Computer Tools to Analyze Lung CT Changes after Radiotherapy
Abstract
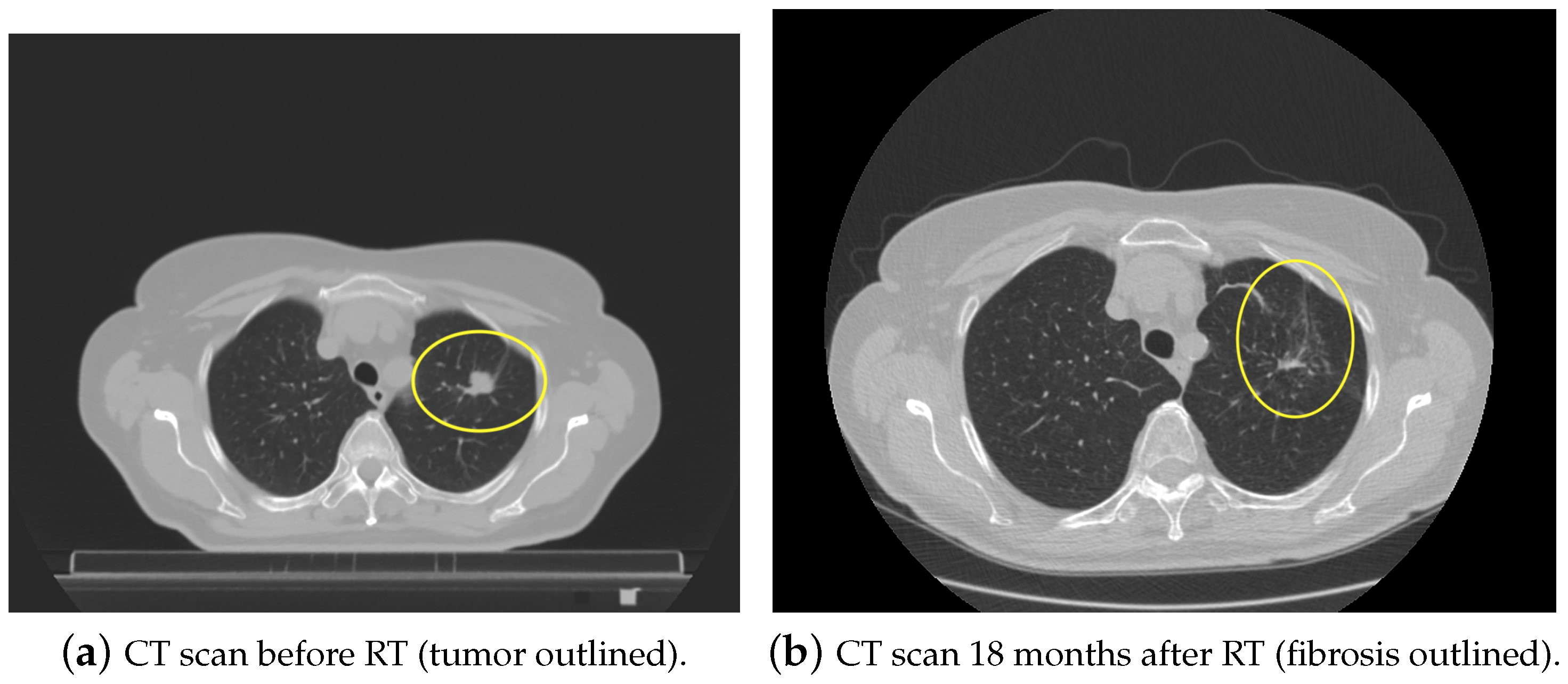
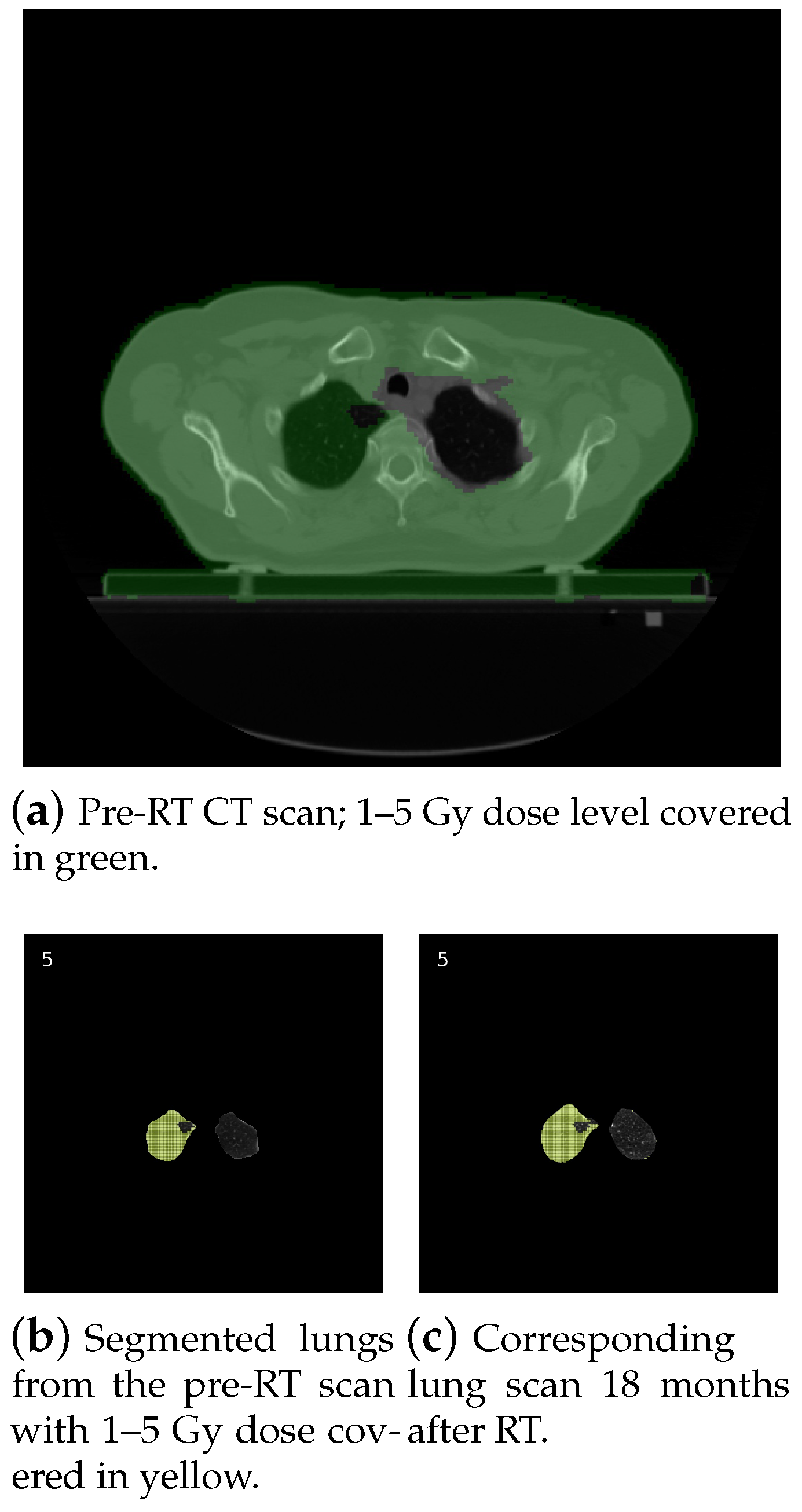
1. Introduction
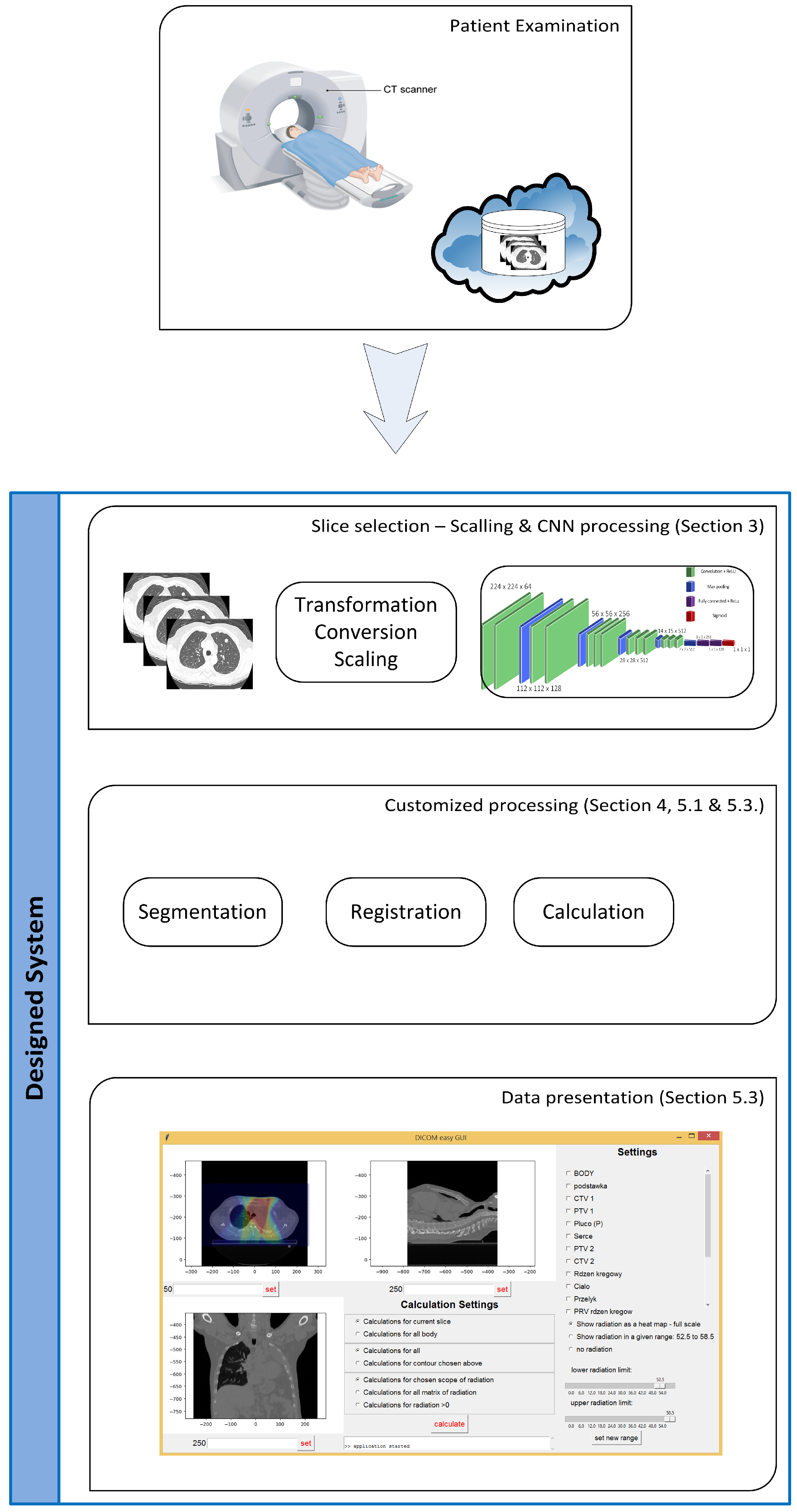
2. Structure of the System
- The import of DICOM (Digital Imaging and Communications in Medicine) files (including radiation doses),
- The election of CT slices containing lungs,
- The reduction of the number of slices in the moving set (after radiation series),
- Lung segmentation,
- Affine lung registration,
- Elastic lung registration,
- Calculations on preprocessed images,
- The export of results to a CSV file.
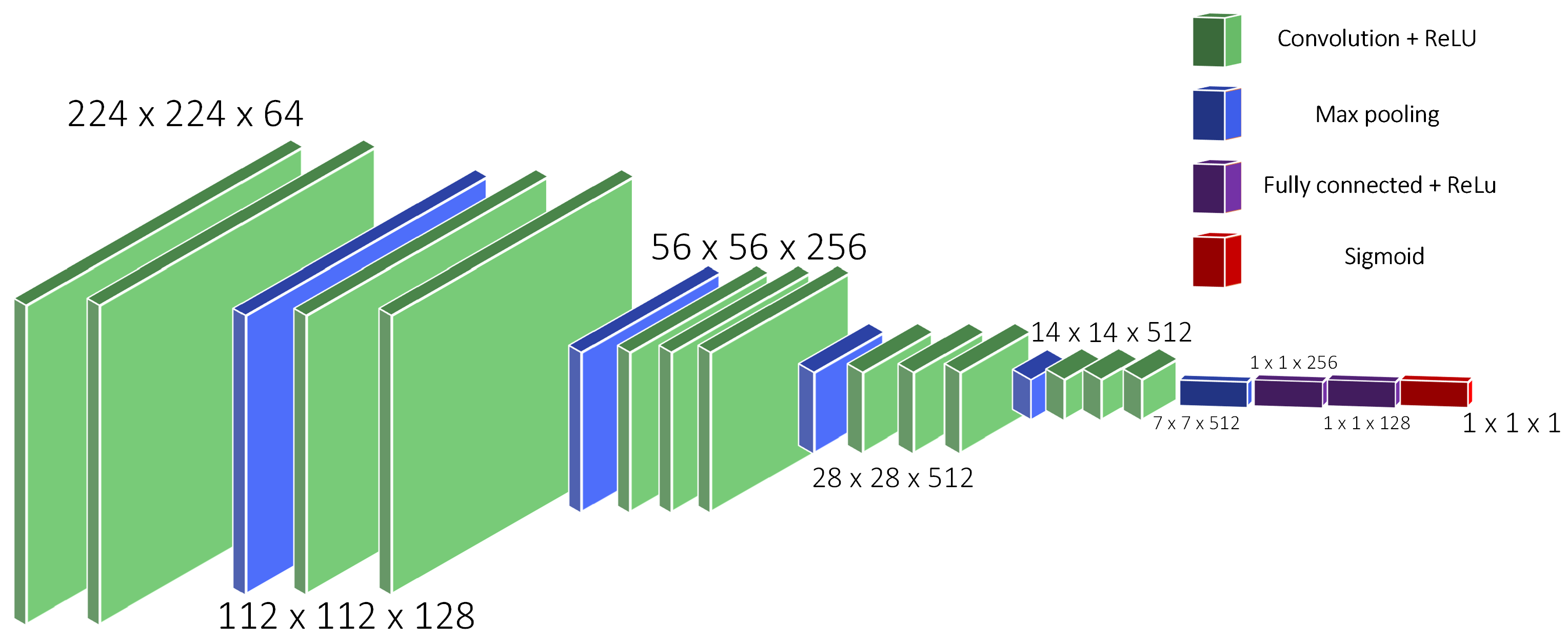
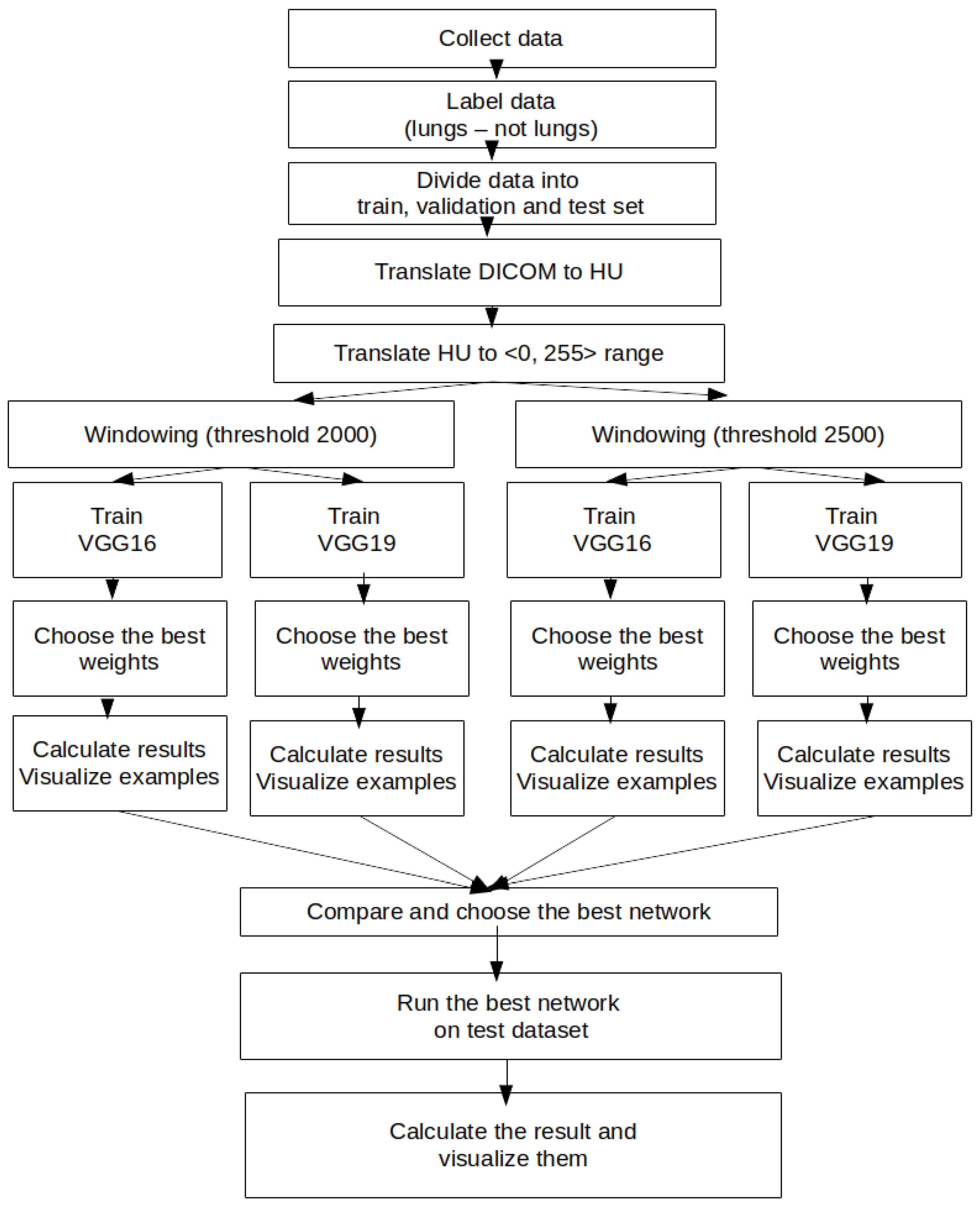
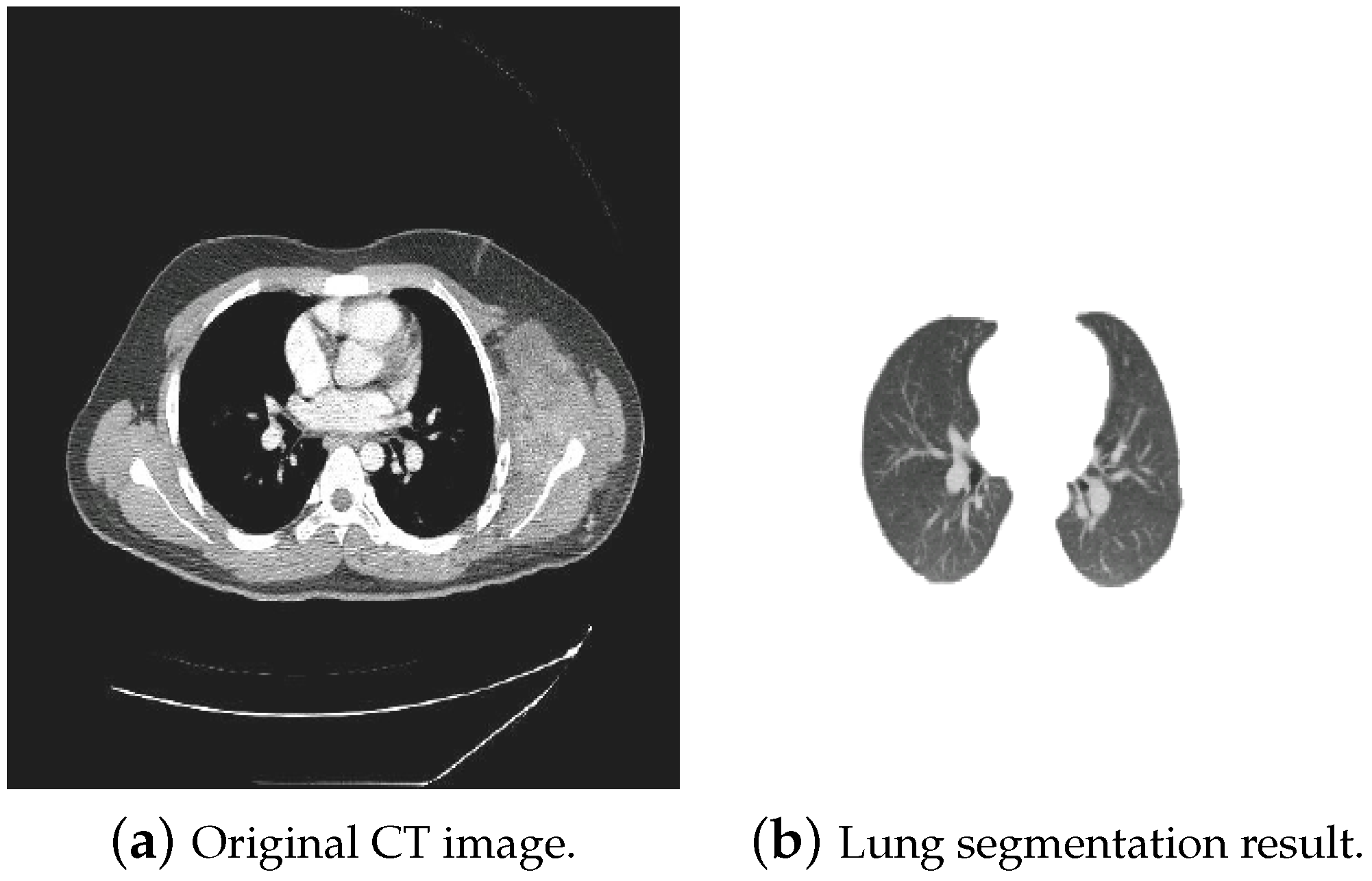
3. CT Images’ Preprocessing Using CNN
- Collections: NSCLC-radiomics
- Image modality: CT
- Anatomical site: lung
4. Lung Images’ Segmentation and Registration
4.1. Segmentation Process
4.1.1. Thresholding-Based Segmentation
4.1.2. Region-Based Segmentation
4.2. Registration Process
- Intensity based—registration relies on statistical criteria for matching different intensities between fixed and moving images.
- Feature based—registration is based on different geometrical structures, i.e., points or curves, very useful while registering pathological lungs.
- Segmentation based—in rigid registration, the same structures from both images are extracted; after that, they become the input parameters for the main registration method; unfortunately, registration accuracy is connected with the segmentation quality.
- Mass-preserving—registration relies on detecting density changes on CT images, which are related to different inhalation volumes (air volume in lungs).
5. System Implementation
5.1. Segmentation
5.1.1. Watershed Segmentation
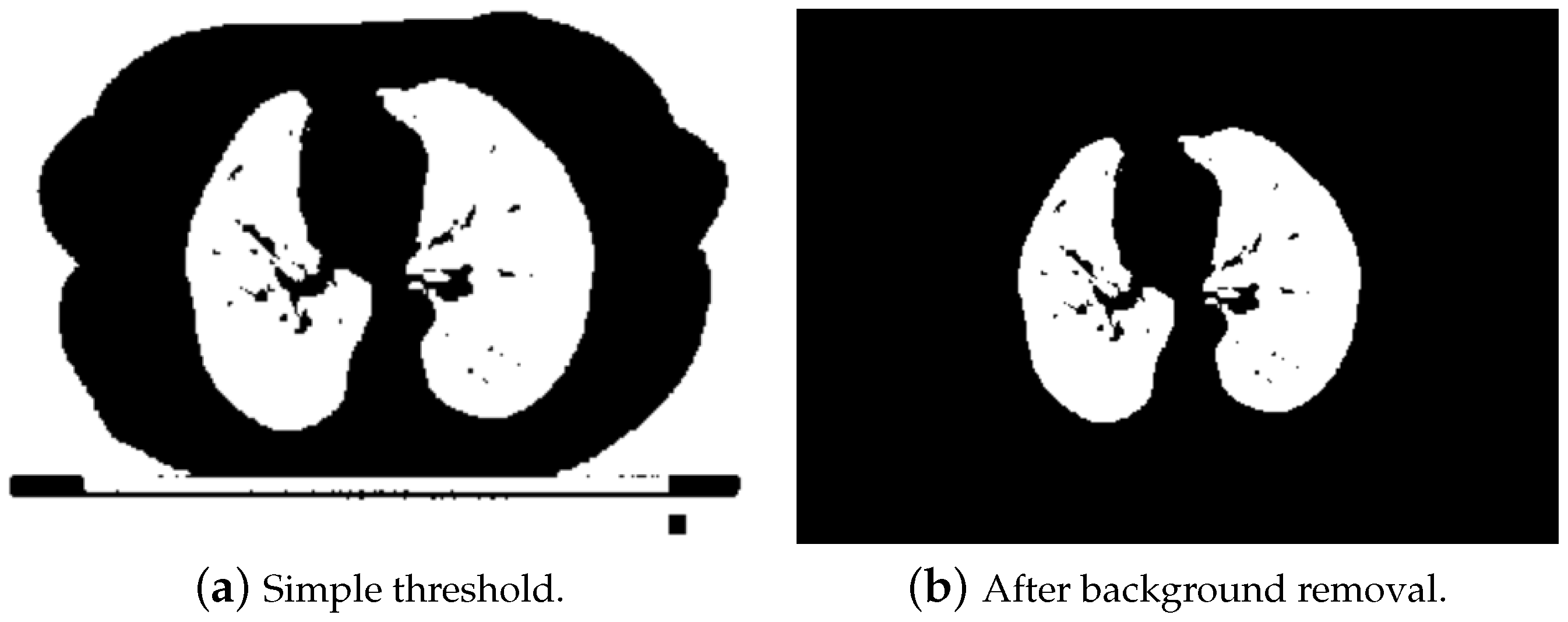
- The first step was aimed at finding internal labels. The threshold filter at the level of −360 HU was applied in order to distinguish lungs. Obviously, as presented in Figure 7a, after thresholding, we obtained not only lungs, but also other elements, e.g., the background, pixel intensity of which was at the level of −1000 HU. In order to remove the background, a clear_border function from the skiimage.segmentation module was used. Thanks to that, only lung pixels were left, as shown in Figure 7b.Afterwards a labeling function from the scikit.measure package was used, in order to mark pixels that belonged to the same groups with unique labels. Unfortunately, this solution worked well only for the slices from the middle part of the series. The first ones contained organs like trachea or other significant airways, which were also detected using the threshold (due to the air inside them; a similar Hounsfield value), as shown in Figure 8. In further steps, these areas could be mistakenly treated as internal markers.To solve this problem for the first 9% of the slices (which very likely contained trachea), additional steps were implemented in order to properly detect only lungs.
- All regions with areas smaller than 0.00012 of the whole image size were removed. This was necessary in order to get rid of noise and very small insignificant objects, but at the same time, this value cannot be too high, because it would reject regions with small sections of lungs (beginning slices). As a result of this step, we were able obtain two or three segmented regions.
- If only two regions were left, that meant that one of them was lung, and the other was trachea. We could assume that due to the fact that in all images containing lungs, lungs were bigger than trachea. We found the minimum y value of pixels in both areas (located at the top of each area) and removed this region, which had the smaller value (trachea is above lungs on the first slices). This analysis is illustrated in Figure 9.
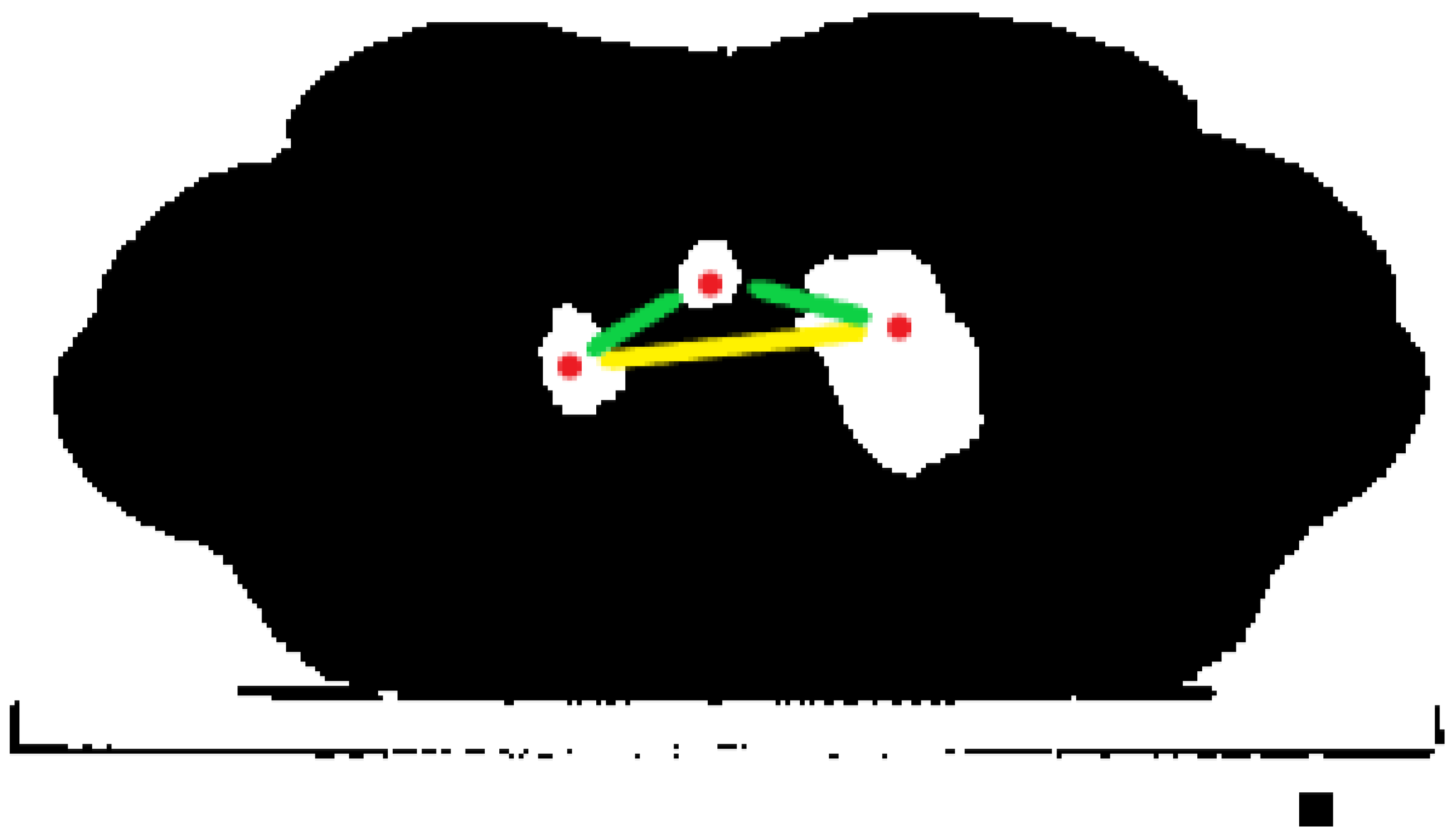
- If in one of the first slices, three regions were detected, that meant that two of them were lungs, and one was trachea. Unfortunately, we could not simply preserve the two largest ones, because we were not able to ensure that lungs were bigger than trachea (usually, it is the opposite). That is why for this purpose, we detected the centroids of each region (they had x and y coordinates). Next, we made a list of all possible pairs of centroids and calculated the Euclidean distance between each pair. As a result, we obtained a list of three distances, sorted them, and kept the two regions between which the distance was largest. A simple visualization is shown in Figure 10.
- The next step was finding external labels. This was done by dilating the internal marker, creating two temporary matrices, and taking regions where they were different from each other (XOR operation). By changing the dilation iterations, we were able to control the distance between those two markers. The external marker looked like a wide border surrounding the internal one. The border had to be wide enough to cover all minima, which could be located in the neighborhood. The final watershed marker containing internal and external ones is presented in Figure 11.
- Afterwards, the Sobel filter along both axes (x and y) was applied in order to detect edges on the input image (Figure 11).
- The watershed algorithm was executed with the sobel_gradient and watershed_ markers as inputs. In order to find the border of the result of the watershed algorithm, morphological_gradient was applied (with the difference between the dilation and erosion of the watershed as the input). In order to re-include significant nodules near the border, a black-hat operation was applied. Thanks to that, the areas that might carry significant information about the treatment were still in the segmented lungs. We decided that it was better to include an area that normally would not be treated as lung in the segmentation process. If we wanted to omit it during the calculations, we would just contour it with an ROI and exclude it from the calculation area. Removing it during segmentation would lead to a loss of the significant information that it carried.
- Finally, binary opening, closing, and then, small erosion operations were applied in order to remove noise and fill the holes, which were inside lungs after thresholding. Examples of segmented lungs are presented in Figure 12.
5.1.2. Segmentation Using the SITK Library
- In order to denoise and smooth the image, we used a CurvatureFlow image filter. Smoothed images are better for further processing.
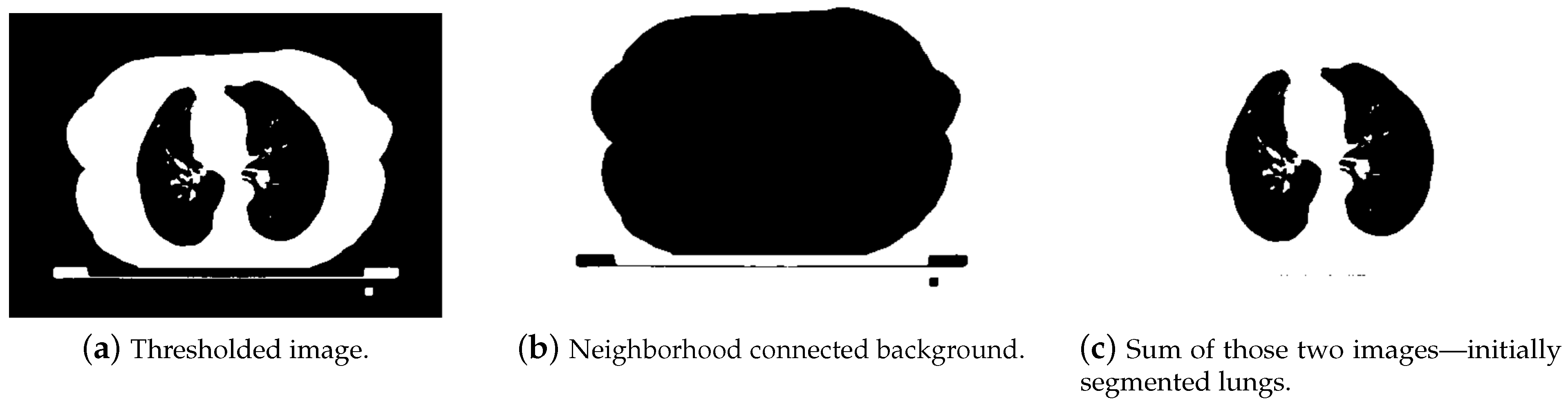
- In order to separate lungs from the background and other organs, a threshold value at −280 HU was set. It does not strictly correspond to the theoretical Hounsfield range of lungs (form −400 to −600 HU), but the acquired images came from different modalities, so we wanted to cover all lung areas, with pixels in a close neighborhood.
- The next challenge was to remove the background. We decided to use a NeighborhoodConneted function. It labeled pixels connected to an initially selected seed and checked if all pixel neighbors were within the threshold range. The initial seed was chosen as point (10, 10), which was located near the upper left image corner. Pixels with a value equal to zero in the thresholded image (background) were set to one. In the thresholded image, pixels containing body parts other than lungs were also set to one. Adding a thresholded image to the neighborhood connected one gave us a result with segmented lungs and some noise. In Figure 13, the first segmentation steps are depicted.
- After detecting lungs, a ConnectedComponents filter was applied to label the object on the input binary image. Pixels equal to zero were labeled as background, whereas those equal to one were treated as objects. Different groups of connected pixels were labeled with unique labels.
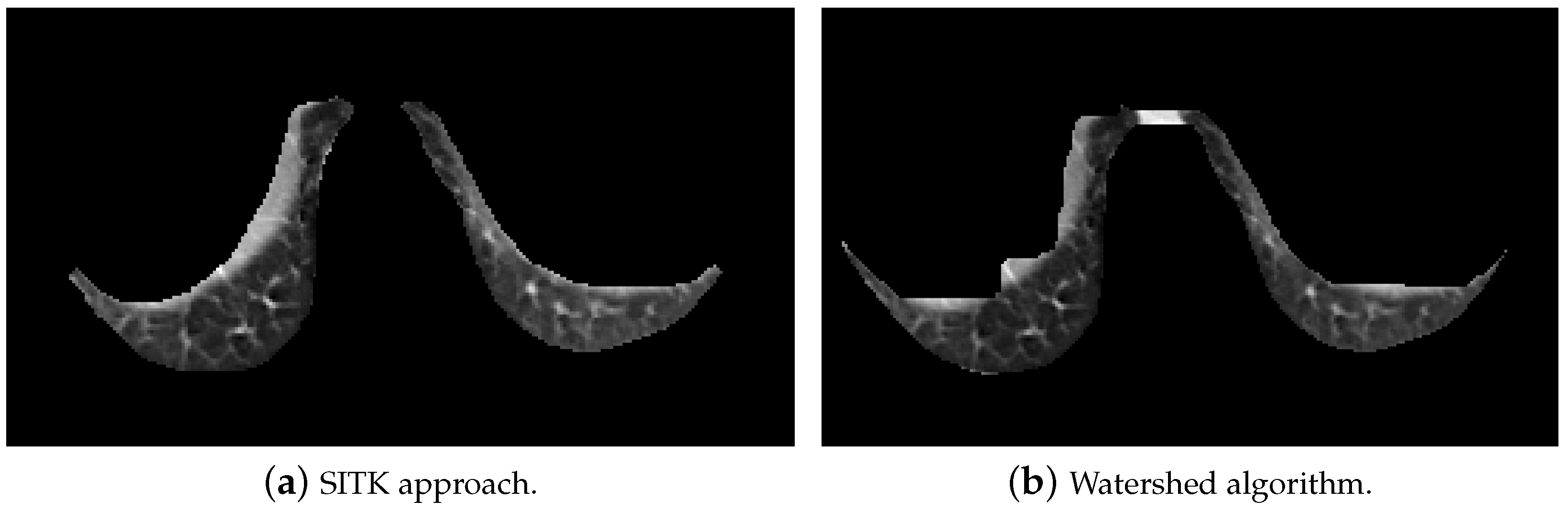
- In the next step, the area of detected objects was calculated, and the largest one or two, whose size was larger than 0.002 of the whole image, were chosen. Due to this condition, right lung in the third slice was not detected by this algorithm, but it was in the watershed. In order to fix this issue, additional restrictions for the extreme slices should be added.
- On such a segmented lung binary image, opening and closing were applied. Binary opening was responsible for removing small structures (smaller than the radius) from the image. It consisted of two functions executed on the output of each other: Dilatation(Erosion(image)). On the other hand, binary closing removed small holes present in the image.
- Sometimes, after applying those filters, still, some large holes were left inside the images. Usually, they should stay there, because the area did not match the conditions. However, in our lung segmentation approach, there should not be any holes left inside the lung area. In order to satisfy this requirement, a VotingBinaryHoleFilling filter was applied. It also filled in holes and other cavities. The voting operation on each pixel was applied. If the result was positive, it was set to one. We chose a large radius for this operation (16) in order to ensure that no holes would be left inside.
- Finally, we applied a small erosion filter with the radius set to one. We excluded the lung border from the segmented image. Tissue located in that area was irrelevant for the analysis and could even falsify the results.
5.2. Registration
5.2.1. Affine Slice Registration
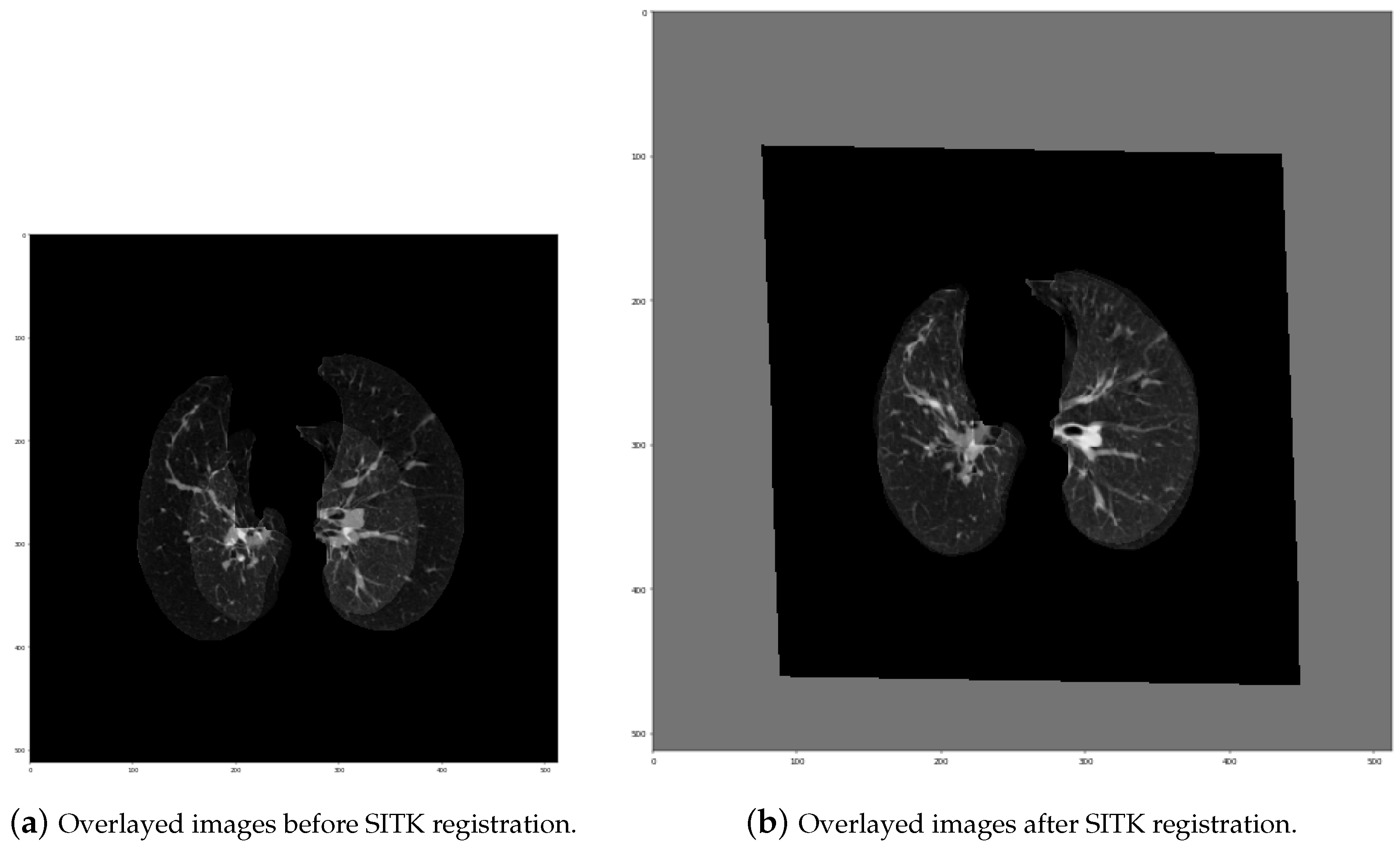
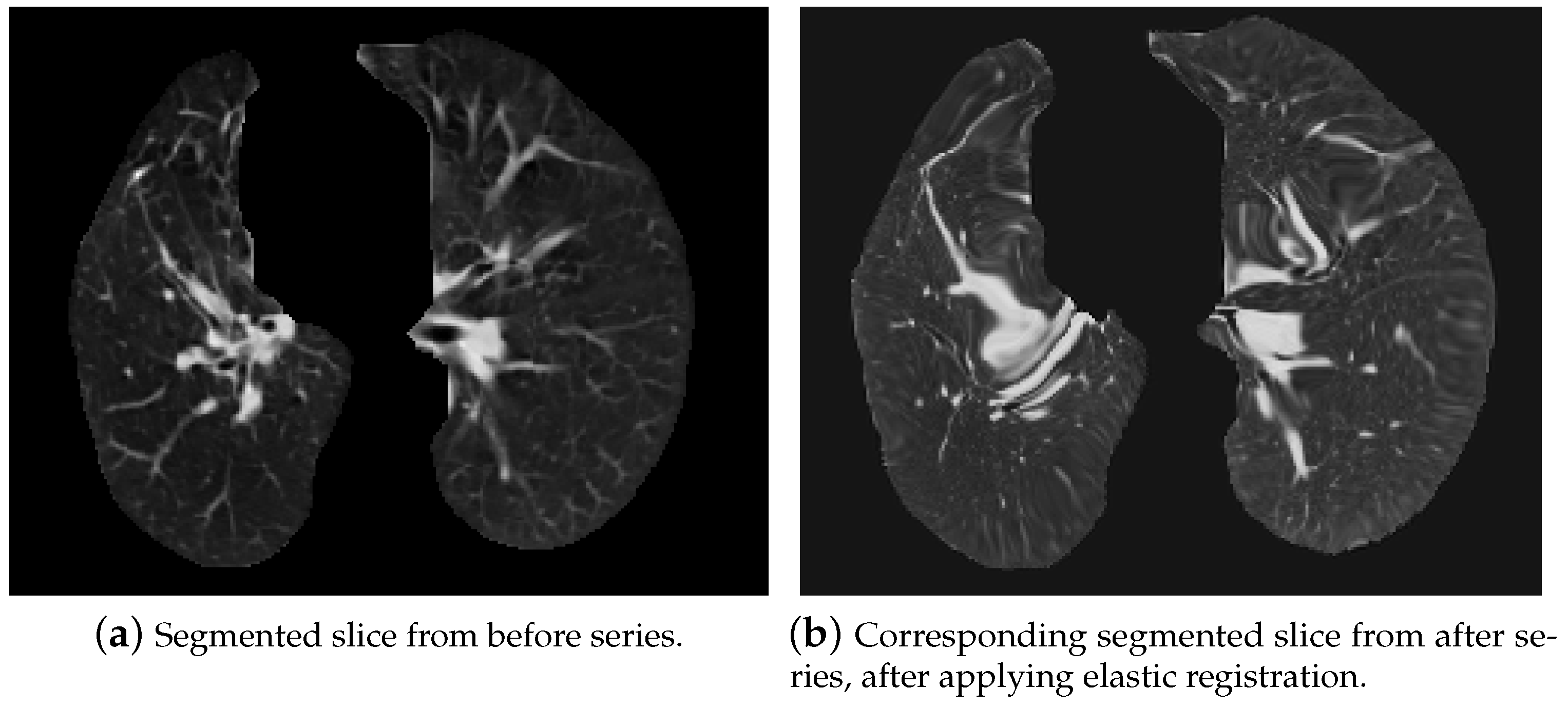
- Initial transformation: In this step, we used CenteredTransformInitializer in order to align the centers of both slices. At the same time, the AffineTransform transformation type was chosen. Transformations like Euler2DTransform would not be proper in this case, because both series were acquired by different devices, which means they had various scales. Euler transformation is specified as rotation and translation, but without scaling. As seen in Figure 15, lungs on the “before” series were much smaller those on the “after” series, so scaling was one of the most significant needed transformations.
- Measure metric: We used MattesMutualInformation. The metric sampling strategy was set to REGULAR, and the metric sampling percent was set to 100%. That means that to calculate the current measure, all pixels were taken into consideration. Due to the fact that lung segmentation was performed before registration, we could not compute the metric using a small percent of random points, as was proposed in [31]. If only black points (outside the lungs) had been randomly chosen from both images, then the cost function would have been very low, but it would not mean that the lungs were properly registered.
- Interpolator: We set it as Linear. In most cases, linear interpolation is the default setting. It gives a weighted average of surrounding voxels with the usage of distances as weights.
- Optimizer: The gradient descent optimizer was chosen for affine registration. It has many parameters to be set, and we used the following:
- learningRate = 1,
- numberOfIterations = 255.
- Multi-resolution framework: The last set parameters were associated with the multi-resolution framework, which the SITK library provides. Due to this strategy, we were able to improve the robustness and the capture range of the registration process. The input image was firstly significantly smoothed and had a low resolution. Then, during the next iterations, it became less smoothed, and the resolution increased. This additional multi-resolution utility was realized with two functions: SetShrinkFactorsPerLevel (shrink factors applied at the change of level) and SetSmoothingSigmasPerLevel (smoothing sigmas applied also at the change of level—in voxels or physical units). In affine registration, where the images differed significantly from each other, we decided to set four multi-resolution levels. The chosen parameters were as follows:
- shrinkFactors = [6,4,2,1],
- smoothingSigmas = [3,2,1,0].
5.2.2. SimpleITK 3D Affine Registration
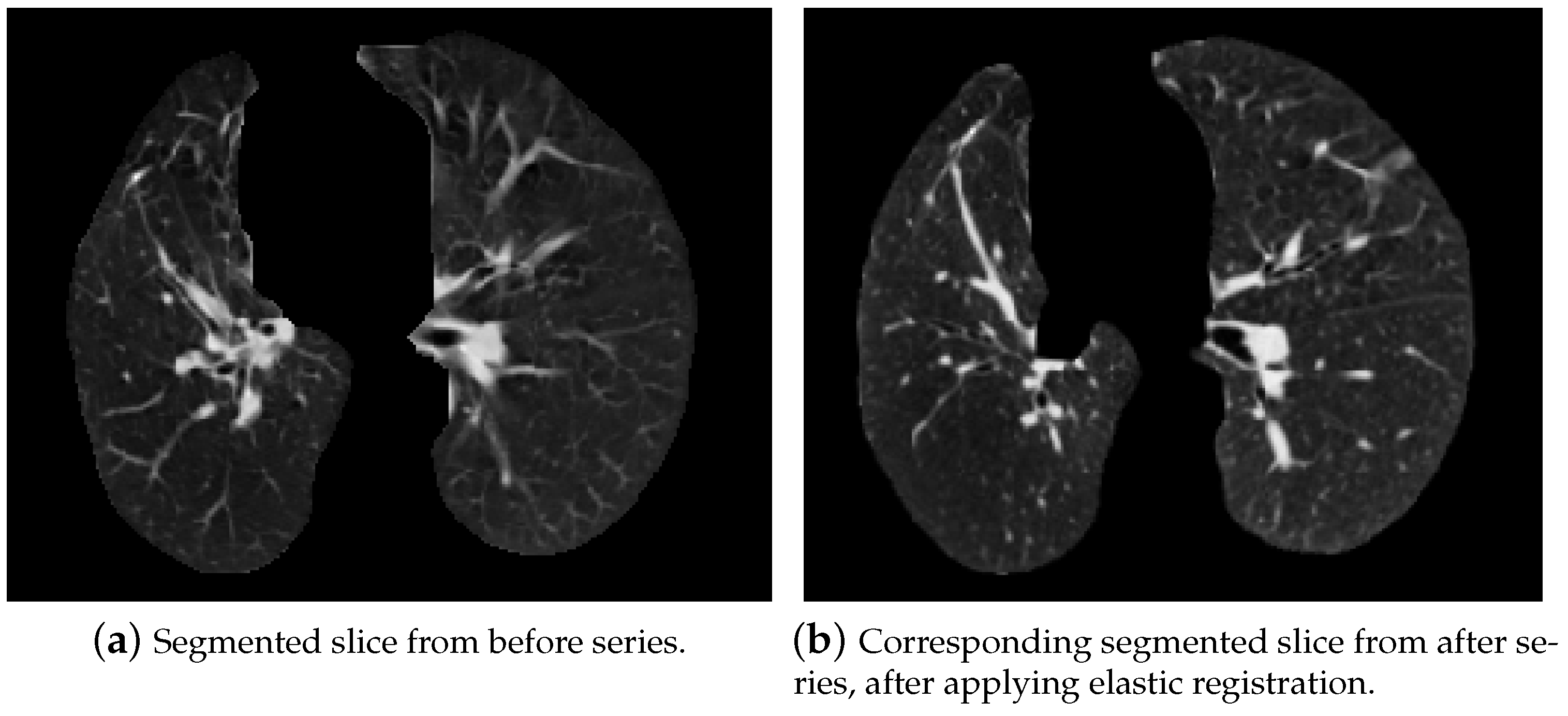
5.2.3. Elastic Lung Registration
- Initial transformation: In this step, we chose the BSplineTransformation type.
- Measure metric: It is analogous to the affine one.
- Interpolator: It was also set as Linear.
- Optimizer: The gradient descent optimizer was also chosen for elastic registration; however, its parameters slightly changed:
- learningRate = 1,
- numberOfIterations = 255.
- Multi-resolution framework: The parameters of the multi-resolution framework were also changed:
- shrinkFactors = [2,1],
- smoothingSigmas = [1,0].
5.3. Data Presentation Module
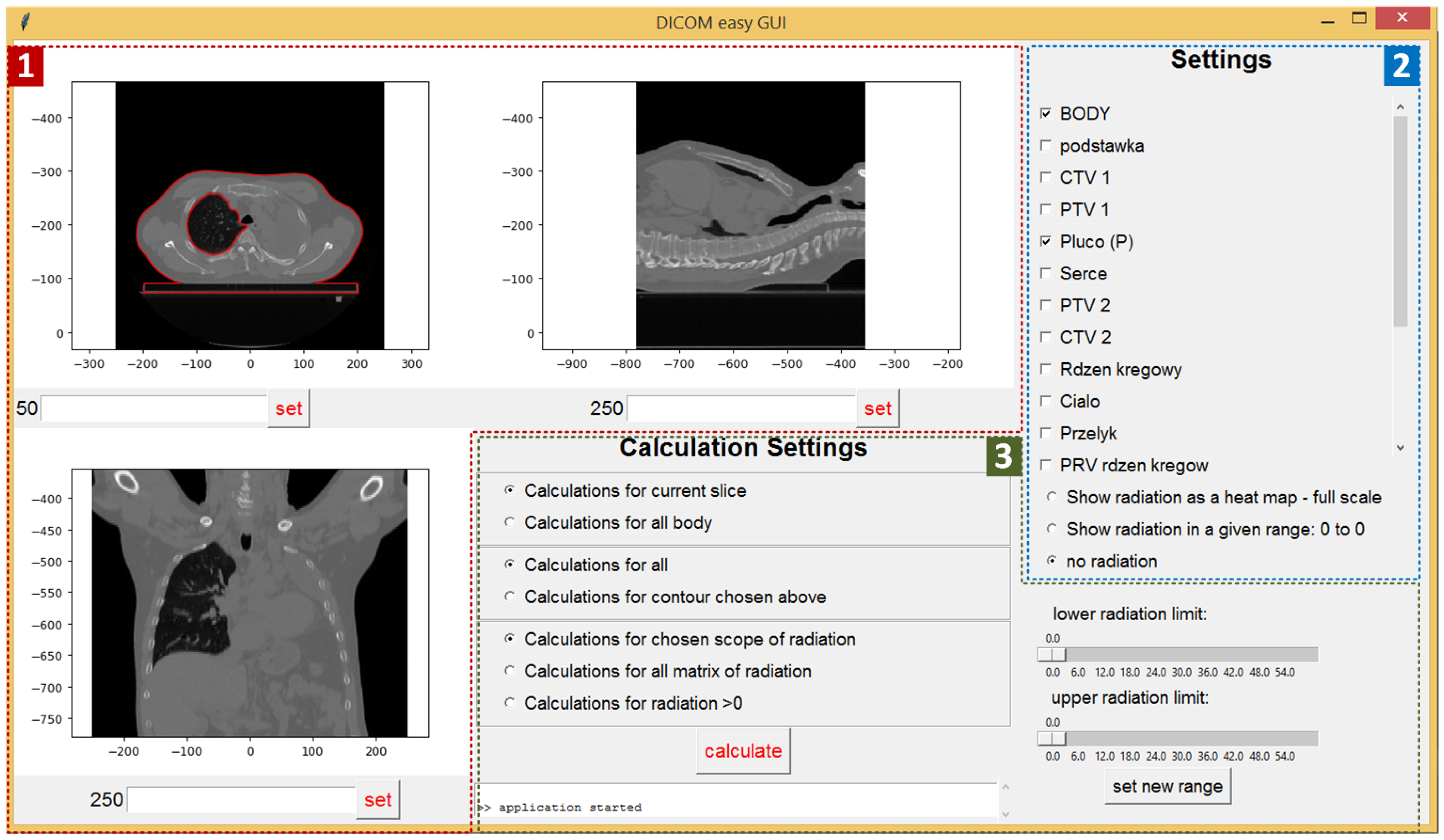
- Current CT slice (displayed for three directions) (red area—1): The slice can be selected by setting its number or by using a mouse scroll wheel. By moving the mouse over the upper left slice, we can read the pointed pixel’s Hounsfield scale value and the dose density value in gray units. The small console at the bottom of the screen is used to display messages to the user (errors, warnings, etc.).
- Input settings (blue area—2): Check boxes allow the selection of all contours loaded from the active files. Radio buttons are used to set the visible range of the radiation dose. There are three possible options:
- Show radiation as a heat map (a “warmer” color corresponds to a higher radiation dose)
- Show radiation in a given range (the range is set by the sliders located below)
- No radiation (the CT slice is displayed without the radiation scheme).
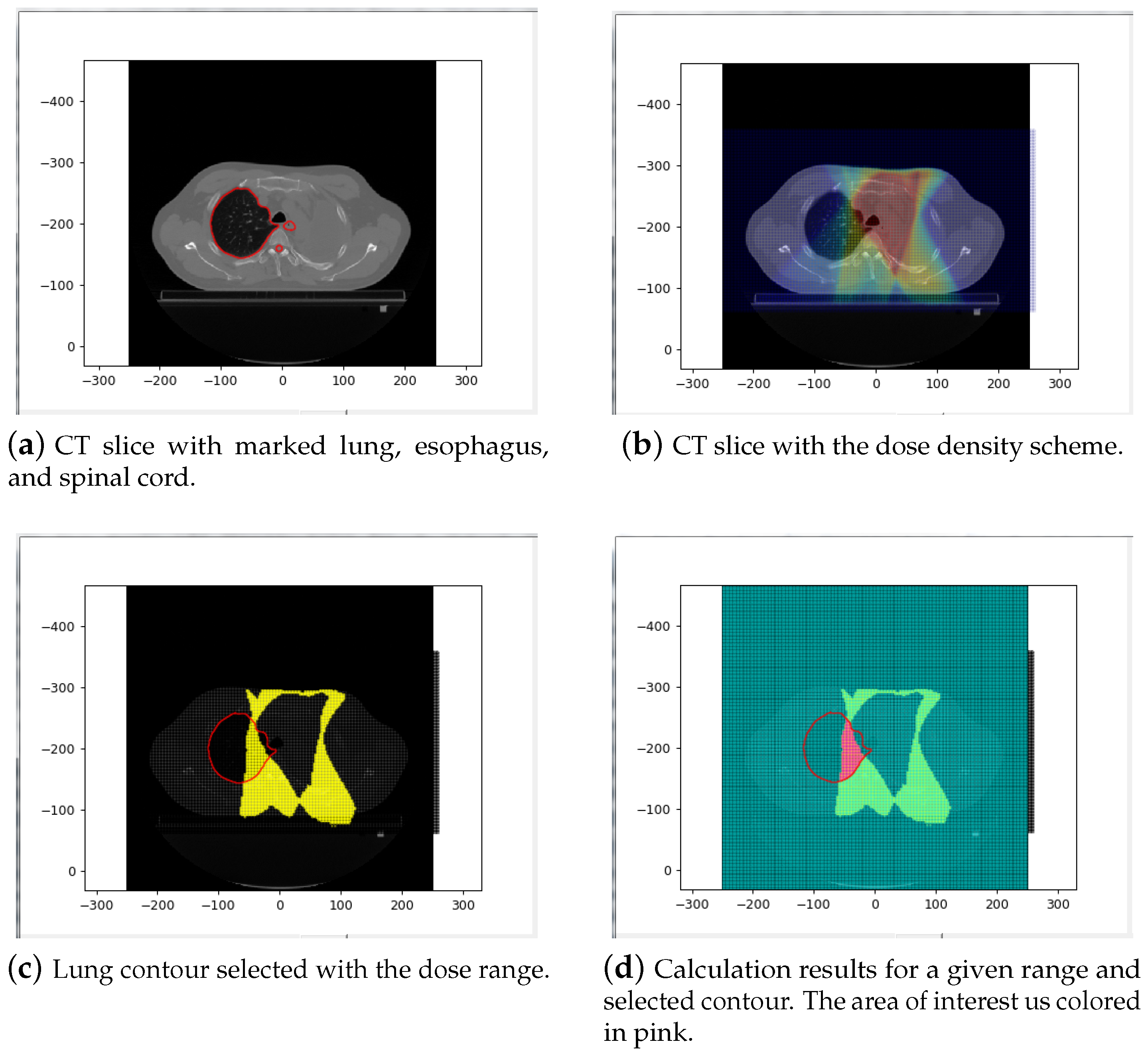
- Calculation parameters (green area—3): The first set of radio buttons specifies whether the results are calculated for the whole body (all slices) or just for the current slice. The second set of radio buttons decides whether the calculations are applied for all pixels or just for pixels within the chosen contour. The last three radio buttons are used to select the settings related to the radiation. Calculation can be done for the chosen radiation range (the scope is set by sliders), for all the matrix of radiation (for all pixels included in the radiation area), or for pixels where the radiation is above zero.
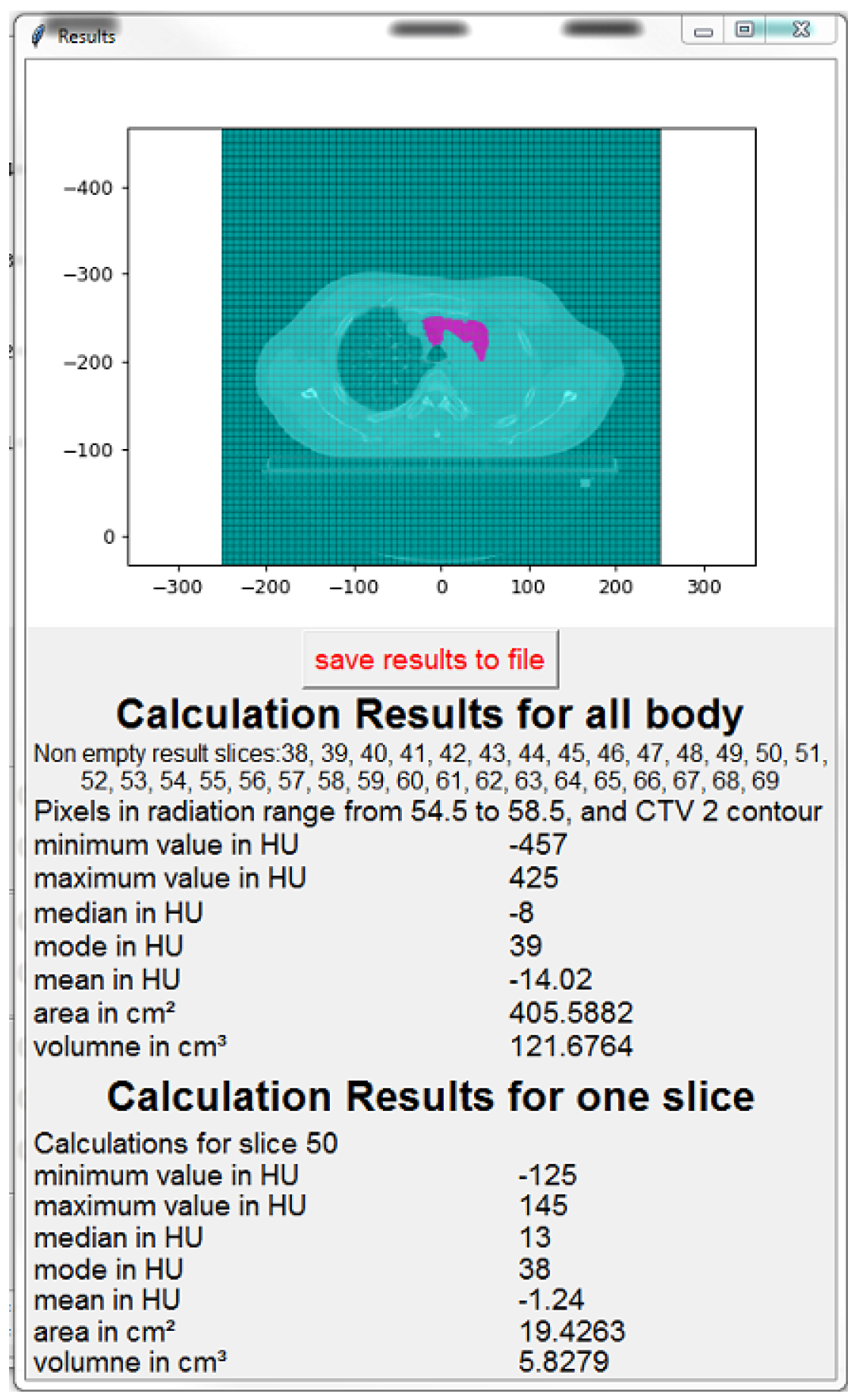
- The image of the current slice with marked pixels selected for calculation,
- The number (indexes) of slices for which the calculation was done,
- The calculation parameters (radiation range, contour type),
- Min, max, median, mode, and average values in HU,
- The volume and area of the selected pixels.
6. Conclusions and Future Works
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Ginneken, B.V.; Romeny, B.M.T.H.; Viergever, M.A. Computer-aided diagnosis in chest radiography: A survey. IEEE Trans. Med. Imaging 2001, 20, 1228–1241. [Google Scholar] [CrossRef] [PubMed]
- Wijsman, R.; Dankers, F.; Troost, E.G.; Hoffmann, A.L.; van der Heijden, E.H.; de Geus-Oei, L.F.; Bussink, J. Comparison of toxicity and outcome in advanced stage non-small cell lung cancer patients treated with intensity-modulated (chemo-) radiotherapy using IMRT or VMAT. Radiother. Oncol. 2017, 122, 295–299. [Google Scholar] [CrossRef]
- Wennberg, B.; Gagliardi, G.; Sundbom, L.; Svane, G.; Lind, P. Early response of lung in breast cancer irradiation: Radiologic density changes measured by CT and symptomatic radiation pneumonitis. Int. J. Radiat. Oncol. Biol. Phys. 2002, 52, 1196–1206. [Google Scholar] [CrossRef]
- Käsmann, L.; Dietrich, A.; Staab-Weijnitz, C.A.; Manapov, F.; Behr, J.; Rimner, A.; Jeremic, B.; Senan, S.; De Ruysscher, D.; Lauber, K.; et al. Radiation-induced lung toxicity–cellular and molecular mechanisms of pathogenesis, management, and literature review. Radiat. Oncol. 2020, 15, 214. [Google Scholar] [CrossRef] [PubMed]
- Warrick, J.H.; Bhalla, M.; Schabel, S.I.; Silver, R.M. High resolution computed tomography in early scleroderma lung disease. J. Rheumatol. 1991, 18, 1520–1528. [Google Scholar] [PubMed]
- Bernchou, U.; Schytte, T.; Bertelsen, A.; Bentzen, S.M.; Hansen, O.; Brink, C. Time evolution of regional CT density changes in normal lung after IMRT for NSCLC. Radiother. Oncol. 2013, 109, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, M.; Kaiser, M.S.; Hussain, A.; Vassanelli, S. Applications of Deep Learning and Reinforcement Learning to Biological Data. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 2063–2079. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Zhao, L.; Luo, X.; Dou, X. Using deep learning for classification of lung nodules on computed tomography images. J. Healthc. Eng. 2017, 2017, 8314740. [Google Scholar] [CrossRef] [PubMed]
- González, G.; Ash, S.Y.; Vegas-Sánchez-Ferrero, G.; Onieva Onieva, J.; Rahaghi, F.N.; Ross, J.C.; Díaz, A.; San José Estépar, R.; Washko, G.R. Disease Staging and Prognosis in Smokers Using Deep Learning in Chest Computed Tomography. Am. J. Respir. Crit. Care Med. 2018, 197, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Kovács, A.; Palásti, P.; Veréb, D.; Bozsik, B.; Palkó, A.; Kincses, Z.T. The sensitivity and specificity of chest CT in the diagnosis of COVID-19. Eur. Radiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Li, Z.; Zhou, J.; Li, H.; Chen, Y.; Huang, Y.; Xie, D.; Zhao, L.; Fan, M.; Hashmi, S.; et al. A Rapid, Accurate and Machine-Agnostic Segmentation and Quantification Method for CT-Based COVID-19 Diagnosis. IEEE Trans. Med. Imaging 2020, 39, 2638–2652. [Google Scholar] [CrossRef]
- Luo, N.; Zhang, H.; Zhou, Y.; Kong, Z.; Sun, W.; Huang, N.; Zhang, A. Utility of chest CT in diagnosis of COVID-19 pneumonia. Diagn. Interv. Radiol. 2020, 26, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Zuo, H. Contribution of CT Features in the Diagnosis of COVID-19. Can. Respir. J. 2020, 2020, 1237418. [Google Scholar] [CrossRef]
- Shah, V.; Keniya, R.; Shridharani, A.; Punjabi, M.; Shah, J.; Mehendale, N. Diagnosis of COVID-19 using CT scan images and deep learning techniques. Radiol. Imaging 2020. [Google Scholar] [CrossRef]
- Cancer Image Archive Website. Available online: https://www.cancerimagingarchive.net/ (accessed on 10 September 2020).
- Mansoor, A.; Bagci, U.; Foster, B.; Xu, Z.; Papadakis, G.Z.; Folio, L.R.; Udupa, J.K.; Mollura, D. Segmentation and Image Analysis of Abnormal Lungs at CT: Current Approaches, Challenges, and Future Trends. Radiographics 2015, 35, 1056–1076. [Google Scholar] [CrossRef] [PubMed]
- Anjna, E.A.; Kaur, E. Review of Image Segmentation Technique. Int. J. Adv. Res. Comput. Sci. 2017, 8. [Google Scholar] [CrossRef]
- Yuheng, S.; Hao, Y. Image Segmentation Algorithms Overview. arXiv 2017, arXiv:1707.02051. [Google Scholar]
- AucklandImageProcessing. Available online: https://www.cs.auckland.ac.nz/courses/compsci773s1c/lectures/ImageProcessing-html/topic3.htm (accessed on 22 May 2019).
- Kaur, D.; Kaur, Y. Various Image Segmentation Techniques: A Review. Int. J. Comput. Sci. Mob. Comput. 2014, 3, 809–814. [Google Scholar]
- Bala, A.; Kumar Sharma, A. Split and Merge: A Region Based Image Segmentation. Int. J. Emerg. Res. Manag. Technol. 2018, 6, 306. [Google Scholar] [CrossRef]
- Rusu, A. Segmentation of Bone Structures in Magnetic Resonance Images (MRI) for Human Hand Skeletal Kinematics Modelling. Ph.D. Thesis, Institute of Robotics and Mechatronics, German Aerospace Center (DLR), Oberpfaffenhofen, Wessling, Germany, 2012. [Google Scholar]
- Yellow Watershed Marked. Available online: http://www.cmm.mines-paristech.fr/~beucher/wtshed.html (accessed on 22 May 2019).
- Shojaii, R.; Alirezaie, J.; Babyn, P. Automatic lung segmentation in CT images using watershed transform. In Proceedings of the IEEE International Conference on Image Processing 2005, Genova, Italy, 14 September 2005; Volume 2, p. II-1270. [Google Scholar] [CrossRef]
- Elsayed, O.; Mahar, K.; Kholief, M.; Khater, H.A. Automatic detection of the pulmonary nodules from CT images. In Proceedings of the 2015 SAI Intelligent Systems Conference (IntelliSys), London, UK, 10–11 November 2015; pp. 742–746. [Google Scholar] [CrossRef]
- Mistry, D.; Banerjee, A. Review: Image Registration. Int. J. Graph. Image Process. 2012, II, 18–22. [Google Scholar]
- Antoine Maintz, J.B.; Viergever, M.A. An Overview of Medical Image Registration Methods. Utrecht University Repository. 1998. Available online: http://dspace.library.uu.nl/handle/1874/18921 (accessed on 16 June 2019).
- Cavoretto, R.; De Rossi, A.; Freda, R.; Qiao, H.; Venturino, E. Numerical Methods for Pulmonary Image Registration. arXiv 2017, arXiv:1705.06147. [Google Scholar]
- Lis, A. System Wspomagania Oceny Wpływu Rozkładu Dawki w Radioterapii Fotonowej na Późne Zmiany w Obrazie Radiologicznym Płuc. Master’s Thesis, Poznan University of Technology, Poznań, Poland, 2018. [Google Scholar]
- SimpleITK Registration Documentation. Available online: https://simpleitk.readthedocs.io/en/master/Documentation/docs/source/registrationOverview.html/ (accessed on 16 June 2019).
- SimpleITK Registration Introduction. Available online: http://insightsoftwareconsortium.github.io/SimpleITK-Notebooks/Python_html/60_Registration_Introduction.html (accessed on 16 June 2019).
- Wilson, J.D.; Hammond, E.M.; Higgins, G.S.; Petersson, K. Ultra-High Dose Rate (FLASH) Radiotherapy: Silver Bullet or Fool’s Gold? Front. Oncol. 2020, 9, 1563. [Google Scholar] [CrossRef] [PubMed]
- Favaudon, V.; Caplier, L.; Monceau, V.; Pouzoulet, F.; Sayarath, M.; Fouillade, C.; Poupon, M.; Brito, I.; Hupe, P.; Bourhis, J.; et al. Ultrahigh dose-rate FLASH irradiation increases the differential response between normal and tumor tissue in mice. Sci. Transl. Med. 2014, 6, 245–293. [Google Scholar] [CrossRef] [PubMed]







| Matter | Hounsfield Scale Range |
|---|---|
| Bone | +400 → +1000 |
| Soft tissue | +40 → +80 |
| Water | 0 |
| Fat | −60 → −100 |
| Lung | −400 → −600 |
| Air | −1000 |
| Number of CT Slices | |||
|---|---|---|---|
| Not Lungs | Lungs | Total | |
| Training data | 2409 (50 patients) | 3293 (45 patients) | 5702 |
| Validation data | 567 (10 patients) | 887 (12 patients) | 1454 |
| Testing data | 655 (13 patients) | 946 (13 patients) | 1601 |
| Notation | ||
| Real: Lung | False negative (FN) | True positive (TP) |
| Real: Not lung | True negative (TN) | False positive (FP) |
| Predicted: Not lung | Predicted: Lung | |
| VGG16-2000 | ||
| Real: Lung | 23 | 864 |
| Real: Not lung | 558 | 9 |
| Predicted: Not lung | Predicted: Lung | |
| VGG16-2500 | ||
| Real: Lung | 11 | 876 |
| Real: Not lung | 544 | 23 |
| Predicted: Not lung | Predicted: Lung | |
| VGG19-2000 | ||
| Real: Lung | 27 | 860 |
| Real: Not lung | 555 | 12 |
| Predicted: Not lung | Predicted: Lung | |
| VGG19-2500 | ||
| Real: Lung | 25 | 862 |
| Real: Not lung | 554 | 13 |
| Predicted: Not lung | Predicted: Lung | |
| VGG19-2000 | VGG19-2500 | VGG16-2000 | VGG16-2500 | |
|---|---|---|---|---|
| Accuracy = (TP + TN)/All | 0.9732 | 0.9739 | 0.9780 | 0.9766 |
| Sensitivity = TP/(TP + FN) | 0.9696 | 0.9718 | 0.9741 | 0.9876 |
| Specificity = TN/(TN + FP) | 0.9788 | 0.9771 | 0.9841 | 0.9594 |
| Real: Lung | 33 | 913 | Accuracy | 0.9763 |
| Real: Not lung | 650 | 5 | Sensitivity | 0.9651 |
| Predicted: Not lung | Predicted: Lung | Specificity | 0.9924 |
| Parameter | Value |
|---|---|
| Transformation | “Affine” |
| Final metric value | −0.4329 |
| Total registration time | 2.5 s |
| Raw Images | Segmented Images | |
|---|---|---|
| Transformation | “Affine” | “Affine” |
| Sampler | ‘Random’ | “Regular” |
| Final metric value | −0.72 | −0.201 |
| Total registration time | 59.5 s | 157.4 s |
| Parameter | Value |
|---|---|
| Transformation | “Bspline” |
| Final metric value | −0.3012 |
| Total registration time | 21.0 s |
| Parameter | Value |
|---|---|
| Transformation 1 | “Affine” |
| Transformation 2 | “Bspline” |
| Final metric value | −0.5657 |
| Total registration time | 144.9 s |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Konkol, M.; Śniatała, K.; Śniatała, P.; Wilk, S.; Baczyńska, B.; Milecki, P. Computer Tools to Analyze Lung CT Changes after Radiotherapy. Appl. Sci. 2021, 11, 1582. https://doi.org/10.3390/app11041582
Konkol M, Śniatała K, Śniatała P, Wilk S, Baczyńska B, Milecki P. Computer Tools to Analyze Lung CT Changes after Radiotherapy. Applied Sciences. 2021; 11(4):1582. https://doi.org/10.3390/app11041582
Chicago/Turabian StyleKonkol, Marek, Konrad Śniatała, Paweł Śniatała, Szymon Wilk, Beata Baczyńska, and Piotr Milecki. 2021. "Computer Tools to Analyze Lung CT Changes after Radiotherapy" Applied Sciences 11, no. 4: 1582. https://doi.org/10.3390/app11041582
APA StyleKonkol, M., Śniatała, K., Śniatała, P., Wilk, S., Baczyńska, B., & Milecki, P. (2021). Computer Tools to Analyze Lung CT Changes after Radiotherapy. Applied Sciences, 11(4), 1582. https://doi.org/10.3390/app11041582







