Healthy Knee Kinematic Phenotypes Identification Based on a Clustering Data Analysis
Abstract
:1. Introduction
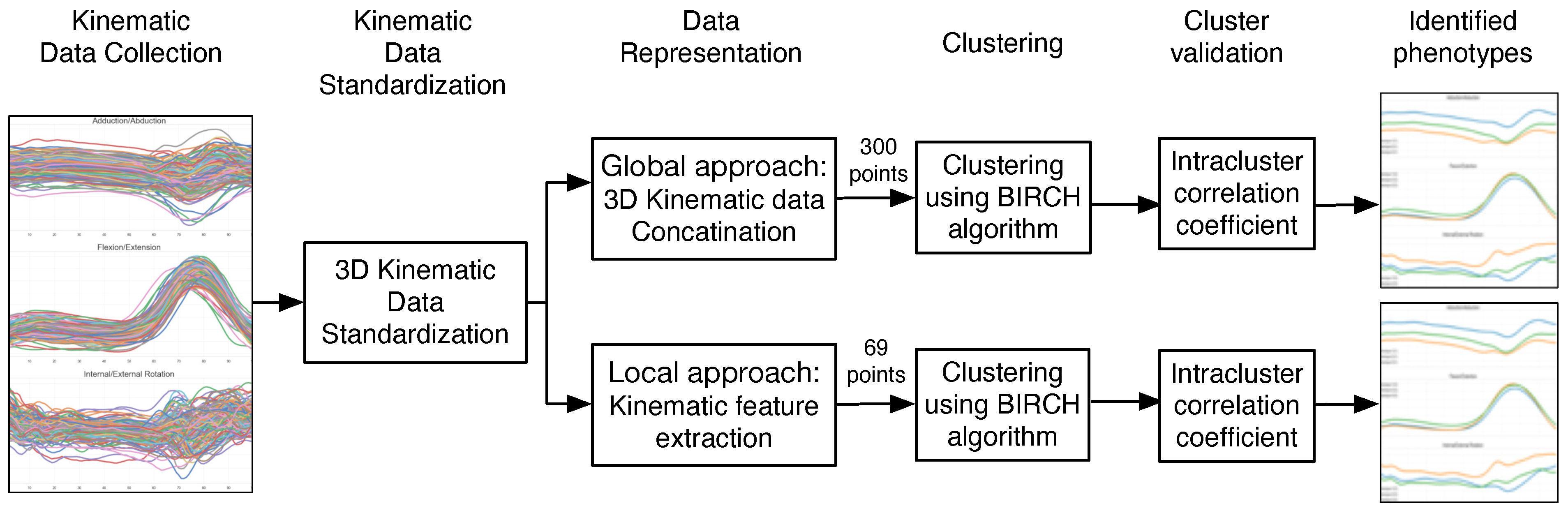
2. Materials and Methods
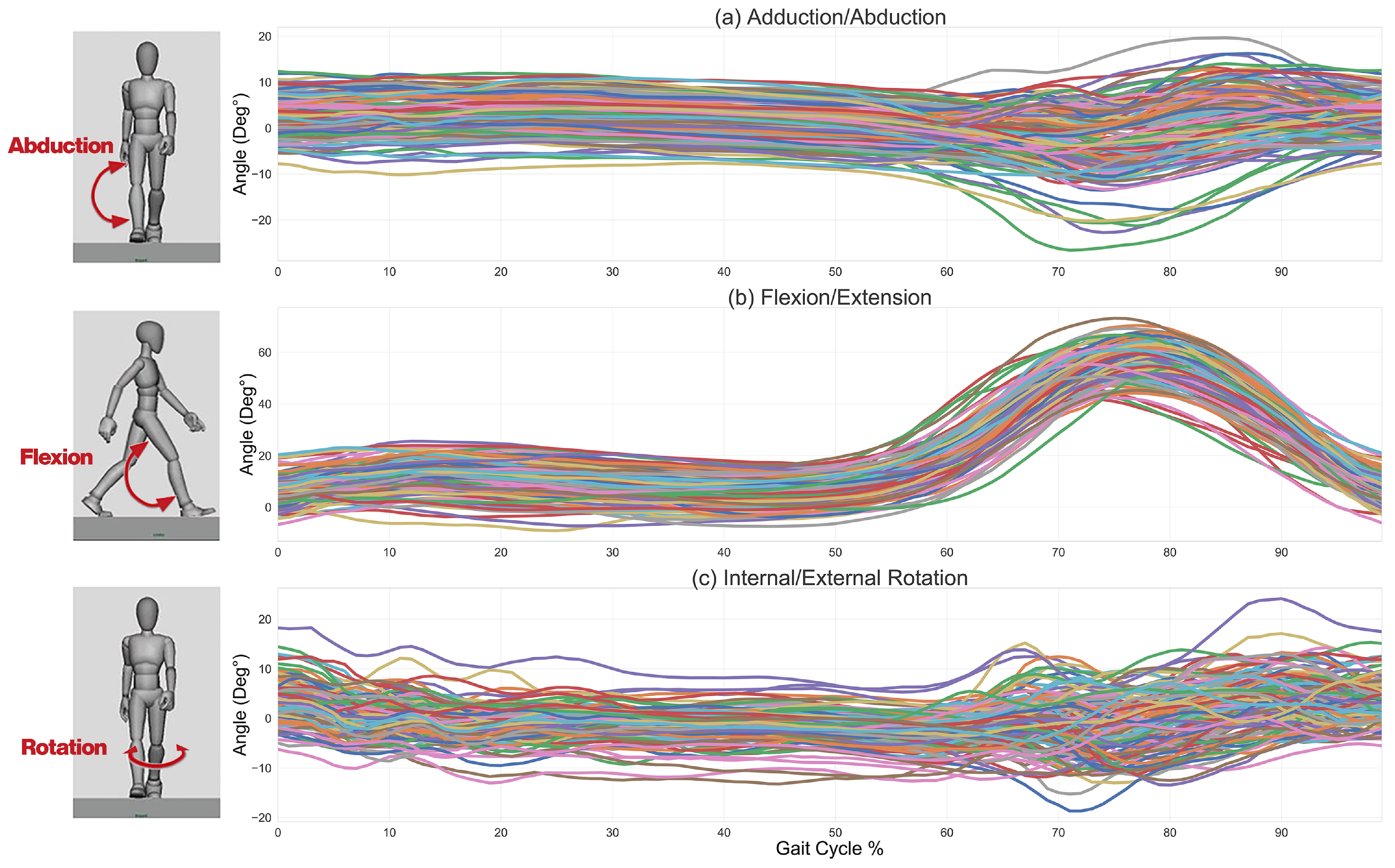
2.1. Kinematic Data Collection
2.2. Kinematic Data Standardization
2.3. Kinematic Data Representation
2.4. Clustering for Phenotype Identification
2.4.1. Clustering Model
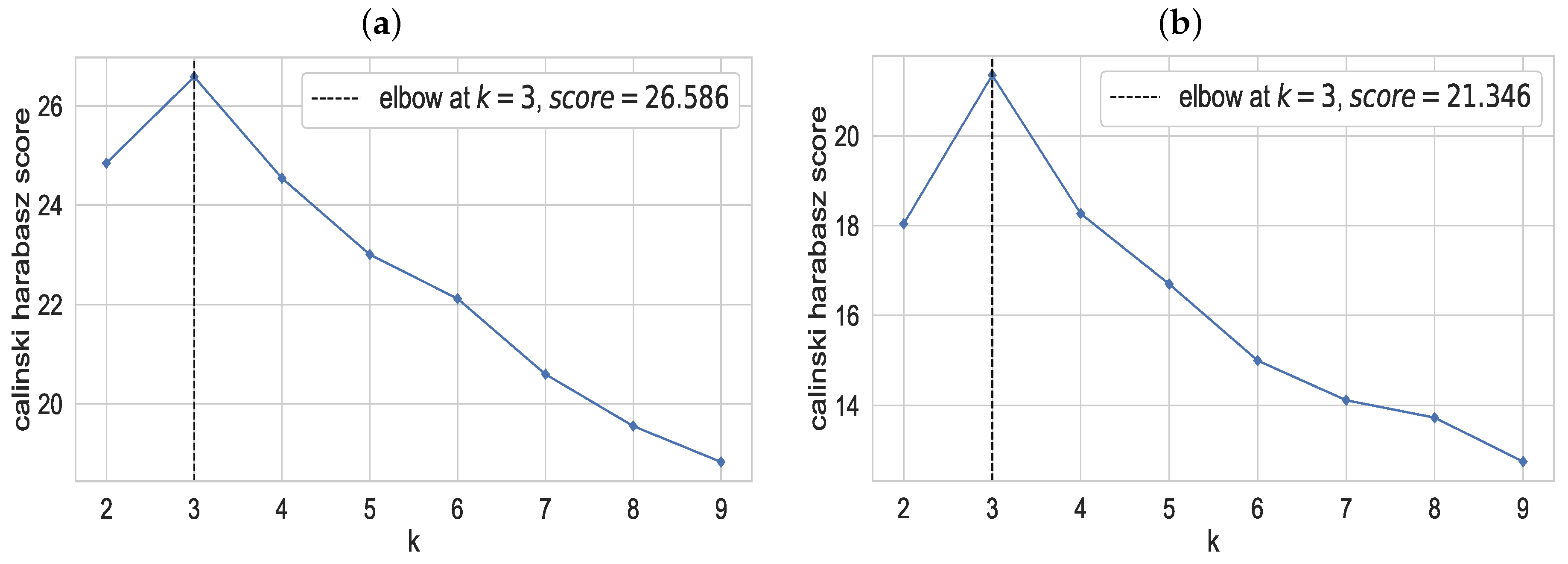
2.4.2. Optimum Number of Clusters
2.4.3. Cluster Evaluation
2.5. Statistical Analysis: Hypothesis Testing and Phenotype Interpretation
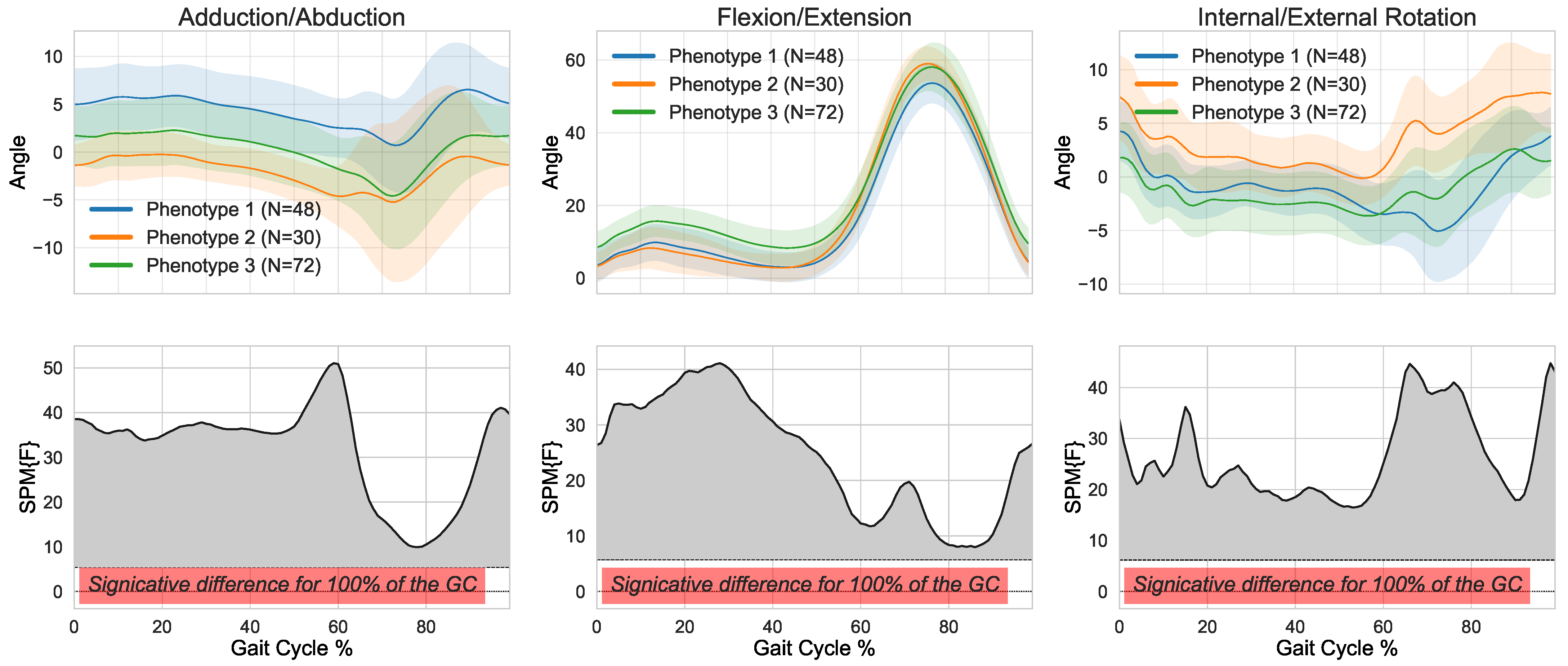
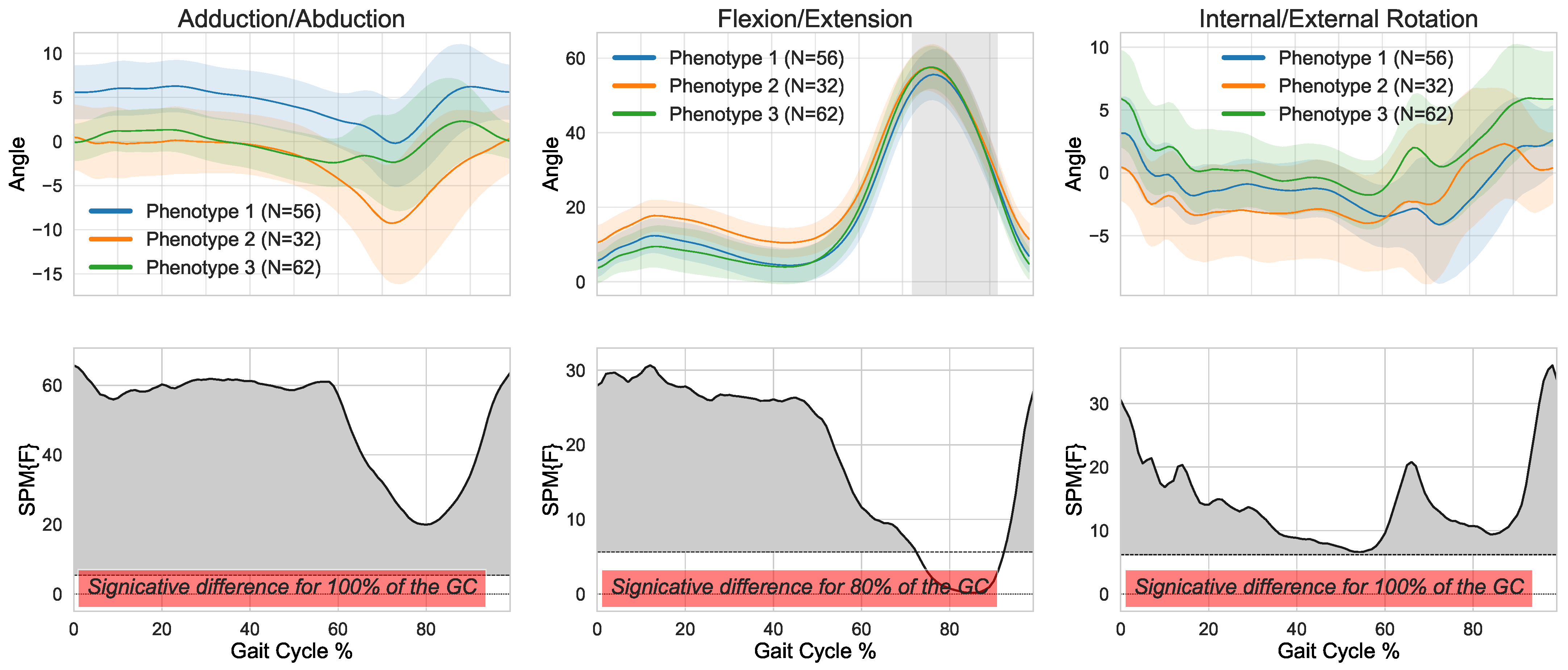
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Phinyomark, A.; Osis, S.; Hettinga, B.A.; Ferber, R. Kinematic gait patterns in healthy runners: A hierarchical cluster analysis. J. Biomech. 2015, 48, 3897–3904. [Google Scholar] [CrossRef] [PubMed]
- Clément, J.; Toliopoulos, P.; Hagemeister, N.; Desmeules, F.; Fuentes, A.; Vendittoli, P.A. Healthy 3D knee kinematics during gait: Differences between women and men, and correlation with X-ray alignment. Gait Posture 2018, 64, 198–204. [Google Scholar] [CrossRef]
- Duda, R.O.; Hart, P.E.; Stork, D.G. Pattern Classification; Wiley-Interscience Publication: New York, NY, USA, 2000. [Google Scholar]
- Phinyomark, A.; Petri, G.; Ibáñez-Marcelo, E.; Osis, S.T.; Ferber, R. Analysis of big data in gait biomechanics: Current trends and future directions. J. Med. Biol. Eng. 2018, 38, 244–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chau, T.; Young, S.; Redekop, S. Managing variability in the summary and comparison of gait data. J. Neuroeng. Rehabil. 2005, 22, 22–42. [Google Scholar] [CrossRef] [Green Version]
- Winter, D. Kinematic and kinetic patterns in human gait: Variability and compensating effects. Hum. Mov. Sci. 1984, 3, 51–76. [Google Scholar] [CrossRef]
- Lafortune, M.A.; Cavanagh, P.; Sommer, H.J.; Kalenak, A. Three-dimensional kinematics of the human knee during walking. J. Biomech. 1992, 25, 347–357. [Google Scholar] [CrossRef]
- Toliopoulos, P. Knee Gait Kinematics: Describing a Normal Cohort; Montreal University: Montreal, QC, Canada, 2018. [Google Scholar]
- Han, S.; Cheng, G.; Xu, P. Three-dimensional lower extremity kinematics of Chinese during activities of daily living. J. Back Musculoskelet. Rehabil. 2015, 28, 327–334. [Google Scholar] [CrossRef]
- Chehab, E.; Andriacchi, T.; Favre, J. Speed, age, sex, and body mass index provide a rigorous basis for comparing the kinematic and kinetic profiles of the lower extremity during walking. J. Biomech. 2017, 58, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wyss, U.; Costigan, P.; Deluzio, K. An integrated procedure to assess knee-joint kinematics and kinetics during gait using an optoelectric system and standardized X-rays. J. Biomed. Eng. 1993, 15, 392–400. [Google Scholar] [CrossRef]
- Chao, E.; Laughman, R.; Schneider, E.; Stauffer, R. Normative data of knee joint motion and ground reaction forces in adult level walking. J. Biomech. 1983, 16, 219–233. [Google Scholar] [CrossRef]
- Benedetti, M.; Catani, F.; Leardini, A.; Pignotti, E.; Giannini, S. Data management in gait analysis for clinical applications. Clin. Biomech. 1998, 13, 204–215. [Google Scholar] [CrossRef]
- Shiavi, R.; Limbird, T.; Frazer, M.; Stivers, K.; Strauss, A.; Abramovitz, J. Helical motion analysis of the knee II. Kinematics of uninjured and injured knees during walking and pivoting. J. Biomech. 1987, 20, 653–665. [Google Scholar] [CrossRef]
- Frigo, C.; Rabuffetti, M.; Kerrigan, D.; Deming, L.; Pedotti, A. Functionally oriented and clinically feasible quantitative gait analysis method. Med. Biol. Eng. Comput. 1998, 36, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Isacson, J.; Gransberg, L.; Knutsson, E. Three-dimensional electrogoniometric gait recording. J. Biomech. 1986, 19, 627–635. [Google Scholar] [CrossRef]
- Kadaba, M.; Ramakrishnan, H.; Wootten, M.; Gainey, J.; Gorton, G.; Cochran, G. Repeatability of kinematic, kinetic, and electromyographic data in normal adult gait. J. Orthop. Res. 1989, 7, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Benoit, D.; Ramsey, D.; Lamontagne, M.; Xu, L.; Wretenberg, P.; Renstrom, P. In vivo knee kinematics during gait reveals new rotation profiles and smaller translations. Clin. Orthop. Relat. Res. 2007, 454, 81–88. [Google Scholar] [CrossRef]
- Watelain, E.; Barbier, F.; Allard, P.; Thévenon, A.; Angué, J. Gait pattern classification of elderly healthy men on the basis of biomechanical data. Arch. Phys. Med. Rehabil. 2000, 81, 579–586. [Google Scholar] [CrossRef]
- Vardaxis, V.; Allard, P.; Lachance, R.; Duhaime, M. Classification of able-bodied gait using 3-D muscle powers. Hum. Mov. Sci. 1998, 17, 121–136. [Google Scholar] [CrossRef]
- Toro, B.; Nester, C.; Farren, P. Cluster analysis for the extraction of sagittal gait patterns in children with cerebral palsy. Gait Posture 2007, 25, 157–165. [Google Scholar] [CrossRef]
- Xu, G.; Zhang, Y.; Begg, R. Mining Gait Pattern for Clinical Locomotion Diagnosis Based on Clustering Techniques. In Advanced Data Mining and Applications; Springer: Berlin/Heidelberg, Germany, 2006; pp. 296–307. [Google Scholar]
- Mulroy, S.; Gronley, J.; Weiss, W.; Newsam, C.; Perry, J. Use of cluster analysis for gait pattern classification of patients in the early and late recovery phases following stroke. Gait Posture 2003, 18, 114–125. [Google Scholar] [CrossRef]
- O’Byrne, J.; Jenkinson, A.; O’Brien, T. Quantitative analysis and classification of gait patterns in cerebral palsy using a three-dimensional motion analyzer. J. Child Neurol. 1998, 13, 101–108. [Google Scholar] [CrossRef]
- Kienast, G.; Bachmann, D.; Steinwender, G.; Zwick, E.; Saraph, V. Determination of gait patterns in children with cerebral palsy using cluster analysis. Gait Posture 1999, 57. [Google Scholar] [CrossRef]
- Zeng, W.; Ma, L.; Yuan, C.; Liu, F.; Wang, Q.; Wang, Y.; Zhang, Y. Classification of asymptomatic and osteoarthritic knee gait patterns using gait analysis via deterministic learning. Artif. Intell. Rev. 2019, 52, 449–467. [Google Scholar] [CrossRef]
- O’Malley, M.; Abel, M.; Damiano, D.; Vaughan, C. Fuzzy clustering of children with cerebral palsy based on temporal-distance gait parameters. IEEE Trans. Rehabil. Eng. 1997, 5, 300–309. [Google Scholar] [CrossRef]
- Mezghani, N.; Mechmeche, I.; Mitiche, A.; Ouakrim, Y.; de Guise, J.A. An analysis of 3D knee kinematic data complexity in knee osteoarthritis and asymptomatic controls. PLoS ONE 2018, 13, e0202348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lustig, S.; Magnussen, R.; Cheze, L.; Neyret, P. The KneeKG system: A review of the literature. Knee Surg. Sports Traumatol. Arthrosc. 2011, 20, 633–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abid, M.; Mezghani, N.; Mitiche, A. Knee Joint Biomechanical Gait Data Classification for Knee Pathology Assessment: A Literature Review. Appl. Bionics Biomech. 2019, 2019, 7472039. [Google Scholar] [CrossRef] [Green Version]
- Chang, A.H.; Chmiel, J.S.; Moisio, K.C.; Almagor, O.; Zhang, Y.; Cahue, S.; Sharma, L. Varus thrust and knee frontal plane dynamic motion in persons with knee osteoarthritis. Osteoarthr. Cartil. 2013, 21, 1668–1673. [Google Scholar] [CrossRef] [Green Version]
- Sharma, L.; Song, J.; Felson, D.T.; Cahue, S.; Shamiyeh, E.; Dunlop, D.D. The role of knee alignment in disease progression and functional decline in knee osteoarthritis. JAMA 2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deluzio, K.J.; Wyss, U.P.; Costigan, P.A.; Sorbie, C.S. Principal component models of knee kinematics and kinetics: Normal vs. pathological gait patterns. J. Hum. Mov. Sci. 1997, 16, 201–217. [Google Scholar] [CrossRef]
- Shabani, B.; Bytyqi, D.; Lustig, S.; Cheze, L.; Bytyqi, C.; Neyret, P. Gait changes of the ACL-deficient knee 3D kinematic assessment. Knee Surg. Sports Traumatol. Arthrosc. 2014, 23, 3259–3265. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Ramakrishnan, R.; Livny, M. BIRCH: A New Data Clustering Algorithm and its Applications. Data Min. Knowl. Discov. 1997, 1, 141–182. [Google Scholar] [CrossRef]
- Caliński, T.; Harabasz, J. A Dendrite Method for Cluster Analysis. Commun. Stat.-Theory Methods 1974, 3, 1–27. [Google Scholar] [CrossRef]
- Riley, P.O.; Paolini, G.; Della Croce, U.; Paylo, K.W.; Kerrigan, D.C. A kinematic and kinetic comparison of overground and treadmill walking in healthy subjects. Gait Posture 2019, 26, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Shetty, N.; Shirwaikar, R. A Comparative Study: BIRCH and CLIQUE. Int. J. Eng. Res. Technol. 2013, 2, 2019–2023. [Google Scholar]
- Bellemans, J.; Colyn, W.; Vandenneucker, H.; Victor, J. The Chitranjan Ranawat award: Is neutral mechanical alignment normal for all patients? The concept of constitutional varus. Clin. Orthop. Relat. Res. 2012, 470, 45–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Phase 1: Build the CF Tree. Load the data into the memory by building a cluster-feature tree (CF tree, defined below). Optionally, condense this initial CF tree into a smaller CF. Phase 2: Global clustering. Apply an existing clustering algorithm on the leaves of the CF tree. Optionally, refine these clusters. |
| Clusters | Frontal (Adduction/Abduction) | Sagittal (Flexion/Extension) | Transverse (Int./External Rotation) |
|---|---|---|---|
| Cluster 1 | 0.74 | 0.54 | 0.39 |
| Cluster 2 | 0.51 | 0.27 | 0.60 |
| Cluster 3 | 0.76 | 0.34 | 0.60 |
| Characteristics | All Participants | Cluster 1 | Cluster 2 | Cluster 3 |
|---|---|---|---|---|
| Number of knees | 150 | 48 | 30 | 72 |
| Age (years) | ||||
| Height (m) | ||||
| Weight (kg) | ||||
| BMI (kg/m2) * | ||||
| Ratio Men/Women * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mezghani, N.; Soltana, R.; Ouakrim, Y.; Cagnin, A.; Fuentes, A.; Hagemeister, N.; Vendittoli, P.-A. Healthy Knee Kinematic Phenotypes Identification Based on a Clustering Data Analysis. Appl. Sci. 2021, 11, 12054. https://doi.org/10.3390/app112412054
Mezghani N, Soltana R, Ouakrim Y, Cagnin A, Fuentes A, Hagemeister N, Vendittoli P-A. Healthy Knee Kinematic Phenotypes Identification Based on a Clustering Data Analysis. Applied Sciences. 2021; 11(24):12054. https://doi.org/10.3390/app112412054
Chicago/Turabian StyleMezghani, Neila, Rayan Soltana, Youssef Ouakrim, Alix Cagnin, Alexandre Fuentes, Nicola Hagemeister, and Pascal-André Vendittoli. 2021. "Healthy Knee Kinematic Phenotypes Identification Based on a Clustering Data Analysis" Applied Sciences 11, no. 24: 12054. https://doi.org/10.3390/app112412054
APA StyleMezghani, N., Soltana, R., Ouakrim, Y., Cagnin, A., Fuentes, A., Hagemeister, N., & Vendittoli, P.-A. (2021). Healthy Knee Kinematic Phenotypes Identification Based on a Clustering Data Analysis. Applied Sciences, 11(24), 12054. https://doi.org/10.3390/app112412054






