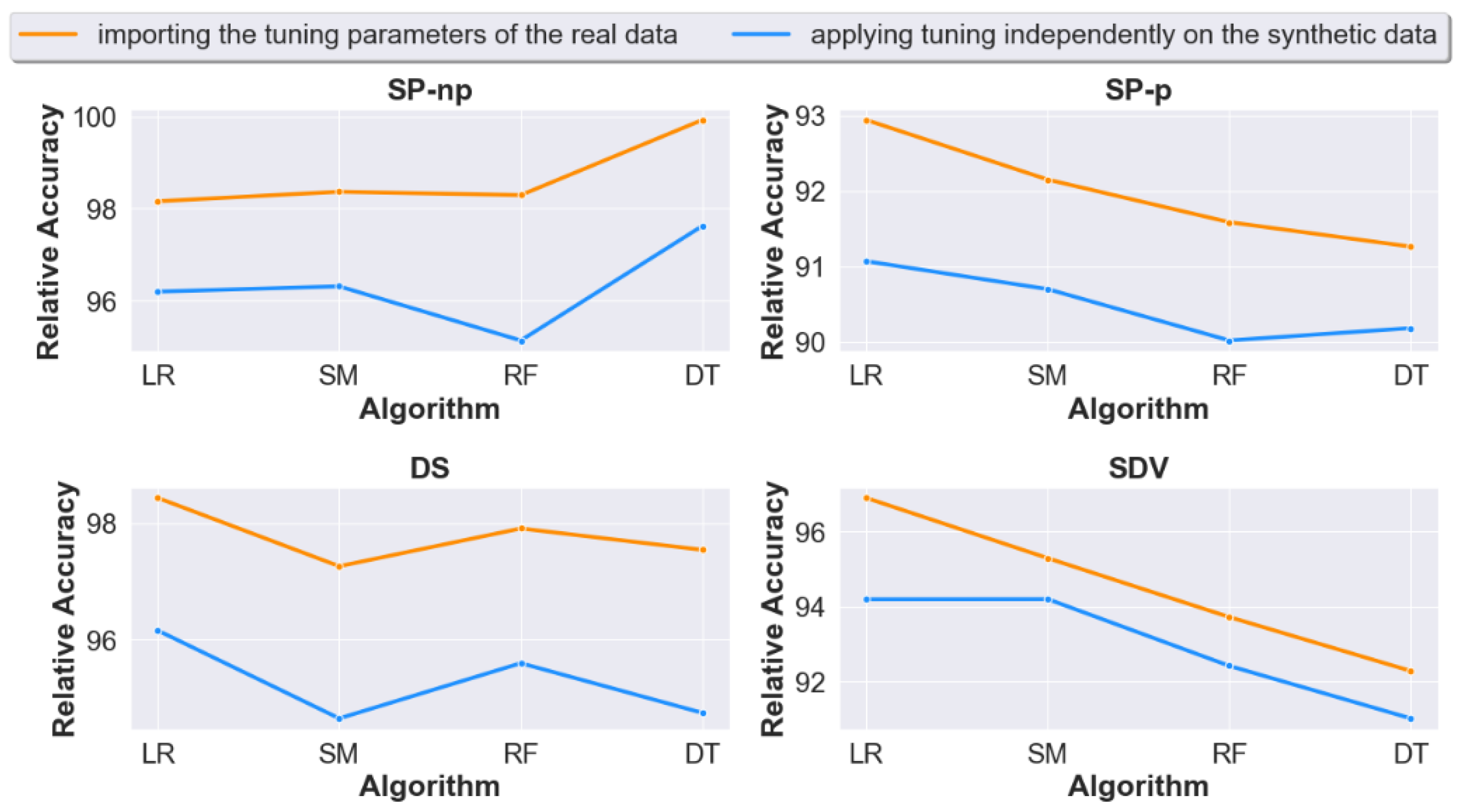
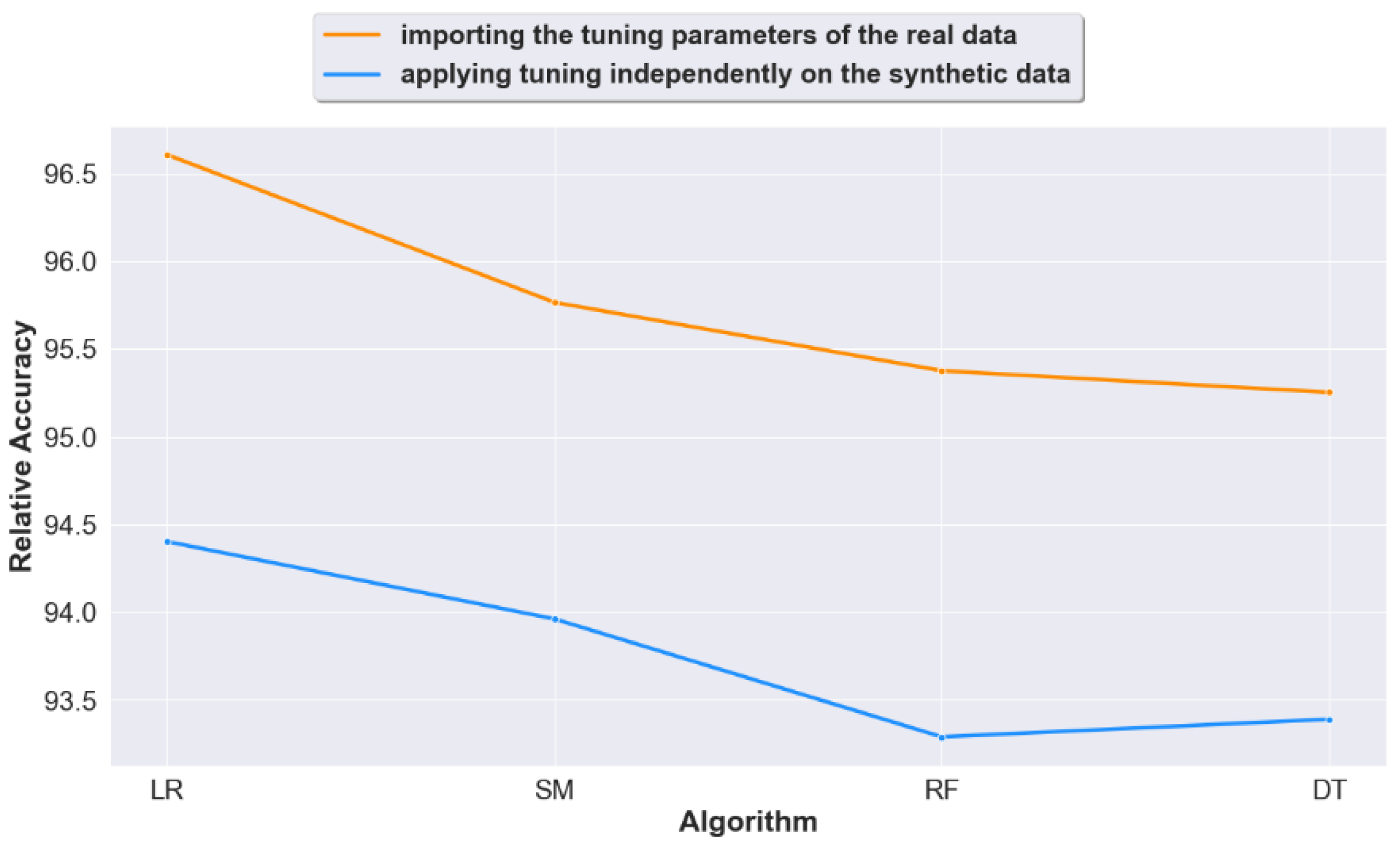
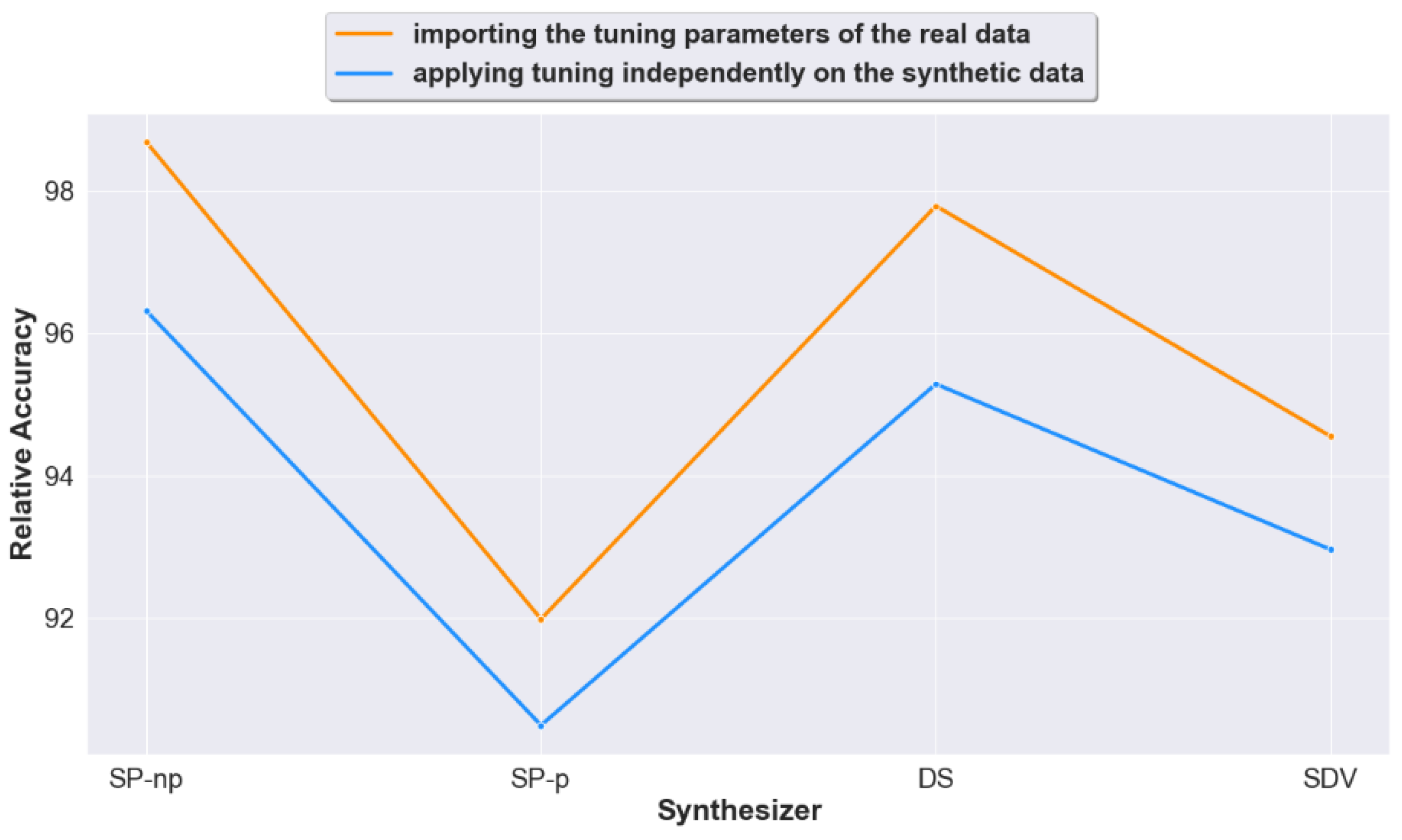
4.2. Question 2: When Calculating Prediction Accuracy, Does Importing the Real Data Tuning Settings to the Generated Synthetic Data Lead to Improved Accuracy Results?
4.3. Question 3: Should We Apply Tuning on the Synthetic Datasets Generated from Raw Unprocessed Data? Or Would Non-Tuning Lead to a Better Accuracy?
4.4. Question 4: Is Propensity Score a Good Indicator for Prediction Accuracy?
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Davenport, T.; Kalakota, R. The potential for artificial intelligence in healthcare. Future Healthc. J. 2019, 6, 94–98. [Google Scholar] [CrossRef]
- Lysaght, T.; Lim, H.Y.; Xafis, V.; Ngiam, K.Y. AI-Assisted Decision-making in Healthcare. Asian Bioeth. Rev. 2019, 11, 299–314. [Google Scholar] [CrossRef]
- McGlynn, E.A.; Lieu, T.A.; Durham, M.L.; Bauck, A.; Laws, R.; Go, A.S.; Chen, J.; Feigelson, H.S.; Corley, D.A.; Young, D.R.; et al. Developing a data infrastructure for a learning health system: The PORTAL network. J. Am. Med. Inform. Assoc. JAMIA 2014, 21, 596–601. [Google Scholar] [CrossRef]
- Use of Artificial Intelligence in Infectious Diseases. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7153335/ (accessed on 18 February 2021).
- Dankar, F.K.; el Emam, K.; Neisa, A.; Roffey, T. Estimating the re-identification risk of clinical data sets. BMC Med. Inform. Decis. Mak. 2012, 12, 66. [Google Scholar] [CrossRef]
- GDPR. General Data Protection Regulation (GDPR). 2018. Available online: https://gdpr-info.eu/ (accessed on 9 December 2018).
- U.S. Department of Health & Human Services. Available online: http://www.hhs.gov/ (accessed on 22 September 2015).
- Dankar, F.K.; Gergely, M.; Dankar, S.K. Informed Consent in Biomedical Research. Comput. Struct. Biotechnol. J. 2019, 17, 463–474. [Google Scholar] [CrossRef] [PubMed]
- Artificial Intelligence In Health Care: Benefits and Challenges of Machine Learning in Drug Development (STAA)-Policy Briefs & Reports-EPTA Network. Available online: https://eptanetwork.org/database/policy-briefs-reports/1898-artificial-intelligence-in-health-care-benefits-and-challenges-of-machine-learning-in-drug-development-staa (accessed on 1 September 2020).
- Howe, B.; Stoyanovich, J.; Ping, H.; Herman, B.; Gee, M. Synthetic Data for Social Good. arXiv 2017, arXiv:171008874. [Google Scholar]
- Mostert, M.; Bredenoord, A.L.; Biesaart, M.C.; van Delden, J.J. Big Data in medical research and EU data protection law: Challenges to the consent or anonymise approach. Eur. J. Hum. Genet. 2016, 24, 956. [Google Scholar] [CrossRef]
- Dankar, F.K.; Ptitsyn, A.; Dankar, S.K. The development of large-scale de-identified biomedical databases in the age of genomics—principles and challenges. Hum. Genom. 2018, 12, 19. [Google Scholar] [CrossRef]
- Dankar, F.K.; Badji, R. A risk-based framework for biomedical data sharing. J. Biomed. Inform. 2017, 66, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Ervine, C. Directive 2004/39/Ec of the European Parliament and of the Council of 21 April 2004. In Core Statutes on Company Law; Macmillan Education: London, UK, 2015; pp. 757–759. [Google Scholar]
- Naveed, M.; Ayday, E.; Clayton, E.W.; Fellay, J.; Gunter, C.A.; Hubaux, J.P.; Malin, B.A.; Wang, X. Privacy in the genomic era. ACM Comput. Surv. CSUR 2015, 48, 6. [Google Scholar] [CrossRef] [PubMed]
- Dankar, F.K.; Gergely, M.; Malin, B.; Badji, R.; Dankar, S.K.; Shuaib, K. Dynamic-informed consent: A potential solution for ethical dilemmas in population sequencing initiatives. Comput. Struct. Biotechnol. J. 2020, 18, 913–921. [Google Scholar] [CrossRef]
- Taub, J.; Elliot, M.; Pampaka, M.; Smith, D. Differential Correct Attribution Probability for Synthetic Data: An Exploration. In Privacy in Statistical Databases; Springer: Cham, Spain, 2018; pp. 122–137. [Google Scholar] [CrossRef]
- Data Synthesis Based on Generative Adversarial Networks|Proceedings of the VLDB Endowment. Available online: https://dl.acm.org/doi/10.14778/3231751.3231757 (accessed on 1 September 2020).
- Rubin, D.B. Statistical disclosure limitation. J. Off. Stat. 1993, 9, 461–468. [Google Scholar]
- Ruiz, N.; Muralidhar, K.; Domingo-Ferrer, J. On the Privacy Guarantees of Synthetic Data: A Reassessment from the Maximum-Knowledge Attacker Perspective. In Privacy in Statistical Databases; Springer: Cham, Spain, 2018; pp. 59–74. [Google Scholar] [CrossRef]
- Hu, J. Bayesian Estimation of Attribute and Identification Disclosure Risks in Synthetic Data. arXiv 2018, arXiv:180402784. [Google Scholar]
- Polonetsky, J.; Elizabeth, R. 10 Privacy Risks and 10 Privacy Technologies to Watch in the Next Decade. Presented at the Future of Privacy Forum. 2020. Available online: https://fpf.org/wp-content/uploads/2020/01/FPF_Privacy2020_WhitePaper.pdf (accessed on 26 February 2021).
- Patki, N.; Wedge, R.; Veeramachaneni, K. The Synthetic Data Vault. In Proceedings of the 2016 IEEE International Conference on Data Science and Advanced Analytics (DSAA), Montreal, QC, Canada, 17–19 October 2016; pp. 399–410. [Google Scholar] [CrossRef]
- Ping, H.; Stoyanovich, J.; Howe, B. Datasynthesizer: Privacy-preserving synthetic datasets. In Proceedings of the 29th International Conference on Scientific and Statistical Database Management, Chicago, IL, USA, 27–29 June 2017; pp. 1–5. [Google Scholar]
- Raab, G.M.; Nowok, B.; Dibben, C. Guidelines for Producing Useful Synthetic Data. arXiv 2017, arXiv:171204078. [Google Scholar]
- Yoon, J.; Drumright, L.N.; van der Schaar, M. Anonymization through Data Synthesis using Generative Adversarial Networks (ADS-GAN). IEEE J. Biomed. Health Inform. 2020, 24, 2378–2388. [Google Scholar] [CrossRef]
- Rankin, D.; Black, M.; Bond, R.; Wallace, J.; Mulvenna, M.; Epelde, G. Reliability of Supervised Machine Learning Using Synthetic Data in Health Care: Model to Preserve Privacy for Data Sharing. JMIR Med. Inform. 2020, 8, e18910. [Google Scholar] [CrossRef] [PubMed]
- Hittmeir, M.; Ekelhart, A.; Mayer, R. Utility and Privacy Assessments of Synthetic Data for Regression Tasks. In Proceedings of the 2019 IEEE International Conference on Big Data (Big Data), Los Angeles, CA, USA, 9–12 December 2019; pp. 5763–5772. [Google Scholar]
- Nowok, B. Utility of Synthetic Microdata Generated Using Tree-Based Methods. UNECE Stat. Data Confidentiality Work Sess. 2015. Available online: https://unece.org/fileadmin/DAM/stats/documents/ece/ces/ge.46/20150/Paper_33_Session_2_-_Univ._Edinburgh__Nowok_.pdf (accessed on 26 February 2021).
- Hittmeir, M.; Ekelhart, A.; Mayer, R. On the utility of synthetic data: An empirical evaluation on machine learning tasks. In Proceedings of the 14th International Conference on Availability, Reliability and Security, Canterbury, UK, 26–29 August 2019; pp. 1–6. [Google Scholar]
- Dandekar, A.; Zen, R.A.; Bressan, S. Comparative Evaluation of Synthetic Data Generation Methods. Available online: https://www.di.ens.fr/~adandekar/files/papers/data_gen.pdf (accessed on 26 February 2021).
- Drechsler, J. Synthetic Datasets for Statistical Disclosure Control: Theory and Implementation; Springer: New York, NY, USA, 2011. [Google Scholar]
- Benaim, A.R.; Almog, R.; Gorelik, Y.; Hochberg, I.; Nassar, L.; Mashiach, T.; Khamaisi, M.; Lurie, Y.; Azzam, Z.S.; Khoury, J.; et al. Analyzing Medical Research Results Based on Synthetic Data and Their Relation to Real Data Results: Systematic Comparison From Five Observational Studies. JMIR Med. Inform. 2020, 8. [Google Scholar] [CrossRef]
- Heyburn, R.; Bond, R.; Black, M.; Mulvenna, M.; Wallace, J.; Rankin, D.; Cleland, B. Machine learning using synthetic and real data: Similarity of evaluation metrics for different healthcare datasets and for different algorithms. Data Sci. Knowl. Eng. Sens. Decis. Support 2018, 1281–1291. [Google Scholar]
- Nowok, B.; Raab, G.M.; Dibben, C. synthpop: Bespoke creation of synthetic data in R. J. Stat. Softw. 2016, 74, 1–26. [Google Scholar] [CrossRef]
- PrivBayes: Private Data Release via Bayesian Networks: ACM Transactions on Database Systems: Vol 42, No 4. Available online: https://dl.acm.org/doi/10.1145/3134428 (accessed on 24 December 2020).
- Trivedi, P.K.; Zimmer, D.M. Copula Modeling: An Introduction for Practitioners; Now Publishers Inc.: Hanover, MA, USA, 2007. [Google Scholar]
- General and Specific Utility Measures for Synthetic Data-Snoke-2018-Journal of the Royal Statistical Society: Series A (Statistics in Society)-Wiley Online Library. Available online: https://rss.onlinelibrary.wiley.com/doi/full/10.1111/rssa.12358 (accessed on 19 November 2020).
- Practical Synthetic Data Generation [Book]. Available online: https://www.oreilly.com/library/view/practical-synthetic-data/9781492072737/ (accessed on 6 September 2020).
- Snoke, J.; Raab, G.; Nowok, B.; Dibben, C.; Slavkovic, A. General and specific utility measures for synthetic data. arXiv 2017, arXiv:160406651. [Google Scholar] [CrossRef]
- Woo, M.-J.; Reiter, J.P.; Oganian, A.; Karr, A.F. Global Measures of Data Utility for Microdata Masked for Disclosure Limitation. J. Priv. Confid. 2009, 1. [Google Scholar] [CrossRef]
- Westreich, D.; Lessler, J.; Funk, M.J. Propensity score estimation: Neural networks, support vector machines, decision trees (CART), and meta-classifiers as alternatives to logistic regression. J. Clin. Epidemiol. 2010, 63, 826–833. [Google Scholar] [CrossRef]
- Rosenbaum, P.R.; Rubin, D.B. Reducing Bias in Observational Studies Using Subclassification on the Propensity Score. J. Am. Stat. Assoc. 1984, 79, 516–524. [Google Scholar] [CrossRef]
- El Emam, K. Seven Ways to Evaluate the Utility of Synthetic Data. IEEE Secur. Priv. 2020, 18, 56–59. [Google Scholar] [CrossRef]
- Reiter, J.P. Using CART to generate partially synthetic public use microdata. J. Off. Stat. 2005, 21, 441. [Google Scholar]
- Raschka, S. Model evaluation, model selection, and algorithm selection in machine learning. arXiv 2018, arXiv:181112808. [Google Scholar]
- Konen, W.; Koch, P.; Flasch, O.; Bartz-Beielstein, T.; Friese, M.; Naujoks, B. Tuned data mining: A benchmark study on different tuners. In Proceedings of the 13th Annual Conference on Genetic and Evolutionary Computation, New York, NY, USA, 9–13 July 2011; pp. 1995–2002. [Google Scholar] [CrossRef]
- Taylor, R. Interpretation of the Correlation Coefficient: A Basic Review. J. Diagn. Med. Sonogr. 1990, 6, 35–39. [Google Scholar] [CrossRef]




| Dataset Name | Short Name | Number of Observations | Number of Attributes (Predictors) | Categorical Predictors | Number of Labels | Total Synthetic Datasets Generated | Origin |
|---|---|---|---|---|---|---|---|
| BankNote | 1372 | 4 | 0 | 2 | 160 | UCI | |
| Titanic | 891 | 7 | 7 | 2 | 160 | Kaggle | |
| Ecoli | 336 | 7 | 0 | 8 | 160 | UCI | |
| Diabetes | 768 | 9 | 2 | 2 | 160 | UCI | |
| Cleveland heart | 297 | 13 | 8 | 2 | 160 | UCI | |
| Adult | 48,843 | 14 | 8 | 2 | 144 1 | UCI | |
| Breast cancer | 570 | 30 | 0 | 2 | 160 | UCI | |
| Dermatology | 366 | 34 | 33 | 6 | 160 | UCI | |
| SPECTF Heart | 267 | 44 | 0 | 2 | 160 | UCI | |
| Z-Alizadeh Sani | 303 | 55 | 34 | 2 | 160 | UCI | |
| Colposcopies | 287 | 68 | 6 | 2 | 160 | UCI | |
| ANALCATDATA | 841 | 71 | 3 | 2 | 160 | OpenML | |
| Mice Protein | 1080 | 80 | 3 | 8 | 160 | UCI | |
| Diabetic Mellitus | 281 | 97 | 92 | 2 | 135 2 | OpenML | |
| Tecator | 240 | 124 | 0 | 2 | 160 3 | OpenML |
| SP-np | SP-p | SDV | DS | |||||
|---|---|---|---|---|---|---|---|---|
| AD | Prop | AD | Prop | AD | Prop | AD | Prop | |
| 1.031553 | 0.074972 | 3.4375 | 0.155349 | 2.038835 | 0.155198 | 0.039442 | 0.099199 | |
| 0.037313 | 0.074571 | 2.103545 | 0.119064 | 4.650187 | 0.132735 | −0.46642 | 0.128916 | |
| 4.009901 | 0.075293 | 3.589109 | 0.108319 | 9.034653 | 0.157922 | 2.487624 | 0.165095 | |
| 2.288961 | 0.08493 | 4.821429 | 0.09705 | 6.504329 | 0.133996 | 2.505411 | 0.098722 | |
| 2.388889 | 0.086021 | 3.583333 | 0.095108 | 2.472222 | 0.152168 | 1.347222 | 0.154571 | |
| −0.41583 | 0.004673 | 4.805833 | 0.031423 | 6.4375 | 0.11616 | 2.659167 | 0.025647 | |
| 2.295322 | 0.128166 | 3.092105 | 0.191422 | 3.79386 | 0.155758 | 3.347953 | 0.159778 | |
| 5.363636 | 0.083088 | 43.65909 | 0.154837 | 8.684211 | 0.129181 | 1.113636 | 0.066643 | |
| 1.095679 | 0.132029 | 1.496914 | 0.144961 | 2.037037 | 0.142773 | 2.052469 | 0.118727 | |
| 10.26099 | 0.124009 | 11.78571 | 0.128848 | 10.6044 | 0.161104 | 8.873626 | 0.151974 | |
| 7.241379 | 0.153457 | 12.52874 | 0.178307 | 8.390805 | 0.234168 | 5.402299 | 0.233867 | |
| −6.03261 | 0.138881 | −7.67787 | 0.146417 | −3.57812 | 0.193994 | 1.215415 | 0.145242 | |
| 5.9825 | 0.171316 | 15.485 | 0.195285 | 25.8875 | 0.194417 | 5.6475 | 0.248655 | |
| 0.735294 | 0.112665 | 14.66176 | 0.143493 | 11.07353 | 0.166697 | 2.566845 | 0.140432 | |
| 10.88542 | 0.121138 | 26.63194 | 0.235745 | 9.809028 | 0.158509 | 14.44444 | 0.157601 | |
| Case | Synthesizer | Rel Accuracy | AD | Prop |
|---|---|---|---|---|
| Synthesized from original real data Accuracy for Case 3 | SP-np | 96.653917 | 3.1445598 | 0.104347 (1) |
| DS | 96.025506 | 3.5491086 | 0.139671 (2) | |
| SDV | 92.117844 | 7.1893318 | 0.158985 (4) | |
| SP-p | 89.770557 | 9.60027587 | 0.141708 (3) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dankar, F.K.; Ibrahim, M. Fake It Till You Make It: Guidelines for Effective Synthetic Data Generation. Appl. Sci. 2021, 11, 2158. https://doi.org/10.3390/app11052158
Dankar FK, Ibrahim M. Fake It Till You Make It: Guidelines for Effective Synthetic Data Generation. Applied Sciences. 2021; 11(5):2158. https://doi.org/10.3390/app11052158
Chicago/Turabian StyleDankar, Fida K., and Mahmoud Ibrahim. 2021. "Fake It Till You Make It: Guidelines for Effective Synthetic Data Generation" Applied Sciences 11, no. 5: 2158. https://doi.org/10.3390/app11052158
APA StyleDankar, F. K., & Ibrahim, M. (2021). Fake It Till You Make It: Guidelines for Effective Synthetic Data Generation. Applied Sciences, 11(5), 2158. https://doi.org/10.3390/app11052158





