1. Introduction
Oral cancer has become an aggressive malignancy with an important impact on the global healthcare system [
1]. Out of all oral malignancies, oral squamous cell carcinoma (OSCC) has been reported in more than 90% of cases [
2]. The current problem related to this disease, besides late diagnosis, is the fact that it has developed a certain resistance to the therapeutic treatment, a fact that influences further prognosis. This tumour, as a matter of fact, has a high mortality rate, and relapses after surgery and other therapies, such as chemotherapy and radiation therapy, are frequent [
3,
4]. An explanation for the unappropriated response of the malignancy to treatment could be the complex and yet not completely known interaction between the environmental variables and the developed genetic variations. Although recent progress has been made concerning diagnosis and treatment options, the survival rate of these patients is still limited, representing ~50% [
5]. In order to improve the prognosis of this type of malignancy, a complete diagnosis needs to also include the concept of existing epigenetic mechanisms that can be negatively influenced by environmental factors. Existing studies suggest that the associated oncogenic factors, such as alcohol, human papillomavirus (HPV), tobacco, exogenous carcinogens and lifestyle habits, can determine the epigenetic alterations that are involved in tumour progression and metastasis incidence [
6].
Approximately 20% of the oral dysplasia cases progress into malignant lesions [
7]. One concern during histopathological examination is the presence or absence of histologically safe margins. Studies reveal that in the case of positive histological margins, the recurrence of OSCC is 70–80%, while in the case of negative margins, recurrence is 30–40% [
8,
9]. In their study, Chen et al. [
10] discussed the fact that local recurrence or the incidence of a second primary tumour can be linked to existent molecular changes in the entire oral mucosa.
An increased understanding of the major role that epigenetic alterations play in the initiation, progression and survival rate of oral cancer patients can aid in developing new approaches in the prevention and early diagnosis field as well as in the management of the at-risk population group. Oral tissue is directly influenced by the presence of various environmental factors that can induce abnormal changes. The disturbance of epigenetic enzymes and signalling pathways promote the growth of malignant cells and facilitates metastasis [
11].
Although the published results of several studies focus on various genes as potential biomarkers for OSCC, these findings have not yet been applied in the diagnostic field. An explanation for this could be that the selection of a biomarker is, in many cases, difficult due the fact that it is linked to a large set of data [
12].
There are multiple studies that have focused on potential oral cancer biomarkers and their involvement in the early diagnosis, progression and aggressiveness of tumours [
10,
13,
14]. An important aspect that needs to be taken into consideration is the determination of the individual risk of developing oral cancer through the quantification of personal genetic mutations and associated environmental risk factors.
Molecular characteristics and alterations encountered in oral squamous cell carcinoma reveal the high heterogenicity of the disease, outlining the occurrence in complex anatomical areas with a high number of molecular changes. The Cancer Genome Atlas (TCGA) published a large database concerning genomic data alterations associated with OSCC, outlining the existence of a high number of cancer driver genes that display specific mutational profiles [
15].
Until now, multiple genetic mutations have been associated with the incidence of OSCC, but the persisting problem is the diversity among different populations and the presence or absence of a certain gene [
16]. Currently, somatic mutations that can be identified by next-generation sequencing and that are listed in the Cancer Genome Atlas have provided an important pathway for the potential diagnosis of OSCC. The somatic mutation of gene COL9A1 (collagen type IX alpha chain I) was previously found to be associated with the occurrence of oral malignancy. COL9A1 is localised in chromosome 16q13. The gene is responsible for the encoding of the three alpha chains of collagen type IX, and several studies have reported that a somatic mutation can be involved in the pathogenesis of OSCC [
16].
The use of a biomarker that can be linked to the presence of OSCC could be a ground-breaking approach for an early diagnosis. Currently, tissue samples and histopathological examination represent the gold standard for diagnosis, focusing on cellular and tissue alterations [
17]. The perspectives that a molecular marker in the body’s fluids have to offer in the screening, diagnosis and prognosis of OSCC are appealing. Blood samples have proved to be a reliable source that can provide valuable information related to genetic and epigenetic changes. The identification of a biomarker in serum samples can provide a fast diagnosis, a monitoring tool and a future prognostic modality [
18]. The incidence of OSCC suggests an important interaction between multiple genes and proteins, being the result of a complex number of factors that determine a series of molecular genetic events [
19].
The aim of our study was to identify the presence of a COL9A1 gene variant in patients diagnosed with OSCC, comparatively using both tissue samples and blood samples. The evaluation and quantification of the results based on the presence or absence of gene variants in the samples provided were also associated with existing risk factors, tumour grading as well as the differentiation status. By also using blood samples to identify the genetic variants in patients diagnosed with OSCC, we targeted a non-invasive approach for an early diagnosis and screening method based on a personalised analysis.
2. Materials and Methods
2.1. Study Population
In our pilot study, 32 patients diagnosed with OSCC were included. The patients were diagnosed, admitted and treated in the Maxillo-Facial Clinic at the Emergency County Hospital in Timisoara during March 2020–October 2020, and they had previously undergone a biopsy for histopathological diagnosis. The inclusion criteria for patients were a confirmed diagnosis of OSCC (based on a previous biopsy result), in either gender, and being over 18 years of age. The exclusion criteria were any previous treatments with chemotherapy or radiotherapy in the head and neck area and OSCC relapse. Data related to medical history, current medication, cigarette smoking, and alcohol consumption were obtained during the anamnesis and documented in the medical chart. We considered substance use as a risk factor when usage was for at least 6 months. Individuals without any substance use were defined as non-drinkers and non-smokers. Our study was approved by the Ethics Committee of “Victor Babes” University of Medicine and Pharmacy Timisoara (no. 12/2020), and patients agreed and signed an informed consent form that followed the guidelines of the Declaration of Helsinki.
2.2. Tissue and Serum Samples
Fresh oral tissue from the tumour was collected during the surgical tumour resection and consisted of a fragment of 0.5/0.5 cm tumoral tissue, which was immediately fixed in RNAlater® solution (Ambion, Austin, TX, USA) at 4 degrees Celsius for 24 h and afterwards at −80 degrees Celsius until extraction. In addition, from each OSCC patient, a sample of 2.5 mL peripheral blood was collected on an EDTA (ethylenediaminetetraacetic acid)-coated tube and kept at a temperature between 2–4 degrees Celsius and processed within the next 24 h. The blood samples were collected before the tumoral removal surgery, the same day of the intervention.
Further histopathological examination was performed by two pathologists in order to ensure the results, and the tumours were grouped in either an early clinical stage (stage I and II) or late clinical stage (stage III and IV). The TNM status was stated according to the International Union Against Cancer. The OSCC tumours were also classified as per the World Health Organization (WHO) according to the loss of differentiation as G1 (well differentiated), G2 (moderately differentiated) and G3 (poorly differentiated).
2.3. DNA Analysis
The allelic discrimination analysis was carried out on a QuantStudio 7 Real-Time PCR (reverse-transcription polymerase chain reaction) instrument (Applied Biosystems, Foster City, CA, USA) using a TaqMan genotyping assay for allelic discrimination C___2989110_1_ (rs550675), a TaqMan® PCR Master Mix, and a DNA probe according to the protocol provided by the producer. Two controls were used: AL-1, corresponding to the wild type (wt), was VIC™ dye-labelled; AL-2, corresponding to COL9A1 C > T polymorphism, was Fluorescein FAM dye-labelled. Genotypes were determined by measuring allele-specific fluorescence using software for allelic discrimination (Applied Biosystems, Foster City, CA, USA). Three genotypes were found in the studied samples: C/C (wild-type homozygotes), C/T (heterozygotes) and T/T (homozygotes for the COL9A1 gene variant).
2.4. Statistical Analysis
The statistical analysis was performed using the SPSS v17 statistics software (IBM SPSS
®, Chicago, IL, USA). For the results in
Section 3, we used frequencies tables, chi-square tests with Yates’s correction, relationship maps and a simple linear regression model. The frequency tables outlined the gender and age distribution as well as the association of the two main risk factors: tobacco and alcohol. The associations between the nominal variables were made with chi-squared tests with Yates’s correction in the case of a value lower than 5. Our attention was directed toward the genotype frequencies (i.e., C/C, C/T/ and T/T) in both types of samples (i.e., tissue and blood samples) as well as their relationship with the stated risk factors. By acknowledging the genotype frequencies, the performed crosstabs analysis outlined the association of the COL9A1 variant in the two categories of samples for both genders. A relationship graphic outlines the existent relationships between the identified SNP in the tissue samples and those in the blood samples by providing a visual graphic. Linear regression analysed whether the histological grading was influenced by different variables.
Taking into consideration the epidemiological situation related to SARS-CoV2 infection and the consequence of restrictions, the number of included subjects was small, and we focused on conducting a preliminary pilot study by genotyping the COL9A1 gene in OSCC patients.
4. Discussion
Over the past years, the incidence of OSCC has become a worldwide concern, and the importance of an early diagnosis, a proper staging, prognosis and further treatment requires an improvement in the current protocols.
The incidence of this pathology is directly linked to exposure to environment mutagens that can determine genetic and epigenetic alterations of oral mucosa cells, with an increased risk for further malignant development [
20,
21]. Recently, more studies have focused on the epigenetic alterations of various genes that have potential as biomarkers and could improve early detection [
22,
23]. Numerous clinical applications of epigenetics have the potential to become new approaches for OSCC therapeutic strategies [
24].
Blood sample analysis has the advantage of providing real-time information through specific oral cancer biomarkers regarding the tumour, tumour recurrence, metastasis, any evidence of drug resistance and the probability of developing OSCC through the presence of certain gene mutations [
25,
26].
Although an important number of genetic alterations has been associated with the incidence of OSCC in numerous studies [
27,
28], only several related mutations could be encountered among the population [
29]. In addition, despite the fact that the P53 gene was strongly associated with OSCC, the results of a recent meta-analysis that focused on 2298 cases of diagnosed OSCC revealed the fact that it cannot be highly associated with the incidence of oral squamous cell carcinoma [
30].
Aiming to find a connection between the initiation and progress of the oral malignant process and gene variants, our attention was gained by a promising gene, COL9A1 through a tagged SNP [
31,
32]. Based on previous research and the Cancer Genome Atlas, the COL9A1 gene is responsible for the encoding of the three alpha chains of type IX collagen, and the targeted SNP (rs550675) in our study was highly associated with the occurrence of oral malignancy. Previous studies have reported that the methylation levels of COL9A1 decreased in association with OSCC incidence [
33]. Different types of alterations in the gene’s expression have been identified in the head and neck area, brain, heart, salivary glands, prostate and testis. Depending on the targeted SNP (single-nucleotide polymorphism), genetic mutations can be connected to different types of malignancies.
This type of collagen is localised in multiple tissues, encoding one of the three alpha chains of collagen. The alpha 1 subunit of the gene is produced by non-cartilaginous tissues, such as skin, bone and arterial smooth muscles cells, and regulates the promoter of collagen type V [
34]. Although collagen type IX is frequently associated with birth anomalies, its influence on adult life is not completely known.
Studies have shown that the lack of the synthesis of the alpha 1 chain influences the assembly of collagen molecules and can be related with the onset of various diseases. It has been reported that mutations in this gene can be associated with ophthalmic, oro-facial disorders and articular and auditory defects [
34]. Existent results have shown that modification of the methylation in the COL9A1 gene can be identified in breast cancer, and in their study, Piotrowski et al. [
35] encountered a general drop in methylation of COL9A1 in the CpG island of tumour breast samples. Fu et al. [
36] analysed the involvement of COL9A1 through miR-9-3 in liver hepatocellular carcinoma. Their results showed that COL9A1 was enriched in focal adhesions, suggesting an important role in the carcinogenesis of liver. High levels of COL9A1 were reported as well in fibroblasts associated with lung cancer patients [
37] and colorectal cancer [
38].
In the case of head and neck cancer, Sok et al. [
34] reported that COL9A1 was present in the tumour tissue and undetectable in the corresponding normal tissue. Their results suggest that the COL9A1 gene contributes to continuous cell proliferation in head and neck cancer and demonstrates a higher expression in the case of the tumours in the setting of metastasis compared to those without dissemination. Moreover, by analysing the expression of COL9A1 in HNSCC cell lines compared to normal mucosal epithelial cell lines, it was concluded that the existence of the overexpression of the gene was due to the fact of increased expression in the transformed epithelial cell components of the tumour [
34]. Another evaluation of increased expression of COL9A1 in tumour-derived fibroblasts from tissue explants compared to fibroblasts obtained from healthy individuals showed higher levels of COL9A1 in the HNSSCC fibroblasts from tissue explants compared to healthy ones. The normal fibroblasts in response to a wound secrete different types of collagens, among which is COL9A1 [
34].
A large number of gene mutations have been associated with OSCC and described in the Cancer Genome Atlas, and several were linked to the initiation process as well as the progress of oral squamous cell carcinoma [
39]. In our study, we targeted the COL9A1 gene that had a mutation rate higher than 20%, with various mutation variants that have been encountered in the developmental process of OSCC [
40].
Unfortunately, OSCC is still diagnosed in advanced stages and associated with a high mortality rate. Early diagnosis in this case and minimal invasive treatment options should be the target of further research. This pathology should be revised using genetic information and associated environmental risk factors. A study conducted by Chuang et al. revealed that early detection of oral premalignant disorders reduced the diagnosis of stage III and IV oral cancer by 21% [
41].
A study performed by Chung et al. [
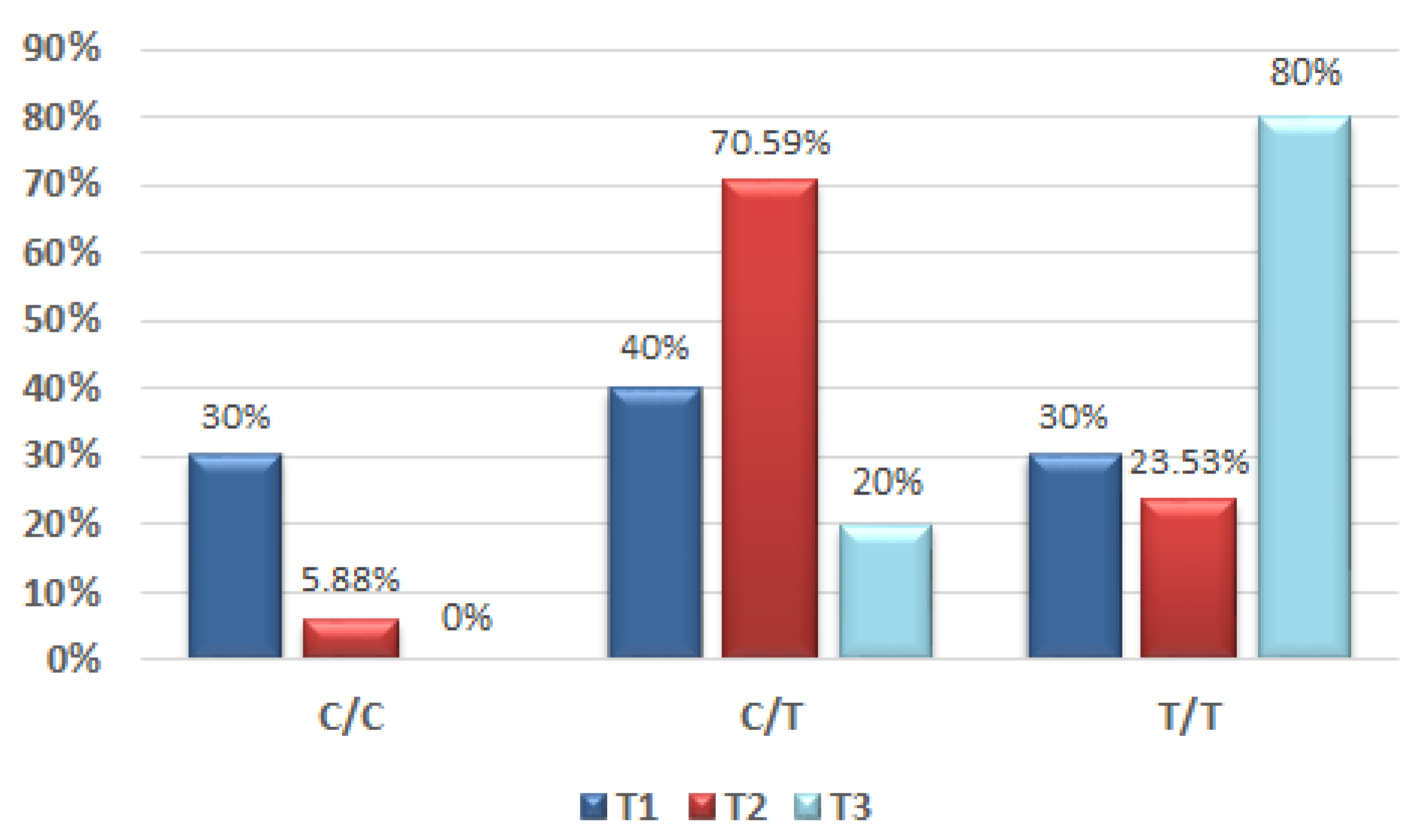
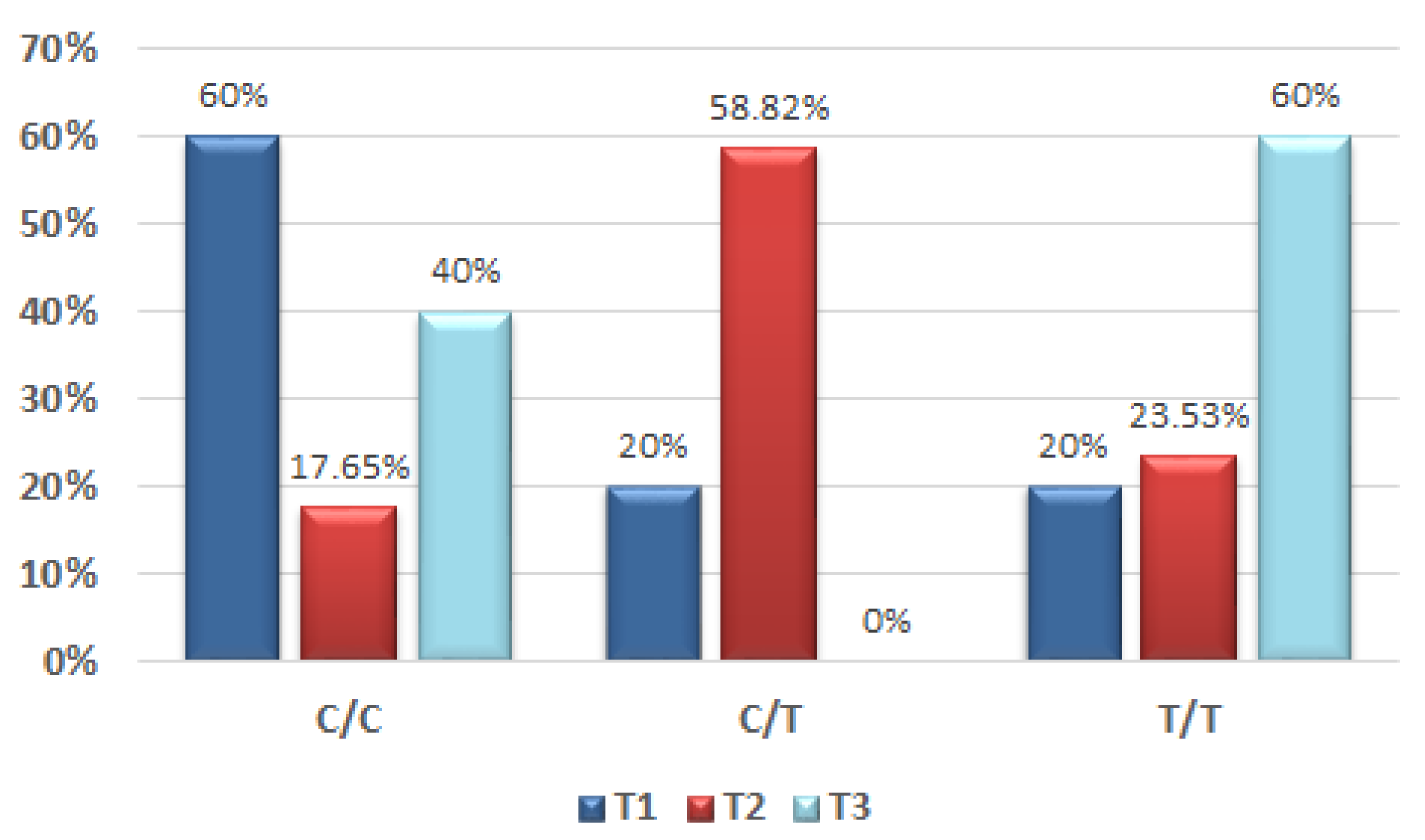
13], which focused on variants of COL9A1 and FAT1 gene variants in the male population and also added risk factors related to oral malignancy, reported that a high genetic risk score was associated with the incidence of OSCC by up to 92%. Their genetic score was based on genotype analysis and the presence or absence of the T allele combined with risk factors (cigarette smoking, betel and alcohol), for both the OSCC group and the controls. This was the first study that focused on the possible association of the COL9A1 gene and the occurrence of OSCC based on tissue sample analysis. The identification of COL9A1 genotypes associated to the OSCC group was 37.8% for the C/T genotype and 18.9% for the T/T genotype, while the C/C genotype was 43.3%. In the case of our pilot study, the results for the male group related to the identification in the tumour sample of the C/T genotype was 55% and for the T/T genotype 35%.
In addition, our results can be explained by the difference in the study group numbers. The habit of smoking in their OSCC group was 86.9%, similar to our percentage among the male group which represented 80%. Related to alcohol consumption in the male population, Chung et al. stated that 68.1% of subjects consumed alcohol, also comparable with the results of our study that reported a 65% incidence of alcohol consumption among the OSCC male group. Compared to their study, we decided to introduce both genders, and the COL9A1 genotypes in the female OSCC group were also present.
The hypothesis of our pilot study gravitated around the theory of an existing relationship between the COL9A1 gene variant C/T encountered in tumour tissue and blood samples of patients. The main limitation of this research consisted of the number of patients, but as a primary attempt to identify a connection between the two types of samples, our results are promising. Our pilot study focused on the correlation and the presence of the gene mutation in blood and tissue samples, with a positive significant statistical correlation (p < 0.001) between the samples. These results are encouraging for a future study including a higher number of subjects.
Arising from the possibility of introducing a liquid biopsy based on serum and by focusing on genetic alterations that are associated with oral cancer, the future horizons for screening and early diagnosis are expanding.
To our knowledge, this is the first study that also focused on using blood samples as a possible screening and diagnostic tool with the help of existent genetic expressions as well as being a non-invasive, faster, easier, cost-effective, and better accepted approach among the patients.
Next-generation sequencing has focused on the use of the “liquid biopsy”, based on blood or saliva samples, that targets the identification of genetic mutations that can be correlated with the development, progression or relapse of OSCC [
19,
20]. These initial findings have shown reliable evidence that their application can be the basis of future disease diagnosis and screening.
The importance of the epigenetic changes that occur in OSCC should not be underestimated, and the mechanisms that trigger them and determine aberrant mutations should be the target of further research [
42].