Abstract
The prevalence of skin diseases has increased dramatically in recent decades, and they are now considered major chronic diseases globally. People suffer from a broad spectrum of skin diseases, whereas skin tumors are potentially aggressive and life-threatening. However, the severity of skin tumors can be managed (by treatment) if diagnosed early. Health practitioners usually apply manual or computer vision-based tools for skin tumor diagnosis, which may cause misinterpretation of the disease and lead to a longer analysis time. However, cutting-edge technologies such as deep learning using the federated machine learning approach have enabled health practitioners (dermatologists) in diagnosing the type and severity level of skin diseases. Therefore, this study proposes an adaptive federated machine learning-based skin disease model (using an adaptive ensemble convolutional neural network as the core classifier) in a step toward an intelligent dermoscopy device for dermatologists. The proposed federated machine learning-based architecture consists of intelligent local edges (dermoscopy) and a global point (server). The proposed architecture can diagnose the type of disease and continuously improve its accuracy. Experiments were carried out in a simulated environment using the International Skin Imaging Collaboration (ISIC) 2019 dataset (dermoscopy images) to test and validate the model’s classification accuracy and adaptability. In the future, this study may lead to the development of a federated machine learning-based (hardware) dermoscopy device to assist dermatologists in skin tumor diagnosis.
1. Introduction
Improving and ensuring healthy lives and promoting wellbeing in the community are some of the United Nations’ Sustainable Development Goals. For that reason, empowering and strengthening health practitioners with sufficient technological advancement (to attain the maximum potential of their health practices) is mandatory. Among other kinds of diseases, skin diseases have increased dramatically in recent decades, and they are now considered a major chronic disease globally [1]. People suffer from a broad spectrum of skin disease types (ranging from low to high severity level), such as eczema, chickenpox, measles, warts, acne, skin tumors, and others [2,3,4]. Skin tumors (melanoma, basal cell carcinoma, squamous cell carcinoma, and others) are the most dangerous type of skin disease and may be fatal if not treated early. For example, melanoma is a form of skin tumor that begins in cells known as melanocytes. Melanoma is more dangerous because of its ability to spread to other organs more rapidly if it is not treated early. To avoid complexities in skin tumors, an initial consultation with a healthcare professional is mandatory. Furthermore, health practitioners must also be equipped with a more accurate, robust, and sustainable solution for diagnosing and treating such diseases. Artificial intelligence can reduce the skin disease epidemic by providing on-time assistance or advice toward the appropriate action for the diagnosis and treatment of skin diseases (assisting the dermatologist) [5]. Some recent studies participated in diagnosing skin diseases using machine learning.
Hyperspectral imaging, laser doppler flow meter [6], and fluorescence spectroscopy have been used to diagnose skin flow oxidation for early diabetes complications [7]. The influence of blood pulsation is essential when analyzing skin disease, and it can be measured by pulse oximetry and photoplethysmography measurements [8]. One study [9] proposed applying mechanical pressure (using a fiber-optic probe) on the diffuse reflectance spectra of human skin measured in vivo [10]. A recent study presented a snapshot from a multiwavelength imaging device for in vivo skin diagnostics [11]. In comparison, laser doppler flowmetry and skin thermometry are also essential for functional diagnosis [12]. Some efforts also have been made to diagnose skin disease using high-dimensional imaging data. One study [13] proposed a compact hyperspectral-based analysis system to visualize skin chromophores and blood oxygenation. Furthermore, polarized hyperspectral imaging and machine learning represent a practical approach to diagnosing skin complications of diabetes mellitus at a very early stage [14].
Another study [15] proposed a transfer learning-based deep convolutional neural network (CNN) to classify skin diseases. The proposed solution is continuously fine-tuned through a weight upgradation approach (using backpropagation) to universally classify skin disease. However, the obtained classification accuracy was not desirable and simulated scenarios could not be applied to clinical trials. The two significant issues primarily reported when identifying skin disease (using machine learning) through dermatological images are data collection [16] and image transformation and feature extraction. Dermatologists are underrepresented in publications describing these technologies. Increased involvement and leadership of dermatologists are paramount to designing clinically relevant and efficacious models [17]. Notably, in clinical trials, dermoscopy, microscopy, or biopsy [18] devices have been used to obtain skin disease for analysis purposes. For example, an early study [19] used image segmentation and feature extraction to diagnose skin diseases. Another study [20] proposed an artificial neural network (ANN) based skin lesion classification model to classify the lesion into melanoma, abnormal, and typical classes. Moreover, [21] proposed a melanoma detection system using multiscale lesion-biased representation (MLR) and joint reverse classification (JRC). However, the abovementioned solutions are extensively expensive in terms of processing time and are not clinically used.
The existing studies are evidence that significant advances have been made in the field of dermatology. The broad implementation of machine learning-based tools is still pending, and prospective clinical trials are lacking. Thus, more appropriate and advanced solutions are required, which could be utilized practically by dermatologists. Therefore, this study proposes an adaptive federated machine learning-based skin disease detection model, which can assist dermatologists in the initial diagnosis of skin tumors and determine their severity level. The proposed model’s adaptive nature also allows the system to improve continuously (by learning on the fly), as well as add new types of skin disease. At the same time, the federated machine learning-based approach can address data privacy concerns.
Contribution
The notable contributions of this study are as follows:
- This study proposes the idea of an adaptive federated machine learning-based skin disease detection system to assist dermatologists.
- This study proposes a federated machine learning-based adaptive framework for skin disease.
- This study validates the proposed model’s classification performance and adaptability at the edge (local device) and cloud (global server) level.
Section 2 of this study surveys the related work and formulates a theoretical foundation. Section 3 proposes the idea of artificial intelligence-based dermoscopy and details the proposed model design. Section 4 presents the experiments and discusses the model validation process. Section 5 highlights the conclusions and future work.
2. Related Work and Theoretical Foundation
Previous research primarily focused on the early screening of skin cancer, whether a lesion was malignant or benign, or whether a lesion was a melanoma. However, upward of 90% of skin problems are not malignant, and addressing these more common conditions is also essential for reducing the global burden of skin disease. Especially in tropical regions, environmental changes (such as urbanization and industrialization) have increased the prevalence of skin diseases [22]. This section discusses the existing dermatology equipment (devices) and existing machine learning-based solutions for skin disease detection. This section also discusses the limitation of current technologies and highlights the motivation for developing adaptive federated machine learning-based dermoscopy devices.
2.1. Existing Devices Used for Skin Disease Analysis
Currently, manual or computer vision-based tools are used to diagnose skin diseases. Figure 1a shows a dermoscopy device widely used in advanced countries to diagnose melanoma cancer. It has demonstrated success in diagnosing other skin conditions such as psoriasis [4], lichen planus [4], and cicatricial alopecia. Dermoscopy visualizes the subtle clinical patterns of skin lesions and subsurface skin structures not typically visible to the unaided eye. However, when using this device, the health professional has to decide the severity and disease condition. Figure 1b presents the dermalite device, a user-friendly dermoscopy device attached to a smartphone. It enables capturing highly magnified dermatologist-grade photos of moles or other skin lesions that can be further shared with professional dermatologists for proper examination and diagnosis.
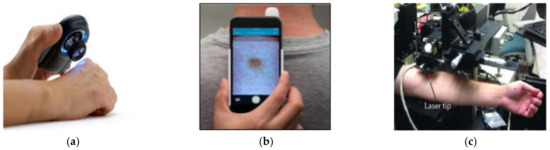
Figure 1.
Existing skin disease detection devices: (a) dermoscopy; (b) dermalite; (c) laser microscopy.
Figure 1c shows a microscope device developed by Stanford scientists to spot the seeds of cancer, diagnose diseases that include skin cancer, and perform exact surgery (without cutting the skin). However, laser-based imaging tools are expensive, slower, bulkier, and less accurate, and they are usually not meant for public adoption [23].
2.2. Current State of Machine Learning in Skin Disease Detection
In recent years, with the progress of machine learning technology, the expectations of artificial intelligence have been increasing, and research on its applications in dermatology has actively progressed [24]. In the literature, several studies proposed shallow learning-based skin disease solutions. For example, one study [25] used the artificial neural network to detect melanoma cancer from color images. The model used a discriminant feature to diagnose the tumor shape, categorize the melanoma skin disease into three main categories, and separate melanoma-like diseases. Another study [26] used the fuzzy c means region segmentation approach based on color discrimination. In this approach, a histogram (two-dimensional) is calculated along with the principal components and the Gaussian low-pass filter analysis. Similarly, an extreme machine learning-based approach was proposed in [27] to detect skin cancer. This approach requires an extensive process for feature extraction and texture analysis; thus, it is not feasible for clinical trials.
In contrast, some other studies worked on optimized feature extraction processes and determined that the spread of chronic skin diseases in different regions may lead to severe consequences. For example, [28] proposed a support vector machine-based model that automatically detects eczema and determines its severity on the basis of three different stages, i.e., effective segmentation, extraction of optimized features (color, texture, and borders), and severity of the disease itself. Another impressive attempt was made by [29] to extract more appropriate features. This study used the joint power of computer vision and machine learning to detect six types of skin disease. However, these studies were not adequate when addressing similar features across multiple diseases (e.g., some types of eczema show similar features to cancer); as such, wrong classifications may be obtained. The feature similarity issue can be overcome using low-level feature extraction techniques. For that reason, deep learning approaches (such as the convolutional neural network) are more desirable due to their pixel-level feature extraction process. Recently, several studies used deep learning-based techniques to classify similar features more accurately. In [30], a hybrid approach was designed by using a combination of shallow learning- and deep learning-based pretrained models, such as AlexNet and support vector machine. A recent study [31] highlighted the enormous potential of deep understanding to detect skin diseases with human-like diagnosis accuracy or better. Furthermore, this study urged the utilization of deep learning-based real-time intelligent healthcare systems for clinical utilization. The current approaches are only based on the batch learning approach and are static; thus, they do not welcome any future change (i.e., the model would need to be retrained if any changes are required). However, clinical procedures require a continuous upgradation to increase their accuracy and add new kinds of skin disease, rendering the current static deep learning approaches inapplicable. Accordingly, new adaptive mechanisms are required to ensure adaptability with high classification accuracy.
After an extensive analysis of the literature, it can be safely concluded that health practitioners usually apply manual or computer vision-based tools for the diagnosis of skin diseases, which may lead to misinterpretation and a longer analysis time. Existing devices such as laser microscopy and multimeter wave devices are only applicable in particular situations and are not soon expected to be implemented in clinical practices. On the contrary, dermoscopy and dermalite are majorly used clinically, but they require extensive health practitioner input to observe skin diseases. Overall, the current technologies are more expensive and need more time to analyze the actual condition and severity level. Deep learning-enabled dermoscopy is an essential approach to diagnose skin diseases and reduce the proliferation of the skin disease epidemic. Research has shown that, with proper training, diagnostic accuracy with dermoscopy is reportedly 75–84% [32,33], which does not meet the desirable level of classification accuracy. Additionally, these devices are static in nature and, thus, do not fulfill the current technological needs. Therefore, it is essential to provide a solution that could help health practitioners (dermatologists) reduce the skin disease epidemic.
3. Methodology
This section elaborates on the proposed solution and presents the developed algorithms. Figure 2 illustrates a system-level diagram of intelligent federated machine learning-based dermoscopy. The proposed solution would be placed on the edge. The master model would be placed in the cloud server. However, the federated machine learning approach can allow continuous upgradation of the master copies by learning the averaging weights from all edges after each classification (classification done by the dermatologist). The proposed model is adaptive with respect to new disease knowledge (to be classified), and it can improve as a function of experience (by learning from recent examples identified by the dermatologist) during deployment.
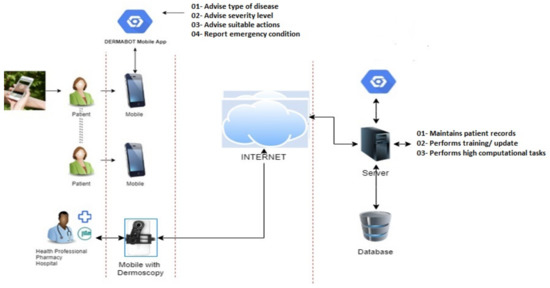
Figure 2.
The system-level architecture of adaptive federated machine learning-based skin disease detection.
Figure 3 depicts the application-level design. Two separate mobile applications are shown. They represent the two kinds of edges, one for the community (this module is not covered in this study) and the other for health practitioners (dermatologists). The community module aims to support patients in getting fundamental knowledge regarding skin diseases, as well as to motivate and assist users in visiting relevant health practitioners (nearest available dermatologists). In comparison, the health practitioner module is equipped with a dermoscopy device, which performs more detailed analysis of the captured dermoscopy images until diagnosis. This module also continuously transfers the updated weights of new samples (after each classification) to its cloud counterpart. Fundamentally, the health practitioner module can work together with the dermoscopy device by providing predictions (to health practitioners) regarding the skin disease type. Figure 4 represents the module and detailed architecture of the proposed model. This prototype was initially developed for the four most common skin diseases (mentioned in the International Skin Imaging Collaboration (ISIC) 2019 dataset). Later, it can incorporate new skin diseases through its adaptability feature. In future work, the provided model will be tuned with a multimodal solution (taking into account the input skin disease image and the patient’s medical history) to detect and monitor the patient’s progress, as depicted in Figure 5.
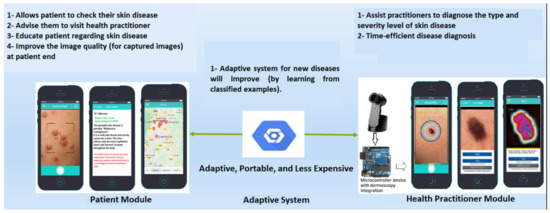
Figure 3.
The application-level architecture of adaptive federated machine learning-based skin disease detection.
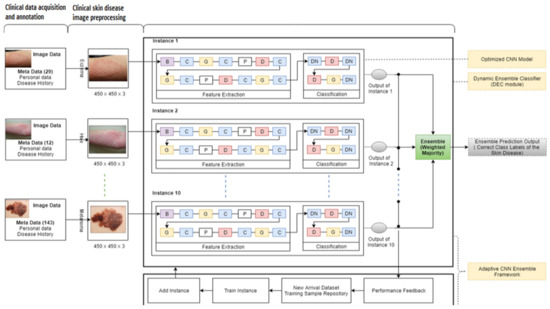
Figure 4.
The detail-level architecture of the proposed model.
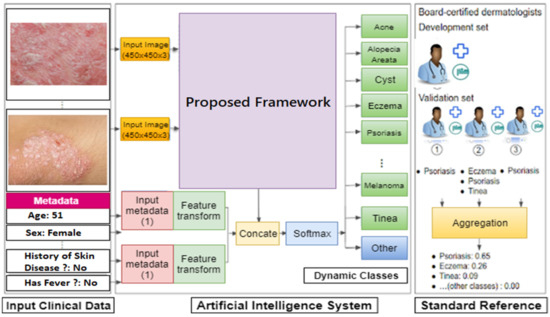
Figure 5.
The module-level architecture of multimodal skin disease detection.
In this proposed approach, the authors used the ensemble mechanism’s diversity from previous models (used to adapt to new spectral bands), which enabled handling the possible arrival of new classes and samples. Remarkably, the proposed approach (ensemble approach) contributes to diversity in a simple yet effective manner. This study also used the single-instance optimized CNN model inspired by [34,35] (which was carefully devised after numerous experiments) as an instance in the cloud server’s ensemble. Furthermore, the authors trained the proposed model using a challenging dataset (the ISIC 2019 dataset). The proposed model contains two core contributions: (1) the model was deployed on the cloud server, and (2) its deployment on the edges majorly contributes toward adaptability by continuously updating. The authors used online training (OT) and online classifier updating (OCU), presented in [36], with some internal tweaking parameters to make the approach suitable for the federated machine learning environment.
Averaging Mechanism: The global model (server) collects the trained weights from all local models (edges). The global model updates the global weight matrix, called G, such that G = G + (Iᵏ * w1ᵏ/S), where Iᵏ is the number of data points used to obtain w1ᵏ, and S is the sum of the number of data points across all local models (edges). Among all the available clients K, the server considers a small fraction of clients (C) in each round to update the global weight. Ic = max(C * K, 1), where Ic is the number of local models (edges).
3.1. Federated Machine Learning-Based Algorithm for Cloud Server
The cloud-based adaptive ensemble CNN was inspired by a previously proposed approach [36]. The primary difference is that this study restructured the previously proposed framework into the federated machine learning-based architecture. Thus, the core dynamic ensemble classifier and other modules such as OT and OCU were used to perform online training and continuously update the new samples. The additional neuron layers and weight update mechanism were mostly used to ensure runtime learning. However, such approaches are yet to be investigated for complex and high-dimensional data streams. In this module, the existing instance classifiers are updated (trained on old data) according to the individual updated weights from unique local models (edges). The primary objective is to update the current classification weight into the newly obtained classification weight to continuously update the global module, as depicted in Figure 6. Similar class weight differences were considered a challenging task, and the maintenance of classification accuracy for the global and local modules was a primary requirement. Algorithm 1 outlines the steps followed in performing cloud-based adaptive ensemble CNN tasks.
| Algorithm 1: Cloud-Based Adaptive Ensemble CNN |
| Input: The proposed model contains the instances I = (I1, I2, …, In), which are trained initial classes of skin disease, contains classes such as Cntrain: (cn1, cn2, …, cni), and classifies the input sample from dermoscopy device samples from edge DS. DS can have multiple samples (dermoscopy images), whereby S = (s1, s2, …, sn) is related to classes such as Cn (cn1, cn2, …, cni, cni + j,) at time interval (t + 1). Samples from cni + j are novel image samples or the same classes with additional complex features. Initialization: Threshold value for performance (Th) = 50 1: Counter (c) = 1 2: While data source > null //validate the input data source 3: Classify (S) using a single instance module (optimized CNN network) [34] 4: Identify the misclassified images using the activate performance feedback module 5: Determine the ensemble accuracies using the majority voting mechanism 6: if (percentage of) % accuracy for S ≥ Th //correctly classify 7: Repeat algorithm steps 3, 4, and 5 8: if % accuracy for S < Th //wrongly classify 9: Save S //save samples 10: Counter++ 11: Repeat algorithm steps 3, 4, and 5 12: if the counter is equal to 100 //number of wrongly classified instances reaches 100 13: Identify possible new classes using Algorithm 2 [36]. 14: Repeat step 3 15: Send the updated model to the edge node 16: End while Output: Module with (in+1) instances and classification using Cni + j. |
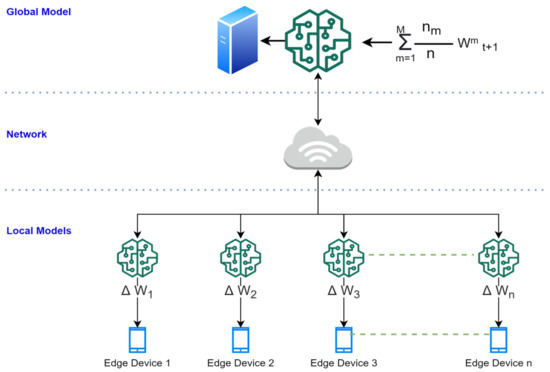
Figure 6.
Illustration of global and local model updates.
3.2. Federated Machine Learning-Based Algorithm for Edges
Fundamentally, this algorithm distributes and enables edges to perform collaborative learning, and it avoids sending the sample data to the cloud server for its upgradation. This process reduces the massive computational complexity and resolves the privacy issues (a critical issue) when using a cloud-based server. The deployed model (global model) is first trained on a server using some initial data. Each edge (smart dermoscopy or mobile) then improves the model using data available on the device (samples of diseases which were correctly tested), i.e., federated data from the device. The edge is trained using the newly observed data and updates the local model’s latest gradient weights. The changes made to the local model are summarized (as an update) and sent to the global model for global upgradation. However, to ensure faster transmission and avoid latency issues, random compression and quantization techniques are used. The process is done after several iterations (until a high-quality global model) is obtained on the cloud server. The edges send their trained models to the global model, which are averaged to obtain a unified cloud service model. TensorFlow federated and federated core application programming interfaces (APIs) are utilized for experimental purposes. A gradient guarantees convergence, whereas the model average cannot. The detailed steps of this algorithm are shown in Algorithm 2.
| Algorithm 2: Edge-Based Adaptive Ensemble CNN |
| Input: Edge receives the computed gradient (model M), ∑W, and computes the new gradient ∆W. Initialization: The edge model downloads the global model 1: Receive the sample data to perform classification//Initial model is received from the server 2: If sample data belong to existing classes, then 3: Perform the classification//regular operation 4: Perform training within the edge device//to compute the updated gradients 5: Update gradient weight to update the global model 6: Send the global updates to all local models 7: If sample data do not belong to existing classes, then 8: Create and train and update the new instance//using Algorithm 1 [36] 9: Update gradient weight to update the global model 10: Send the global updates to all local models Output: The edges send the updated ∆W to the cloud model. |
4. Experimental Results
This section presents three subsections to validate the effectiveness and performance of the proposed ameliorated framework. Section 4.1 details the data preparation and transformation of the datasets. Section 4.2 presents the experimental criteria and experimental setup. Section 4.3 display the obtained results, along with their analysis.
4.1. Data Preparation and Transformation
For evaluating the proposed model, this study used a challenging dataset, i.e., ISIC 2019 to verify the proposed framework (the ISIC skin disease dataset is considered one of the most challenging due to its sophisticated features).
Skin Disease Data Stream Pipeline Preparation to Simulate Concept Drift (CD)
In this study, the authors selected a challenging real dataset created by the International Skin Imaging Collaboration (ISIC). ISIC released this dataset to the research and professional communities for open competition (skin lesion analysis toward melanoma detection) in 2019. The ISIC competition provides a challenging task to the research community with the aim of obtaining optimized solutions worldwide. ISIC is developing proposed standards to address the technologies, techniques, and terminology used in skin imaging with attention to the issues of privacy and interoperability (i.e., the ability to share images across technology and clinical platforms). In ISIC 2019, the skin disease (https://challenge2019.isic-archive.com/) dataset contains imbalanced classes. Furthermore, the samples in the classes have several similar features, which makes this dataset more challenging. ISIC 2019 is an archive repository of dermoscopic images for clinical training and supporting technical research to assist in automated algorithmic analysis. In total, the dataset contains 25,331 dermoscopy image samples consisting of nine classes, namely, melanoma (MEL), melanocytic nevus (NV), basal cell carcinoma (BCC), actinic keratosis (AK), benign keratosis (BKL), dermatofibroma (DF), vascular lesion (VASC), squamous cell carcinoma (SSC), and unknown (UKN). Class UKN is the ninth class of the ISIC 2019 dataset, containing the image samples that are unrelated to the other eight class types, which helps increase the generalization of the developed model. A few random image samples are depicted in Figure 7.
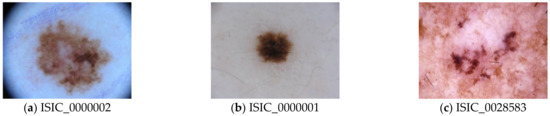
Figure 7.
Random image samples from International Skin Imaging Collaboration (ISIC) dataset; (a) melanoma (MEL); (b) melanocytic nevus; (c) basal cell carcinoma.
The maximum number of samples was 12,875 (for class 1), and the minimum number of samples was 239 (for class 5), which represents the highly imbalanced classes. In addition, for each class, the available number of samples was 4522, 12,875, 3323, 867, 2624, 239, 253, and 628 in classes 0, 1, 2, 3, 4, 5, 6, and 7, respectively, as shown in Figure 8. This class imbalance problem can cause overfitting issue (bias toward classes for which the number of samples is greater). Therefore, image augmentation techniques were used to handle the overfitting issue, such as image flipping, random cropping, random scaling, central zooming, and increasing/decreasing brightness and sharpness, to balance the number of classes in each class. Figure 9 depicts the random samples generated using image augmentation. Python libraries were also used with appropriate parameters to increase each class image sample. The image pixel intensity values were also normalized from 0 to 255 to 0 to 1 to reduce the computational complexity.
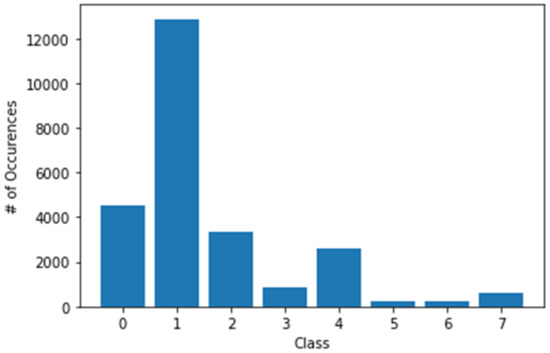
Figure 8.
The number of samples was 4522, 12,875, 3323, 867, 2624, 239, 253, and 628 in classes 0, 1, 2, 3, 4, 5, 6, and 7, respectively.
Figure 9.
Representation of augmented image samples after applying image augmentation techniques; (a) represents the augmented image sample for melanoma skin disease, (b) represents the augmented image sample for melanocytic nevus skin disease.
4.2. Experimental Criteria and Performance Measures
To simulate a federated learning environment, we designed two primary situations: (1) validation of the global model and local models by measuring the classification evaluation measures before and after observing new data samples, and (2) validation of the local models’ overall classification accuracy and histogram clustering gradient for online training dataset formation.
4.2.1. Environment and Libraries
The experiments were carried out on the Google Cloud Platform (GCP) and Google Colaboratory on the GCP server (us-west1-b region) with a computed engine virtual machine and additional machine learning and deep learning libraries. To speed up the complex computing jobs, the authors used 16 virtual central processing units (vCPUs), with 104 GB random-access memory (RAM) and a single NVIDIA graphics processing unit (GPU) Tesla K80. The experiments were implemented using the Python 3 programming language and the libraries below.
Environment setup:
- Python version (Python 3.6.3), installed using PyPI;
- Virtual environment from Anaconda;
- TensorFlow (1.13), Theano, and Keras (as backend) for complicated deep learning classification.
Library setup:
- TensorFlow federated;
- Federated core API;
- Scikit-learn library to perform basic machine learning tasks;
- OpenCV to perform image processing tasks;
- NumPy and Pandas for data manipulation and processing;
- Seaborn and Matplotlib for visualization of the results.
4.2.2. Hyperparameter Optimization and Performance Measures
To select hyperparameters for training the model, the authors of this study used a manual search strategy [37]. The authors acquired the optimized training hyperparameters after various tuning iterations through the manual search strategy, as shown in Table 1. They also followed best practices outlined by the research community, for example, selection of an optimization function (Adam) and cross-entropy selection (one-hot encoded). The classification accuracy is considered the most suitable metric to evaluate model performance in a nonstationary environment [38]. This study has used performance measures recognized as primary classification performance indicators by the research community [39,40].

Table 1.
Training hyperparameters (tuning values and optimized values).
4.3. Experimental Results and Discussion
The authors of this study performed two experiments to analyze the performance of the proposed framework. Initially, experiment 1 was carried out to validate the global and local models by measuring the classification evaluation measures before (case 1) and after (case 2) observing new data samples. Later, in experiment 2, the overall classification accuracy of local models (at edges) was measured. These experiments also allowed validating the histogram of clustering distance during edge training and validating the performance (accuracy and loss) during new sample adaptation.
4.3.1. Experiment 1: Validation of the Global and Local Models by Measuring the Classification Evaluation Measures before (Case 1) and after (Case 2) Observing New Data Samples
The primary intention of this experiment was to evaluate the performance of the proposed framework in a stable condition. The obtained results were promising with a recorded classification accuracy of 95.6%, loss of 2.50 (as shown in Table 2), and 0.95 precision and recall (as shown in Table 3). This study analyzed the proposed framework’s performance with a challenging dataset (complex features), i.e., the ISIC skin disease dataset. Here, the proposed framework was trained using four classes of the skin disease dataset, which were dermatofibroma (DF), vascular lesion (VASC), squamous cell carcinoma (SSC), and unknown (UKN). Despite the complex features and class imbalance problem in the skin disease dataset, the model’s performance was satisfactory, even better than highlighted in the literature. In case 2, the proposed model was trained on the first four classes and correctly classified them. Later, the proposed model incorporated four new classes (which were not trained on the proposed framework), with a subsequent degradation in performance.

Table 2.
Accuracy and loss for ISIC 2019 (skin disease) streams in different cases.

Table 3.
Precision and recall for ISIC skin disease data streams in different cases.
The intent behind this experiment was to determine the accuracy of the proposed model at the cloud level. It was shown that the proposed framework outperformed those in the literature and achieved satisfactory classification accuracy. The reported classification accuracy was 89% (as shown in Table 2). Moreover, a significant level of precision and recall was noted. However, in this case, the loss increased to 3.5 from 2.5 in case 1 (as shown in Table 2). The model’s overall performance after adapting new classes was noticeably lesser, because model training in offline mode is always better than that in online mode (online mode creates online training dataset formation, which might contain noisy data). Thus, more advanced techniques are required to overcome these differences. Additionally, all the individual classification accuracies showed good performance after the arrival of new samples, as shown in Figure 10b.
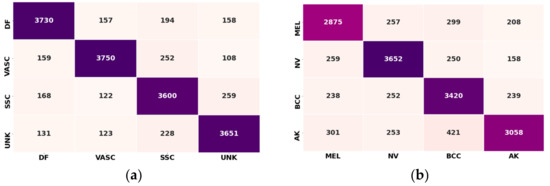
Figure 10.
Confusion matrix: (a) confusion matrix before adaptation (dermatofibroma (DF), vascular lesion (VASC), squamous cell carcinoma (SSC), and unknown (UKN) ISIC dataset classes); (b) confusion matrix after adaptation (class 1: melanoma (MEL), class 2: melanocytic nevus NV, class 3: basal cell carcinoma (BCC), and class 4: actinic keratosis (AK); ISIC dataset classes).
This study also generated receiver operating characteristic (ROC) curve plots to demonstrate the tradeoff between sensitivity and specificity. A possible increase in sensitivity, accompanied by a decrease in particularity was observed in the proposed framework before and after new class arrival, as depicted in Figure 8a,b. The ROC curve was closer to the left-hand border and the top edge of the ROC space. This shows the true positive rate vs. the false positive rate. In model1_SD, the obtained ROC curve was desirable in both cases, as shown in Figure 11.
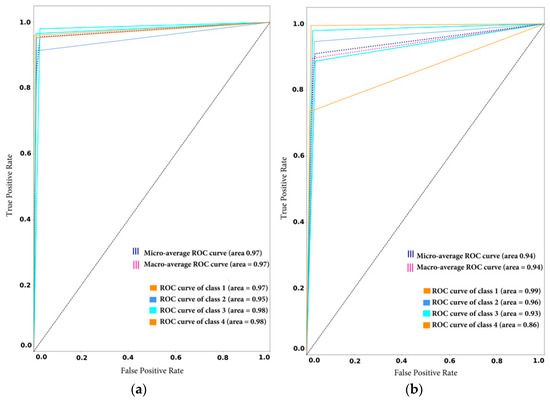
Figure 11.
Receiver operating characteristic (ROC) curve with sensitivity on the x-axis (true positive rate) and specificity on the y-axis (false positive rate): (a) before adaptation; (b) after adaptation.
4.3.2. Experiment 2: Local Model Overall Classification Accuracy Performance, the Histogram of Clustering Distance during Edge Training and Testing, and Validation of Performance with New Samples
The intent behind this experiment was to test the overall classification accuracies of our deployed model, as well as apply its feature extraction technique to prepare for online training dataset formation. Lastly, the updated training and validation accuracies and loss are presented to validate the model’s successful incorporation of the new dataset at the edges and on the cloud server.
Overall Classification Performance of the Cloud Models
Figure 12 shows the confusion matrix for all trained eight classes. It can be noted that the global model performed well in the stable scenario, with classification accuracy above 90%. Simultaneously, some classes performed exceptionally, such as class 5 and class 1, with maximum correct predictions of 4005 and 3900 samples, respectively.

Figure 12.
Confusion metrics representing the correct prediction rate of the cloud server (global) model.
Histogram of Clustering Distance during Edge Model Training for New Samples
The histogram of clustering distances identifies how correctly the new samples are extracted and clustered for new dataset formation. New dataset formation is an essential procedure for training edges with newly collected samples. Table 4 represents the four classes of updated samples that were collected and clustered after the feature extraction process. The feature extraction process was done using the pretrained network. Later, the extracted features were clustered using the k-means clustering algorithm. In Table 4, it can be noted that class 6 means clustered well, showing a mean values of 99.85, variance of 160.55, and standard deviation of 12.6. Class 4 showed worse performance with a mean value of 78.5, variance of 144.34, and standard deviation of 12.10. Well-clustered features ensure better training and validation accuracies when updating the edge models. Furthermore, they reduce the overfitting issue by neglecting the condition of bias toward a particular class.

Table 4.
Precision and recall for ISIC skin disease data streams in different cases.
Figure 13 represents each prediction input used for computing the cluster distance, assigned into histogram bins. Here, the y-axis is the number of predictions and the x-axis represents the cluster distance. Notably, the histogram represents how frequently the prediction/classification falls within a particular range of cluster distances.
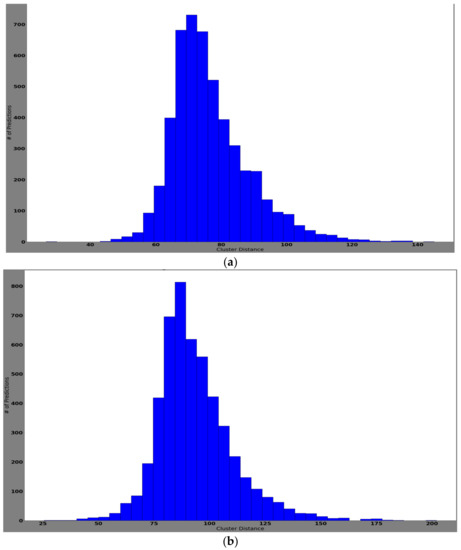
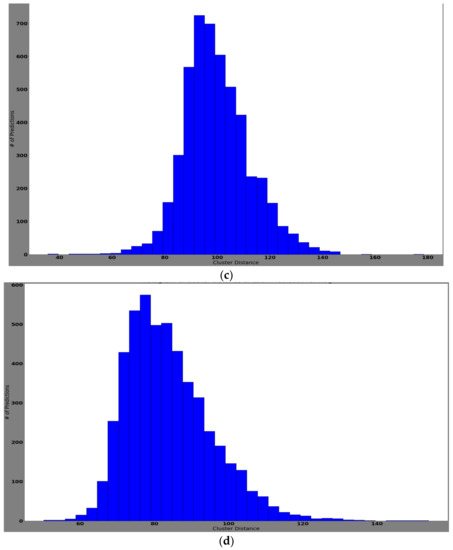
Figure 13.
The histogram of cluster distances representing the model that successfully clustered the features of the classes: (a) histogram of cluster distance of each prediction for class AK; (b) histogram of cluster distance of each prediction for class BCC; (c) histogram of cluster distance of each prediction for class NV; (d) histogram of cluster distance of each prediction for class MEL.
Training Performance of Edge Models with New Samples
Figure 14 illustrates the training and validation accuracy and loss for the new sample adaptation at the edges. Figure 14a shows the correlation reported for training and validation accuracy and loss during the retraining of new samples at epoch 3. Figure 14b depicts the correlation reported for training and validation accuracy and loss during the retraining of new samples at epoch 6. Interestingly, in the presented results, it can be noted that the edge model retrained the new samples by continuously increasing its training and validation accuracy and minimizing loss after each epoch. Furthermore, it can be noted that the observed loss and accuracy were stable at every epoch.
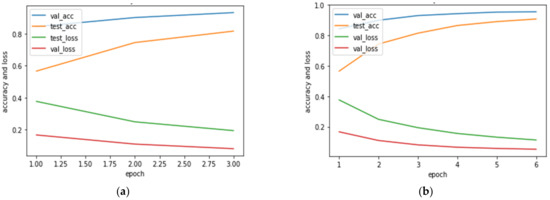
Figure 14.
Training and validation accuracy and loss for new sample training in edge models; (a) validation accuracy (val_acc), testing accuracy (test_acc), validation loss (val_loss), and testing loss (test_loss) when new samples were trained at epoch 3; (b) validation accuracy (val_acc), testing accuracy (test_acc), validation loss (val_loss), and testing loss (test_loss) when new samples were trained at epoch 6.
5. Conclusions and Future Work
The majority of diagnoses in dermatology are based on visual pattern recognition of morphological features. Skin imaging technology currently involves dermoscopy devices, very-high-frequency (VHF) ultrasound, and reflectance confocal microscopy (RCM). Each method of skin imaging has its advantages and limitations. Dermatologists need to choose different imaging methods according to varying conditions of skin lesions. Skin imaging technology has become a vitally important tool for the clinical diagnosis of skin diseases, and it is widely accepted and applied in the world. At the same time, machine learning-based dermoscopy is exceedingly suitable for improving the diagnosis capabilities of dermatologists. Accordingly, this study proposed an intelligent dermoscopy device, which can be used by health practitioners for the clinical diagnosis of skin tumors. This study offers a continuous improvement in classification accuracy by developing a more robust solution through the adaptability mechanism. To ensure adaptability, this study proposed an adaptive federated machine learning-based model that can correctly classify the dermoscopy images for skin disease classification, capable of learning new features (new samples acquired during the classification task through the dermoscopy device). This study used the previously proposed online training and online classifier update (k-means clustering method for new training dataset formation). However, this study found that using the clustering-based mechanism to distinguish different classes for similar features resulted in some classification degradation after adaptation. Hence, a supervised learning mechanism should be used for new dataset formation. The proposed framework showed satisfactory performance for both the cloud and the edge models. The results demonstrated adequate classification performance (in terms of accuracy) for the edge models, which is essential for clinical trials.
Furthermore, a patient-level mobile application is also offered to help patients locate the nearest dermatologist and to provide the necessary information regarding the skin disease to prevent negligence. However, this module was not practically tested herein and will be addressed in a future study. The authors also aim to develop a prototype (hardware) intelligent dermoscopy device for dermatologists (using the proposed federated machine learning-based model), which will be tested clinically.
Author Contributions
Conceptualization, M.A.H. and S.M.J.; methodology, M.A.H. and S.M.J.; software, S.M.J.; validation, M.A.H. and S.M.J. and S.S.; formal analysis, M.A.H. and S.M.J.; investigation, M.A.H. and S.M.J.; resources, Manzoor Ahmed Hashmani and S.S.H.R.; data curation, S.M.J.; writing—original draft preparation, S.M.J.; writing—review and editing, M.A.H. and S.S.H.R.; visualization, S.M.J.; supervision, M.A.H.; project administration, M.A.H. and S.M.J.; funding acquisition, M.A.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research study was conducted at the Universiti Teknologi PETRONAS (UTP), Malaysia, as a part of the research project “A novel approach to mitigate the performance degradation in big data classification model” under the matching grant scheme (Cost Centre: 015ME0-057).
Informed Consent Statement
Not applicable.
Data Availability Statement
The ISIC 2019 challenge dataset is available at https://challenge2019.isic-archive.com.
Acknowledgments
The authors acknowledge the Department of Computer and Information Sciences and High-Performance Cloud Computing Center (HPC3), Universiti Teknologi PETRONAS, for their support in completing this research study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pezzolo, E.; Naldi, L. Epidemiology of major chronic inflammatory immune-related skin diseases in 2019. Expert Rev. Clin. Immunol. 2020, 16, 155–166. [Google Scholar] [CrossRef]
- Tizek, L.; Schielein, M.; Seifert, F.; Biedermann, T.; Böhner, A.; Zink, A. Skin diseases are more common than we think: Screening results of an unreferred population at the Munich Oktoberfest. J. Eur. Acad. Dermatol. Venereol. 2019, 33, 1421–1428. [Google Scholar] [CrossRef]
- Han, W.H.; Yong, S.S.; Tan, L.L.; Toh, Y.F.; Chew, M.F.; Pailoor, J.; Kwan, Z. Characteristics of skin cancers among adult patients in an urban Malaysian population. Australas. J. Dermatol. 2019, 60, e327–e329. [Google Scholar] [CrossRef] [PubMed]
- Hay, R.J.; Johns, N.E.; Williams, H.C.; Bolliger, I.W.; Dellavalle, R.P.; Margolis, D.J.; Marks, R.; Naldi, L.; Weinstock, M.A.; Wulf, S.K.; et al. The global burden of skin disease in 2010: An analysis of skin conditions’ prevalence and impact. J. Investig. Dermatol. 2014, 134, 1527–1534. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhang, C.J.P.; Jiang, B.; Chen, J.; Song, J.; Liu, Z.; He, Z.; Krittanawong, C.; Fang, P.-H.; Ming, W.-K. Artificial intelligence versus clinicians in disease diagnosis: Systematic review. JMIR Med. Inform. 2019, 7, e10010. [Google Scholar] [CrossRef] [PubMed]
- Mizeva, I.; Makovik, I.; Dunaev, A.; Krupatkin, A.; Meglinski, I. Analysis of skin blood microflow oscillations in patients with rheumatic diseases. J. Biomed. Opt. 2017, 22, 070501. [Google Scholar] [CrossRef] [PubMed]
- Zharkikh, E.; Dremin, V.; Zherebtsov, E.; Dunaev, A.; Meglinski, I. Biophotonics methods for functional monitoring of complications of diabetes mellitus. J. Biophotonics 2020, 13, e202000203. [Google Scholar] [CrossRef] [PubMed]
- Dremin, V.; Zherebtsov, E.; Bykov, A.; Popov, A.; Doronin, A.; Meglinski, I. Influence of blood pulsation on diagnostic volume in pulse oximetry and photoplethysmography measurements. Appl. Opt. 2019, 58, 9398–9405. [Google Scholar] [CrossRef]
- Popov, A.P.; Bykov, A.V.; Meglinski, I.V. Influence of probe pressure on diffuse reflectance spectra of human skin measured in vivo. J. Biomed. Opt. 2017, 22, 110504. [Google Scholar] [CrossRef]
- Ahmed, I.; Halder, S.; Bykov, A.; Popov, A.; Meglinski, I.V.; Katz, M. In-body Communications Exploiting Light: A Proof-of-concept Study using ex vivo Tissue Samples. IEEE Access 2020, 8, 190378–190389. [Google Scholar] [CrossRef]
- Spigulis, J.; Rupenheits, Z.; Matulenko, M.; Oshina, I.; Rubins, U. A snapshot multi-wavelengths imaging device for in-vivo skin diagnostics. In Multimodal Biomedical Imaging XV; International Society for Optics and Photonics: Bellingham, WA, USA, 2020; Volume 11232, p. 112320I. [Google Scholar]
- Zherebtsov, E.A.; Zherebtsova, A.I.; Doronin, A.; Dunaev, A.V.; Podmasteryev, K.V.; Bykov, A.; Meglinski, I. Combined use of laser Doppler flowmetry and skin thermometry for functional diagnostics of intradermal finger vessels. J. Biomed. Opt. 2017, 22, 040502. [Google Scholar] [CrossRef][Green Version]
- Zherebtsov, E.; Dremin, V.; Popov, A.; Doronin, A.; Kurakina, D.; Kirillin, M.; Meglinski, I.; Bykov, A. Hyperspectral imaging of human skin aided by artificial neural networks. Biomed. Opt. Exp. 2019, 10, 3545–3559. [Google Scholar] [CrossRef] [PubMed]
- Dremin, V.; Marcinkevics, Z.; Zherebtsov, E.; Popov, A.; Grabovskis, A.; Kronberga, H.; Geldnere, K.; Doronin, A.; Meglinski, I.; Bykov, A. Skin complications of diabetes mellitus revealed by polarized hyperspectral imaging and machine learning. IEEE Trans. Med. Imaging 2021. [Google Scholar] [CrossRef] [PubMed]
- Liao, H. A Deep Learning Approach to Universal Skin Disease Classification. Available online: https://www.cs.rochester.edu/u/hliao6/projects/other/skinprojectreport.pdf (accessed on 2 August 2020).
- Masood, A.; Al-Jumaily, A.A. Computer-aided diagnostic support system for skin cancer: A review of techniques and algorithms. Int. J. Biomed. Imaging 2013, 2013, 323268. [Google Scholar] [CrossRef]
- Zakhem, G.A.; Fakhoury, J.W.; Motosko, C.C.; Ho, R.S. Characterizing the role of dermatologists in developing artificial intelligence for assessment of skin cancer: A systematic review. J. Am. Acad. Dermatol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Binder, M.; Kittler, H.; Seeber, A.; Steiner, A.; Pehamberger, H.; Wolff, K. Epiluminescence microscopy-based classification of pigmented skin lesions using computerized image analysis and an artificial neural network. Melanoma Res. 1998, 8, 261–266. [Google Scholar] [CrossRef]
- Burroni, M.; Corona, R.; Dell’Eva, G.; Sera, F.; Bono, R.; Puddu, P.; Perotti, R.; Nobile, F.; Andreassi, L.; Rubegni, P. Melanoma computer-aided diagnosis: Reliability and feasibility study. Clin. Cancer Res. 2004, 10, 1881–1886. [Google Scholar] [CrossRef] [PubMed]
- Ozkan, I.A.; Koklu, M. Skin Lesion Classification using Machine Learning Algorithms. Int. J. Intell. Syst. Appl. Eng. 2017, 4, 285–289. [Google Scholar] [CrossRef]
- Bi, L.; Kim, J.; Ahn, E.; Feng, D.; Fulham, M. Automatic melanoma detection via multi-scale lesion-biased representation and joint reverse classification. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2006; pp. 1055–1058. [Google Scholar]
- Hay, R.; Bendeck, S.E.; Chen, S.; Estrada, R.; Haddix, A.; McLeod, T.; Mahé, A. Skin diseases. In Disease Control Priorities in Developing Countries, 2nd ed.; The International Bank for Reconstruction and Development/The World Bank: Washington, DC, USA, 2006. [Google Scholar]
- Author of Star Media Group. (2012, May 06, Oct 29). Itchy skin. Available online: https://www.thestar.com.my/lifestyle/health/2012/05/06/itchy-skin (accessed on 15 June 2020).
- Jinnai, S.; Yamazaki, N.; Hirano, Y.; Sugawara, Y.; Ohe, Y.; Hamamoto, R. The development of a skin cancer classification system for pigmented skin lesions using deep learning. Biomolecules 2020, 10, 1123. [Google Scholar] [CrossRef] [PubMed]
- Ercal, F.; Chawla, A.; Stoecker, W.V.; Lee, H.-C.; Moss, R.H. Neural Network Diagnosis of Malignant Melanoma From Color Images. IEEE Trans. Biomed. Eng. 1994, 41, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Schmid, P. Segmentation of Digitized Dermatoscopic Images by Two-Dimensional Color Clustering. IEEE Trans. Med. Imaging 1999, 18, 164–171. [Google Scholar] [CrossRef]
- Hoshyar, A.N.; Al-Jumaily, A.; Sulaiman, R. Review on Automatic Early skin Cancer Detection. In Proceedings of the International Conference in Computer science and Service System (CSSS), Nanjing, China, 27–29 June 2011; pp. 4036–4039. [Google Scholar]
- Alam, N.; Munia, T.; Tavakolian, K.; Vasefi, V.; MacKinnon, N.; Fazel-Rezai, R. Automatic Detection and Severity Measurement of Eczema Using Image Processing. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016. [Google Scholar] [CrossRef]
- Kumar, V.; Kumar, S.; Saboo, V. Dermatological Disease Detection Using Image Processing and Machine Learning. In Proceedings of the 2016 Third International Conference on Artificial Intelligence and Pattern Recognition (AIPR), Lodz, Poland, 19–21 September 2016. [Google Scholar] [CrossRef]
- Soliman, N.; ALEnezi, A. A Method of Skin Disease Detection Using Image Processing and Machine Learning. Procedia Comput. Sci. 2019, 163, 85–92. [Google Scholar]
- Bajwa, M.N.; Muta, K.; Malik, M.I.; Siddiqui, S.A.; Braun, S.A.; Homey, B.; Dengel, A.; Ahmed, S. Computer-Aided Diagnosis of Skin Diseases Using Deep Neural Networks. Appl. Sci. 2020, 10, 2488. [Google Scholar] [CrossRef]
- Codella, N.; Rotemberg, V.; Tschandl, P.; Celebi, M.E.; Dusza, S.; Gutman, D.; Helba, B.; Kalloo, A.; Liopyris, K.; Marchetti, M.; et al. Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (ISIC). arXiv Prepr. 2019; arXiv:1902.03368. [Google Scholar]
- Vestergaard, M.; Macaskill, P.; Holt, P.; Menzies, S. Dermoscopy compared with naked eye examination for the diagnosis of primary melanoma: A meta-analysis of studies performed in a clinical setting. Br. J. Dermatol. 2008, 159, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Jameel, S.M.; Hashmani, M.A.; Rehman, M.; Budiman, A. Adaptive CNN Ensemble for Complex Multispectral Image Analysis. Complexity 2020, 2020, 8361989. [Google Scholar] [CrossRef]
- Jameel, S.M.; Hashmani, M.A.; Alhussain, H.; Rehman, M.; Budiman, A. An optimized deep convolutional neural network architecture for concept drifted image classification. In Intelligent Systems and Applications. IntelliSys 2019. Advances in Intelligent Systems and Computing; Bi, Y., Bhatia, R., Kapoor, S., Eds.; Springer: Cham, Switzerland, 2020; Volume 1037. [Google Scholar] [CrossRef]
- Jameel, S.M.; Hashmani, M.A.; Rehman, M.; Budiman, A. An Adaptive Deep Learning Framework for Dynamic Image Classification in the Internet of Things Environment. Sensors 2020, 20, 5811. [Google Scholar] [CrossRef]
- Gama, J.; Sebastião, R.; Rodrigues, P.P. On evaluating stream learning algorithms. Mach. Learn. 2013, 90, 317–346. [Google Scholar] [CrossRef]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Iwashita, A.S.; Papa, J.P. An Overview on Concept Drift Learning. IEEE Access 2019, 7, 1532–1547. [Google Scholar] [CrossRef]
- Hashmani, M.A.; Muslim, S.; Alhussain, H.; Rehman, M.; Budiman, A. Accuracy Performance Degradation in Image Classification Models due to Concept Drift. Int. J. Adv. Comput. Sci. Appl. 2019, 10. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).