Common Data Model and Database System Development for the Korea Biobank Network
Abstract
:1. Introduction
2. Motivation
3. Related Work
3.1. Related Concepts
3.2. Related Research
4. Proposed System
4.1. KBN-CDM Development
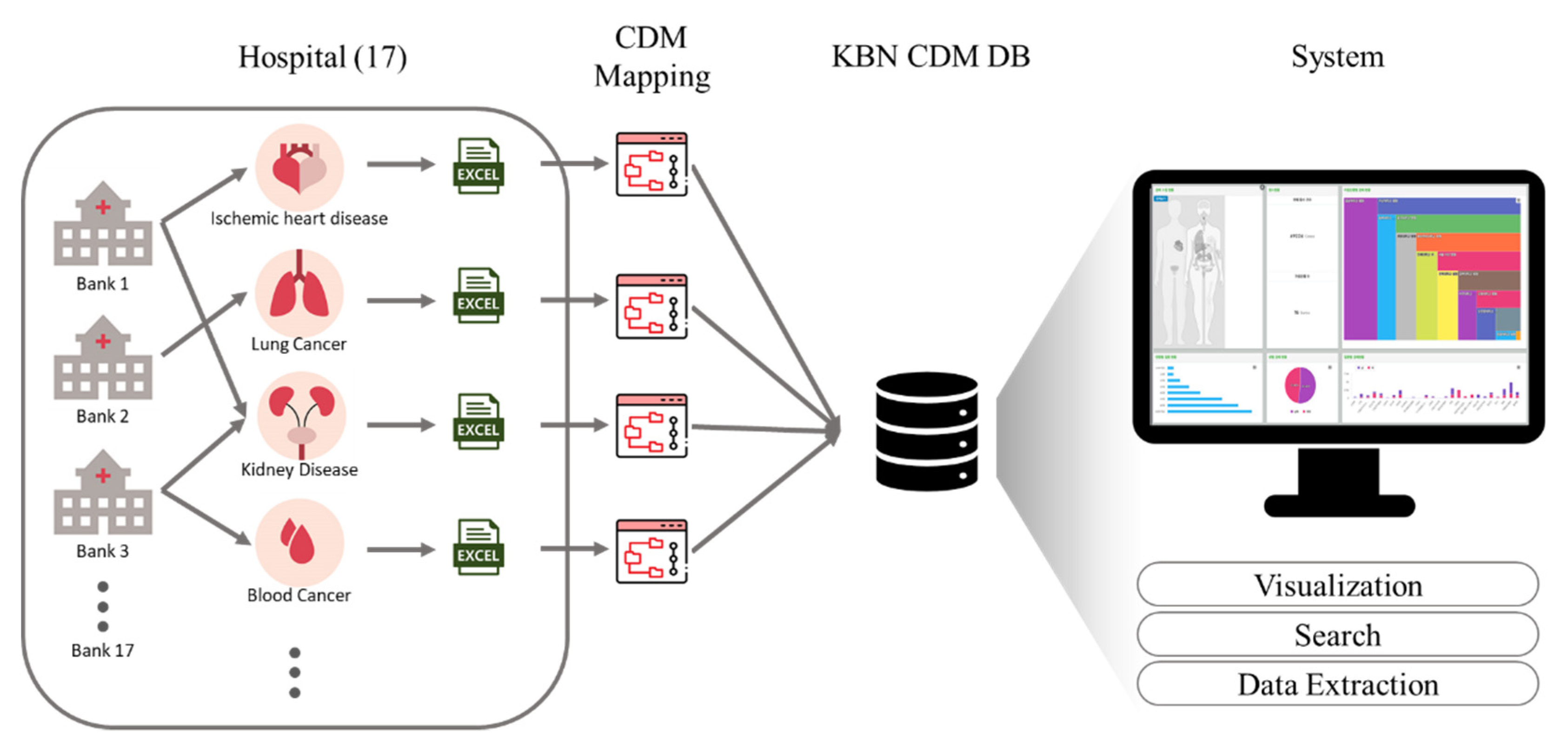
4.2. Migration to the KBN CDM and System Architecture
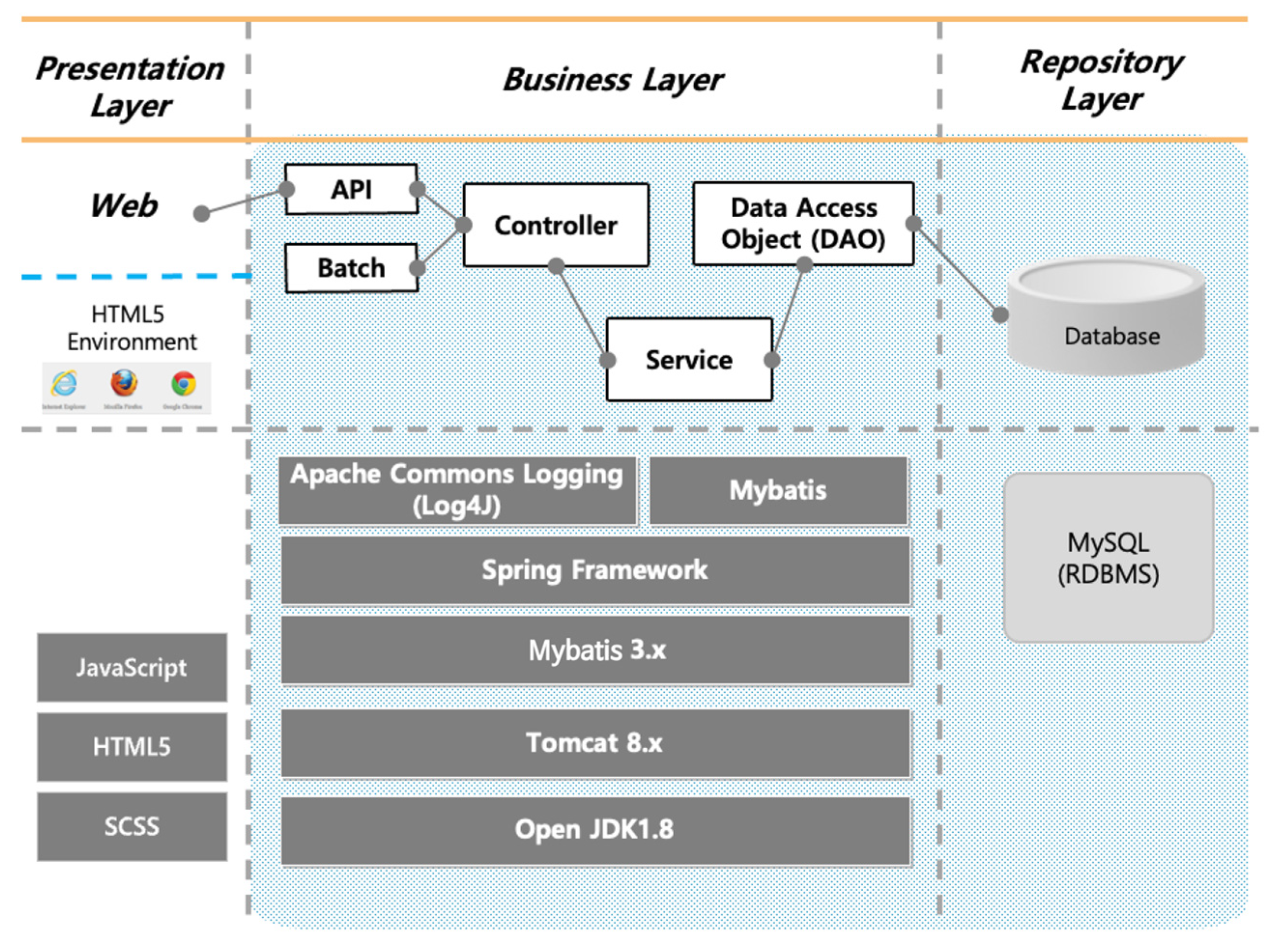
4.3. System Development
4.4. Implementation
5. Evaluation
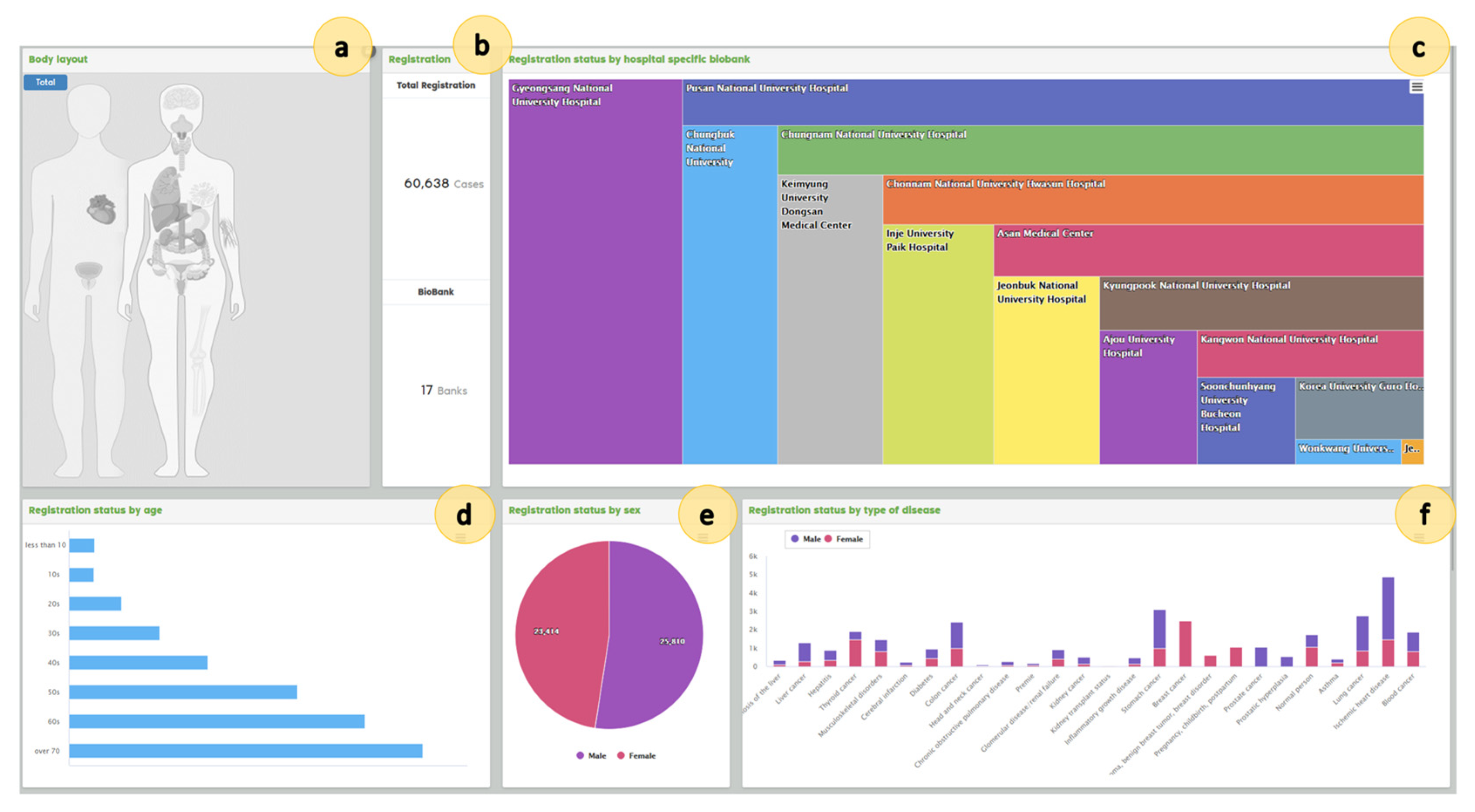
5.1. Results
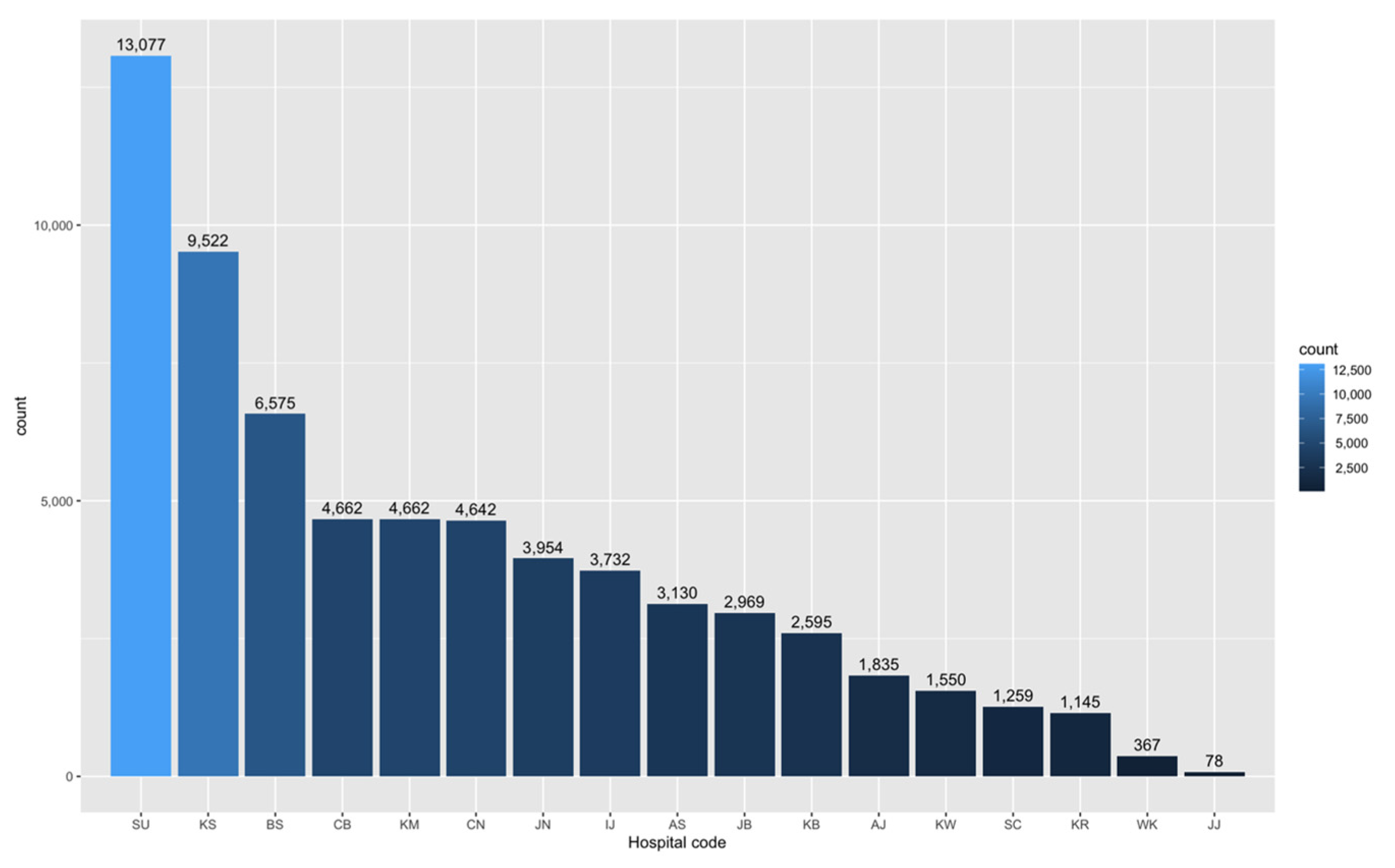
5.1.1. Participating Hospitals
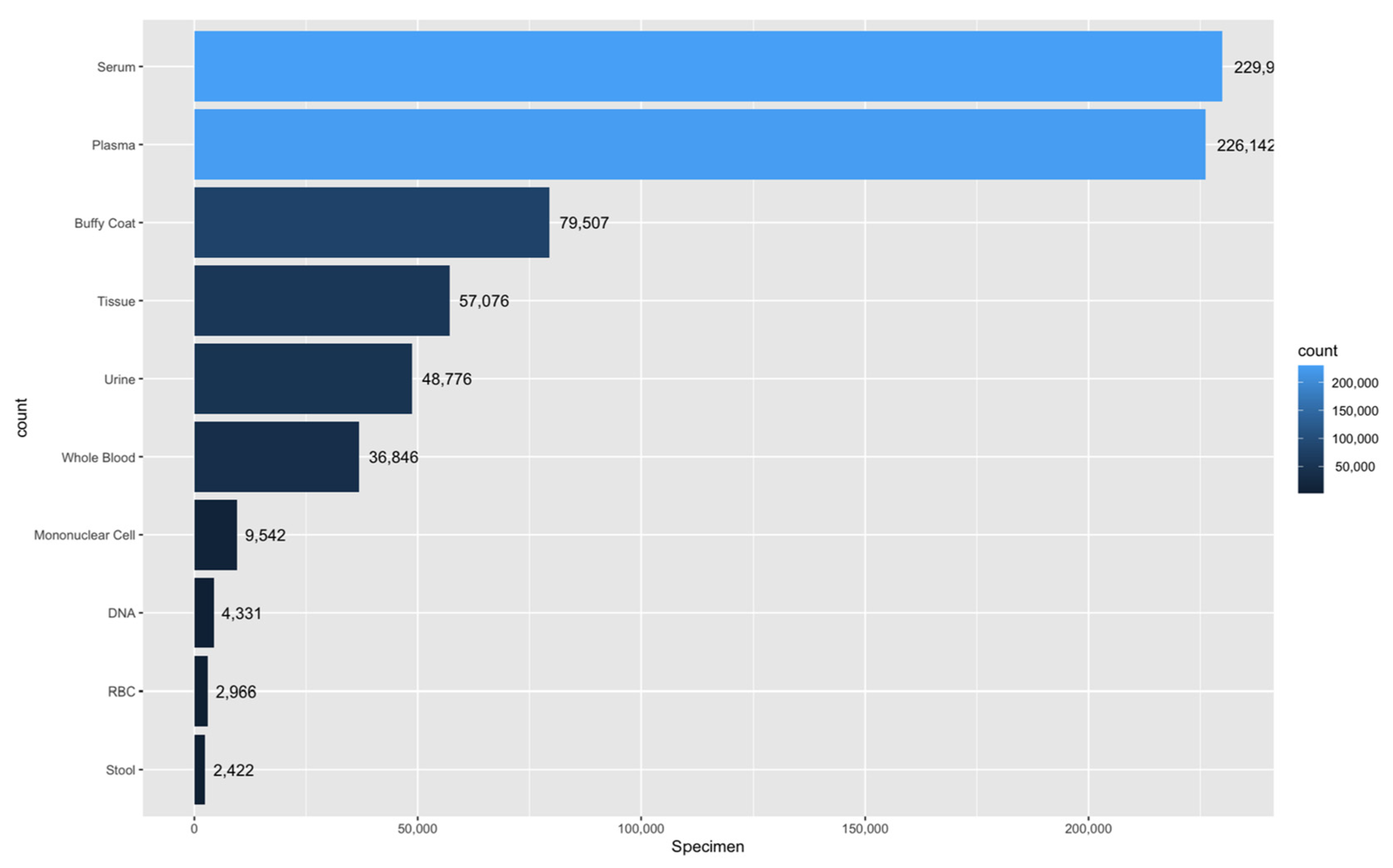
5.1.2. Banking Samples
6. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, X.; Du, L.; Qiao, Y.; Zhang, X.; Zheng, W.; Wu, Q.; Chen, Y.; Zhu, G.; Liu, Y.; Bian, Z.; et al. Ferroptosis is governed by differential regulation of transcription in liver cancer. Redox Biol. 2019, 24, 101211. [Google Scholar] [CrossRef] [PubMed]
- Bellos, I.; Pergialiotis, V.; Perrea, D.N. Kidney biopsy findings in vancomycin-induced acute kidney injury: A pooled analysis. Int. Urol. Nephrol. 2021, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Mecatti, G.C.; Sánchez-Vinces, S.; Fernandes, A.M.A.P.; Messias, M.C.F.; de Santis, G.K.D.; Porcari, A.M.; Marson, F.A.L.; Carvalho, P.O. Potential lipid signatures for diagnosis and prognosis of sepsis and systemic inflammatory response syndrome. Metabolites 2020, 10, 359. [Google Scholar] [CrossRef]
- Yu, Y.; Ruddy, K.J.; Hong, N.; Tsuji, S.; Wen, A.; Shah, N.D.; Jiang, G. ADEpedia-On-OHDSI: A next generation pharmacovigilance signal detection platform using the OHDSI common data model. J. Biomed. Inform. 2019, 91, 103119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, L.; Miao, S.; Xu, H.; Yin, Y.; Zhu, Y.; Dai, Z.; Shan, T.; Jing, S.; Wang, J.; et al. Analysis of treatment pathways for three chronic diseases using OMOP CDM. J. Med. Syst. 2018, 42, 260. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.A.; Kim, H.; Kim, S.; Yoo, S.; Yi, S.; Jeon, Y.; Hwang, H.; Kim, K.J. Analysis of antiseizure drug-related adverse reactions from the electronic health record using the common data model. Epilepsia 2020, 61, 610–616. [Google Scholar] [CrossRef]
- Steven, V.V.A.; Singer, M.; Fink, M.Y. A call to standardize preanalytic data elements for biospecimens. Physiol. Behav. 2019, 176, 139–148. [Google Scholar] [CrossRef]
- Grizzle, W.E.; Bledsoe, M.J.; Al Diffalha, S.; Otali, D.; Sexton, K.C. The utilization of biospecimens: Impact of the choice of biobanking model. Biopreserv. Biobank. 2019, 17, 230–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, A.R.; Kim, K.M.; Park, H.S.; Jang, K.Y.; Moon, W.S.; Kang, M.J.; Lee, Y.C.; Kim, J.H.; Chae, H.J.; Chung, M.J. Association between expression of 8-OHdG and cigarette smoking in non-small cell lung cancer. J. Pathol. Transl. Med. 2019, 53, 217–224. [Google Scholar] [CrossRef]
- Byun, J.K.; Choi, Y.K.; Kang, Y.N.; Jang, B.K.; Kang, K.J.; Jeon, Y.H.; Lee, H.W.; Jeon, J.H.; Koo, S.H.; Jeong, W.I.; et al. Retinoic acid-related orphan receptor alpha reprograms glucose metabolism in glutamine-deficient hepatoma cells. Hepatology 2015, 61, 953–964. [Google Scholar] [CrossRef] [Green Version]
- Yun, D.; Jang, M.J.; An, J.N.; Lee, J.P.; Kim, D.K.; Chin, H.J.; Kim, Y.S.; Lee, D.S.; Han, S.S. Effect of steroids and relevant cytokine analysis in acute tubulointerstitial nephritis. BMC Nephrol. 2019, 20, 88. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Shin, Y.; Kim, T.H.; Kim, D.H.; Lee, A. Plasma metabolites as possible biomarkers for diagnosis of breast cancer. PLoS ONE 2019, 14, e0225129. [Google Scholar] [CrossRef]
- Cho, S.Y.; Hong, E.J.; Nam, J.M.; Han, B.; Chu, C.; Park, O. Opening of the national biobank of korea as the infrastructure of future biomedical science in Korea. Osong Public Health Res. Perspect. 2012, 3, 177–184. [Google Scholar] [CrossRef] [Green Version]
- Park, O.; Cho, S.Y.; Shin, S.Y.; Park, J.S.; Kim, J.W.; Han, B.G. A strategic plan for the second phase (2013–2015) of the Korea biobank project. Osong Public Health Res. Perspect. 2013, 4, 107–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitzhenry, F.; Resnic, F.S.; Robbins, S.L.; Denton, J.; Nookala, L.; Meeker, D.; Ohno-Machado, L.; Matheny, M.E. Creating a common data model for comparative effectiveness with the observational medical outcomes partnership. Appl. Clin. Inform. 2015, 6, 536–547. [Google Scholar] [CrossRef] [Green Version]
- Cambon-Thomsen, A.; Rial-Sebbag, E.; Knoppers, B.M. Trends in ethical and legal frameworks for the use of human biobanks. Eur. Respir. J. 2007, 30, 373–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norlin, L.; Fransson, M.N.; Eriksson, M.; Merino-Martinez, R.; Anderberg, M.; Kurtovic, S.; Litton, J.E. A minimum data set for sharing biobank samples, information, and data: MIABIS. Biopreserv. Biobank. 2012, 10, 343–348. [Google Scholar] [CrossRef] [Green Version]
- Merino-Martinez, R.; Norlin, L.; van Enckevort, D.; Anton, G.; Schuffenhauer, S.; Silander, K.; Mook, L.; Holub, P.; Bild, R.; Swertz, M.; et al. Toward global biobank integration by implementation of the minimum information about biobank data sharing (MIABIS 2.0 Core). Biopreserv. Biobank. 2016, 14, 298–306. [Google Scholar] [CrossRef]
- Isabelle, M.; Teodorovic, I.; Morente, M.M.; Jaminé, D.; Passioukov, A.; Lejeune, S.; Therasse, P.; Dinjens, W.N.; Oosterhuis, J.W.; Lam, K.H.; et al. TuBaFrost 5: Multifunctional central database application for a European tumor bank. Eur. J. Cancer 2006, 42, 3103–3109. [Google Scholar] [CrossRef]
- David, G.A.; Matthew, P.A.; Olaf, A.; Steven, C.; Carolyn, H.; Patrick, R.H.; Melissa, M.; Ikue, N.; Jane, P.; Cynthia, S.; et al. Chapter 3—Autism BrainNet: A network of postmortem brain banks established to facilitate autism research. In Handbook of Clinical Neurology; Huitinga, L., Webster, M.J., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; Volume 150, pp. 31–39. [Google Scholar]
- Patil, S.; Majumdar, B.; Awan, K.H.; Sarode, G.S.; Sarode, S.C.; Gadbail, A.R.; Gondivkar, S. Cancer oriented biobanks: A comprehensive review. Oncol. Rev. 2018, 12, 357. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, Y.M.; Wei, M.Y.; Wu, Y.F.; Gao, J.H.; Liu, L.; Zhou, W.P.; Wang, H.Y.; Wu, M.C. The liver tissue bank and clinical database in China. Front. Med. China 2010, 4, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Trouillon, T.; Dance, C.R.; Gaussier, E.; Welbl, J.; Riedel, S.; Bouchard, G. Knowledge graph completion via complex tensor factorization. J. Mach. Learn. Res. 2017, 18, 1–38. [Google Scholar]
- Xia, S.; Zhang, Z.; Li, W.; Wang, G.; Giem, E.; Chen, Z. GBNRS: A novel rough set algorithm for fast adaptive attribute reduction in classification. IEEE Trans. Knowl. Data Eng. 2020, 1, 1. [Google Scholar] [CrossRef]
- Xia, S.; Zheng, Y.; Wang, G.; He, P.; Li, H.; Chen, Z. Random space division sampling for label-noisy classification or imbalanced classification. IEEE Trans. Cybern. 2021, 51, 1–14. [Google Scholar] [CrossRef] [PubMed]
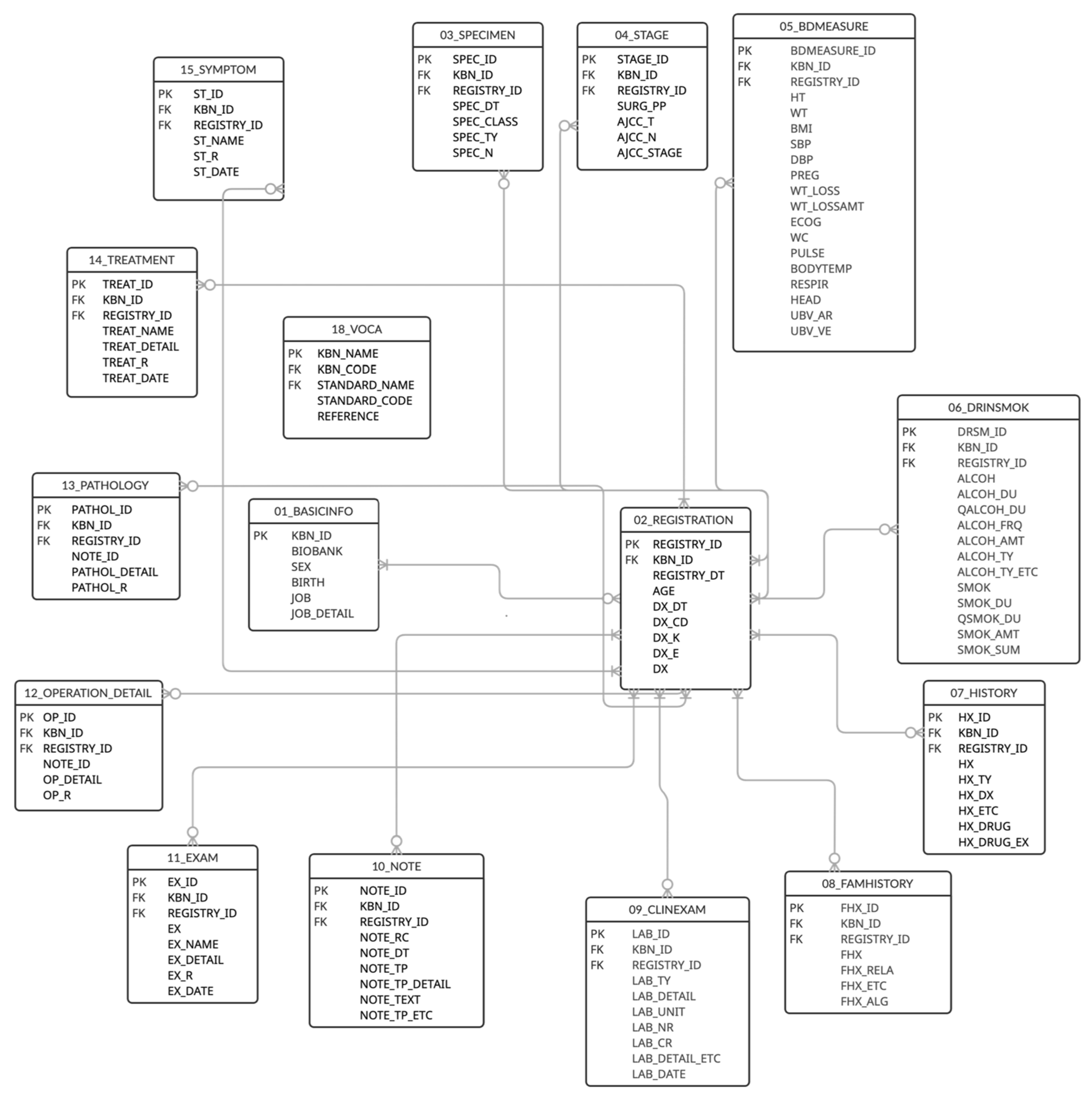
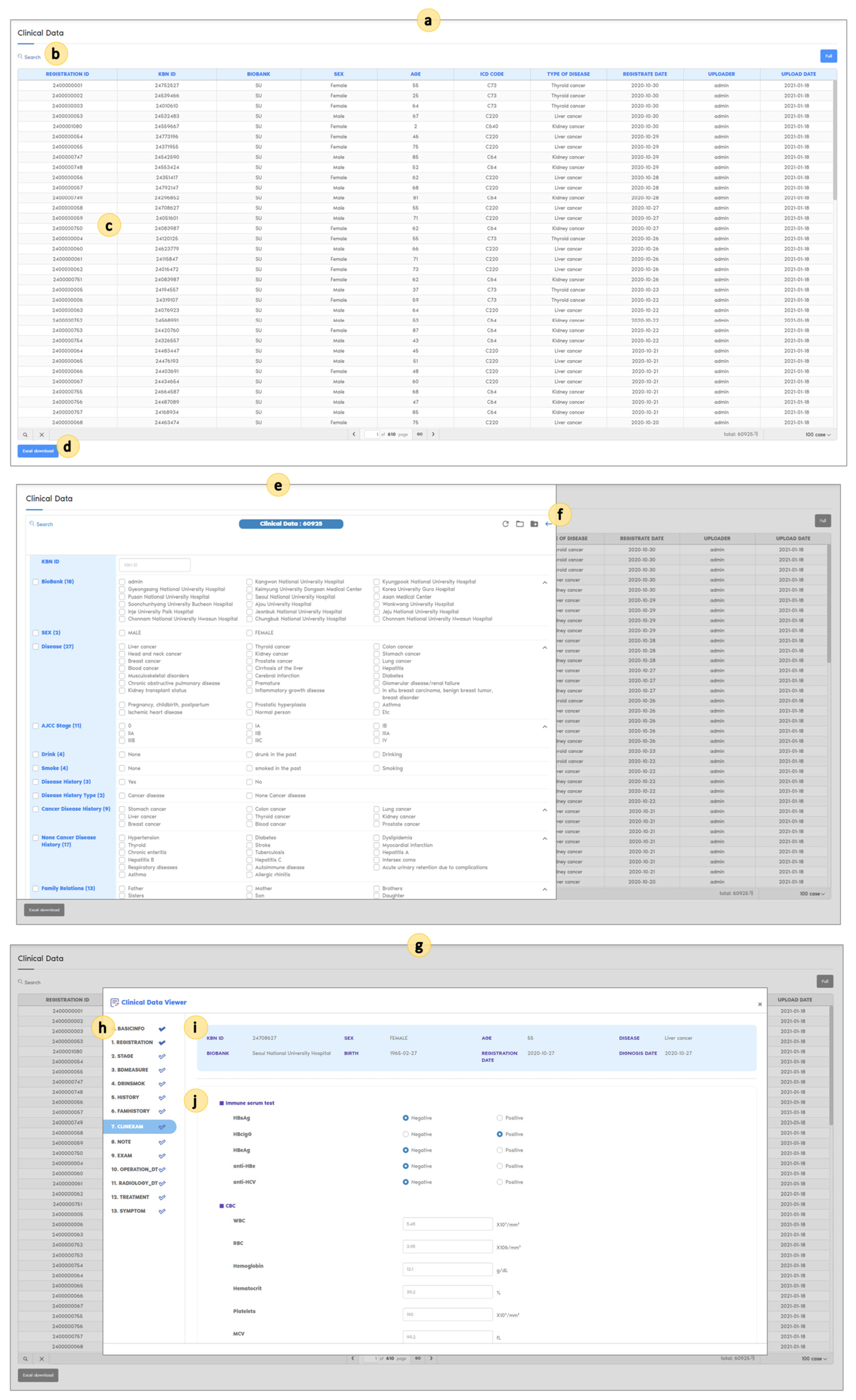
| Table | Columns | Description |
|---|---|---|
| BASICINFO | 6 | Donor ID, bank name, sex, birth, job |
| REGISTRATION | 9 | Age at registration, disease date, disease diagnosis |
| SPECIMEN | 7 | Specimen collection date, specimen type, number of specimens |
| STAGE | 8 | AJCC_T, AJCC_N, AJCC_M, AJCC_stage, AJCC pre-op, AJCC post-op |
| BDMEASURE | 19 | Height, weight, BMI, blood pressure, pregnancy status, weight loss, ECOG, waist circumference, pulse rate, body temperature, respiration rate, head circumference, UBV_Ar (umbilical cord blood arterial), UBV_Ve (umbilical cord blood vessels) |
| DRINSMOK | 16 | Drinking/smoking experience, duration, quit duration, frequency, amount |
| HISTORY | 10 | History disease, drug history |
| FAMHISTORY | 7 | Family history of the same disease, family relations, family history of allergic reactions |
| CLINEXAM | 10 | Laboratory examination name, unit, result of numeric type, result of character type, date |
| NOTE | 10 | Pathology record, surgical record, radiology report, functional test record, record date, record free text |
| EXAM | 8 | Examination name, results, date |
| OPERATION_DETAIL | 6 | Operation record detail, operation detail results, operation date |
| PATHOLOGY | 6 | Pathology record detail, pathology detail results, pathology date |
| TREATMENT | 7 | Treatment name, treatment detail, treatment date |
| SYMPTOM | 6 | Symptom name, symptom results, symptom onset date |
| SURVEY | 8 | Survey name, survey date, survey result |
| FOLLOWUP | 20 | Renal replacement therapy, recurrence, metastasis, death, cause of death |
| VOCA | 5 | KBN name, KBN code, standard code, standard name, reference |
| Variable | Category | Total N (%) |
|---|---|---|
| Sex | Male | 34,531 (52.52%) |
| Female | 31,223 (47.48%) | |
| Age | Less than 10 | 2953 (4.49%) |
| 10 to 19 | 1603 (2.44%) | |
| 20 to 29 | 2976 (4.53%) | |
| 30 to 39 | 5866 (8.92%) | |
| 40 to 49 | 7411 (11.27%) | |
| 50 to 59 | 12,010 (18.27%) | |
| 60 to 69 | 15,563 (23.67%) | |
| 70 years or older | 17,352 (26.39%) | |
| Unknown | 20 (0.03%) | |
| Disease | Ischemic heart disease | 5473 (8.32%) |
| Stomach cancer | 3223 (4.90%) | |
| Blood cancer | 3155 (4.80%) | |
| Lung cancer | 3061 (4.66%) | |
| Colon cancer | 2611 (3.97%) | |
| Breast cancer | 2511 (3.82%) | |
| Pregnancy, childbirth, postpartum care | 2389 (3.63%) | |
| Thyroid cancer | 2220 (3.38%) | |
| Liver cancer | 2129 (3.24%) | |
| Healthy individuals | 1758 (2.67%) | |
| Prostate cancer | 1548 (2.35%) | |
| Glomerular disease/renal failure | 1541 (2.34%) | |
| Musculoskeletal disorders | 1538 (2.34%) | |
| Diabetes | 1165 (1.77%) | |
| Kidney transplant status | 1096 (1.67%) | |
| Hepatitis | 907 (1.38%) | |
| Kidney cancer | 901 (1.37%) | |
| In situ breast carcinoma, benign breast tumor, breast disorder | 648 (0.99%) | |
| Prostatic hyperplasia | 570 (0.87%) | |
| Cirrhosis of the liver | 539 (0.82%) | |
| Inflammatory growth disease | 528 (0.80%) | |
| Asthma | 455 (0.69%) | |
| Cerebral infarction | 300 (0.46%) | |
| Chronic obstructive pulmonary disease | 272 (0.41%) | |
| Premature birth | 161 (0.24%) | |
| Head and neck cancer | 104 (0.16%) | |
| Other diseases * | 24,951 (37.95%) | |
| Total | 65,754 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ko, S.-J.; Choi, W.; Kim, K.-H.; Lee, S.-J.; Min, H.; Oh, S.-W.; Choi, I.Y. Common Data Model and Database System Development for the Korea Biobank Network. Appl. Sci. 2021, 11, 11825. https://doi.org/10.3390/app112411825
Ko S-J, Choi W, Kim K-H, Lee S-J, Min H, Oh S-W, Choi IY. Common Data Model and Database System Development for the Korea Biobank Network. Applied Sciences. 2021; 11(24):11825. https://doi.org/10.3390/app112411825
Chicago/Turabian StyleKo, Soo-Jeong, Wona Choi, Ki-Hoon Kim, Seo-Joon Lee, Haesook Min, Seol-Whan Oh, and In Young Choi. 2021. "Common Data Model and Database System Development for the Korea Biobank Network" Applied Sciences 11, no. 24: 11825. https://doi.org/10.3390/app112411825
APA StyleKo, S.-J., Choi, W., Kim, K.-H., Lee, S.-J., Min, H., Oh, S.-W., & Choi, I. Y. (2021). Common Data Model and Database System Development for the Korea Biobank Network. Applied Sciences, 11(24), 11825. https://doi.org/10.3390/app112411825






