Abstract
The assessment of rice panicle initiation is crucial for the management of nitrogen fertilizer application that affects yield and quality of grain. The occurrence of panicle initiation could be determined via either green ring, internode-elongation, or a 1–2 mm panicle, and was observed through manual dissection. The quadratic polynomial regression model was used to construct the model of the trend of normalized difference vegetation index-based vegetation indexes (NDVI-based VIs) between pre-tillering and panicle differentiation stages. The slope of the quadratic polynomial regression model tended to be alleviated in the period in which the panicle initiation stage should occur. The results indicated that the trend of the NDVI-based VIs was correlated with panicle initiation. NDVI-based VIs could be a useful indicator to remotely assess panicle initiation.
1. Introduction
Rice (Oryza sativa L.) is one of the most important staple foods for more than half of the world population. Due to the rapid growing of food demand and limited arable land, improving yield potential to boost up future rice production is an urgent need. Rice yield is known to be increased by the nitrogen topdressing at the panicle initiation (PI) stage that is the beginning of the reproductive stage [1,2,3]. According to the recommendation of Agricultural Improvement Committee in Taiwan, when the length of the panicle is found to be 2 mm, the nitrogen fertilizer should be applied within two days. Applying a large amount of nitrogen fertilizer before PI would easily cause excessive stem elongation and thus tend to increase lodging risk. On the contrary, applying nitrogen fertilizer after PI is less effective on rice yield improvement. Therefore, accurate determination of the PI stage is crucial for rice production.
The PI is generally considered as the turning point between the vegetative phase and the reproductive phase. When a rice plant has reached maximum tillering, the internodes of the rice stem start elongating and subsequently panicle initiating. The overlapping period between maximum tillering and PI is also termed as vegetative-lag phase. The differences of the rice appearance between the vegetative-lag and PI are obscure to the naked eye. Therefore, they are difficult to be distinguished directly by human observation.
The general methods to assess PI in the farm are identifying the internode-elongation and green-ring [4,5], length of young panicle (1–2 mm) in the cross section of dissected stem [5], and leaf number index/leaf appearance [6]. When 30% of the main culms have panicles 2 mm or longer, it is considered as the panicle differentiation (PD) stage [3] that is late for nitrogen topdressing. Those methods are inconvenient and inefficient for large-scale estimation. Some other convenient, large-scale, and non-destructive approaches that help to monitor the plant growth stages, such as modified-calendar days and heat units, are potential candidates to be used for the precise estimation of PI. The calendar days method is the easiest approach but is not reliable enough as it is largely affected by the weather variability during the cultivation period [7,8,9]. Growing degree day (GDD) is an excellent heat unit that has been widely applied in corn production [10] since it was first proposed to describe the timeline of biological development [7]. However, these methods are rudimentary and are not able to distinguish the variability between fields. Nowadays, quantitative assessment, real-time and site-specific management of precision agriculture are the objectives of community.
Spectral remote sensing is another potential approach for the estimation of various variables that are correlated to plant architecture and physiology, providing high-throughput information non-destructively and rapidly for precision agriculture. The arithmetic combinations of vegetation spectral reflectance, which usually termed as vegetation indexes (VIs), became useful indicators for studying plant health and status. The NDVI (normalized difference vegetation index), which is calculated through a normalization procedure [11], seems to be the most popular and long-established VI. The NDVI is sensitive to responses on the green vegetation [12] as it correlates with the biophysical and physiological changes in plants [13,14,15,16,17,18]. Moreover, the NDVI has been subsequently developed to the other NDVI-based VIs, such as NDRE (normalized differential red-edge), GNDVI (green NDVI), NDSI (normalized difference spectral index), etc. [19,20,21]. Recent studies put efforts on the quantification of nitrogen contents in plants through remote sensing to optimize the fertilization efficiency and increased the yield [22,23,24,25,26]. In the field of rice research, NDVI is one of the most frequently used vegetation indexes. For example, preceding studies revealed that the NDVI is useful for in the research of rice breeding, nitrogen use efficiency monitoring, and rice yield prediction [27,28,29]. Furthermore, NDVI has also been used to monitor the rice growth stages, including the panicle development stage [30]. The preceding studies revealed the trend of NDVI changes during the rice growth cycle; however, the evaluation of PI prediction is currently unavailable.
The main objective of this study, therefore, is to investigate the relationship between NDVI and PI occurrence. Meanwhile, a non-destructive and high temporal resolution approach was also expected to be established for PI assessment in this study.
2. Materials and Methods
2.1. Field Experiment Design and Management
Pot experiments were conducted in 2019 at the experimental field (22°38′ N, 120°36′ E) of National Pingtung University of Science and Technology, Pingtung, Taiwan, to examine the canopy reflectance behavior as a function of panicle initiation on rice (Oryza sativa L.). A japonica cultivar, Kaohsiung147 (KH147), was planted into 4 groups with 5 replications. Ammonium sulphate was applied at a rate of 150 kg/ha N in each group (20% for basal, 20% for 1st tillering topdressing, 30% for 2nd tillering topdressing, and 30% for panicle initiation topdressing).
2.2. Determination of PI through Dissection
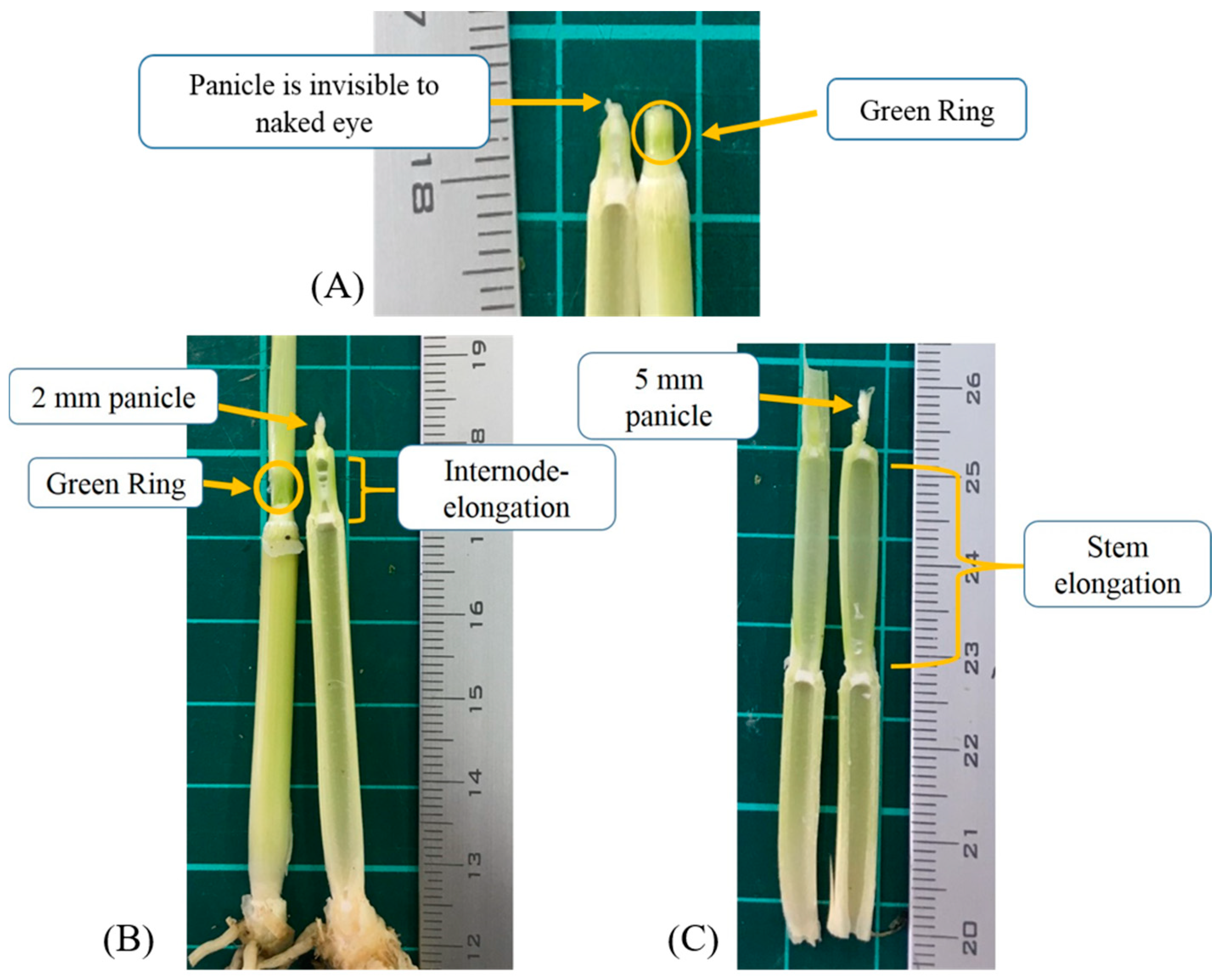
The PI stage was determined through manual dissection and features observation. The entrance of the PI stage was verified via either the green ring (Figure 1A), internode-elongation, or when 1–2 mm panicle was observed (Figure 1B). The ideal timing for nitrogen topdressing is the interval of Figure 1A,B. When the length of the panicle was over 2 mm, it was considered as the panicle differentiation (PD) stage (Figure 1C) that was not included in PI determination. The PI occurrence of each group was determined if more than 50% of dissected samples were verified to PI.
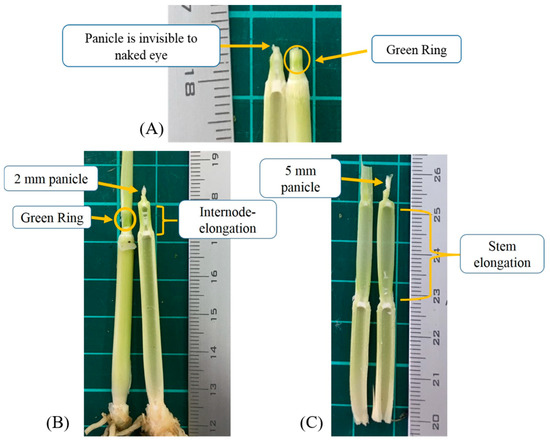
Figure 1.
The early stage of panicle initiation (A), and the optimum stage for topdressing (B). The green ring disappeared, significant stem elongation, panicle above 2 mm, is considered as panicle differentiation stage (C).
2.3. Spectral Measurement and NDVI Calculations
The spectral data of the rice canopy was collected by using SpectraPen SP-100 (range 640–1050 nm, 2 nm scan-range, Photon Systems Instruments, Czech), which is a non-imaging, handheld hyperspectral sensor. The data collection period was between 10 DAT (days after transplanting) and 80 DAT. The acquisition time was between 10:00 a.m. and 2:00 p.m., and the sensor was held horizontally at nadir view in the position about 3–5 cm above the highest leaf of the plants. The integration time of spectrum collection was set to auto-sensitivity to minimize the interference of sunlight intensity variability. The collected spectrum data were used to calculate NDVI-based VIs, which can be expressed as follows:
where the λa and λb respectively denote the reflectance of near-infrared (NIR) and red wavelengths.
2.4. Reference Wavelengths and Recombined NDVI-Based VIs
The key wavelengths were selected by some physiologically related reference wavelengths with the equation of NDVI that have been used in previous studies (Table 1). Those reference wavelengths were recombined and recalculated as the normalized difference procedure with all samples.

Table 1.
Reference wavelengths correlating with plant physiological traits.
2.5. Estimation of PI Occurrence through First-Order Differentiation
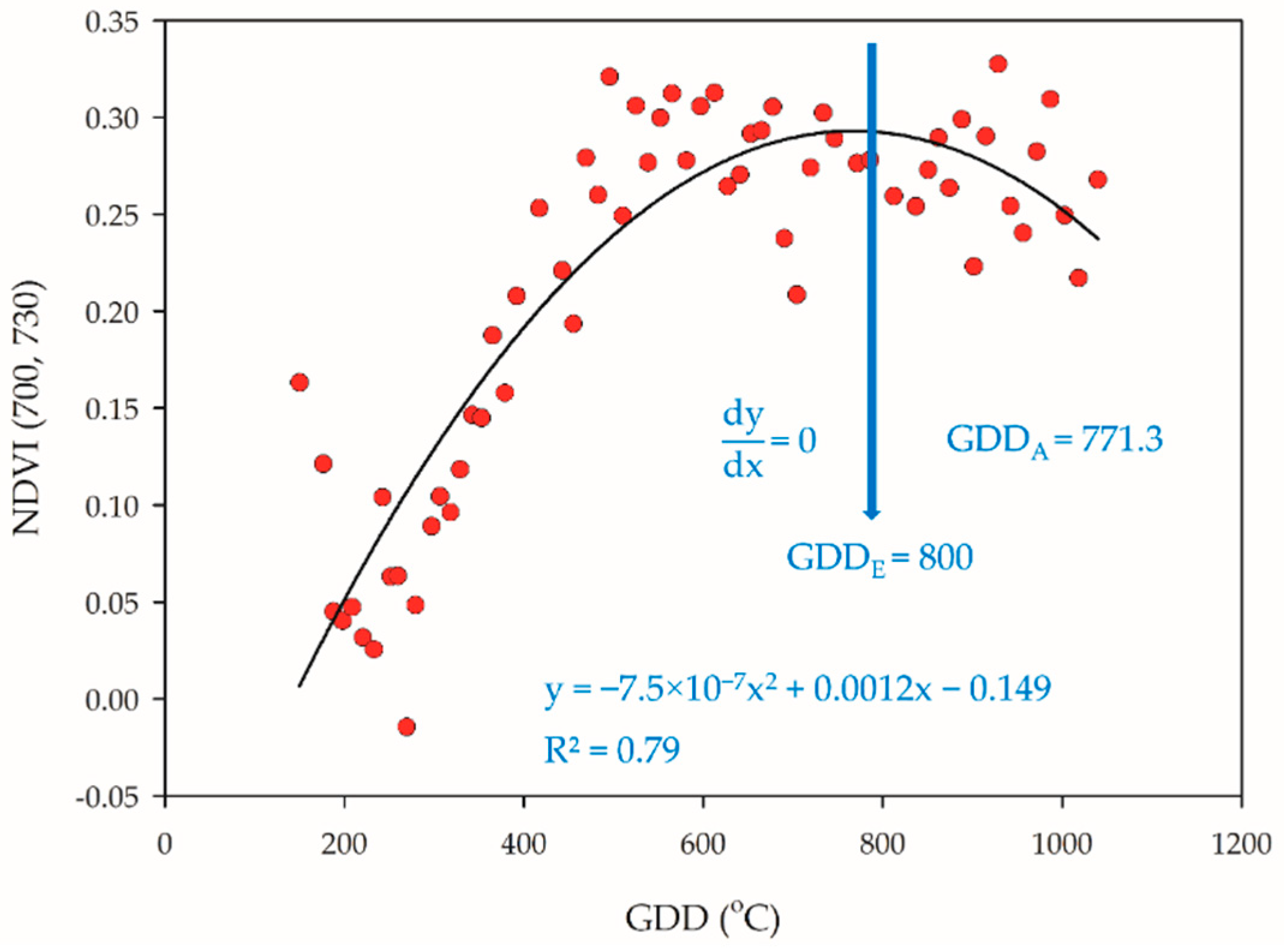
The trends of NDVI-based VIs scatter plots were changed along the rice plant development, and the quadratic slope always gradually decreased after reaching the reproductive phase. We assumed that the NDVI might have reached maxima during PI, and therefore, the slope would be zero (Figure 2).
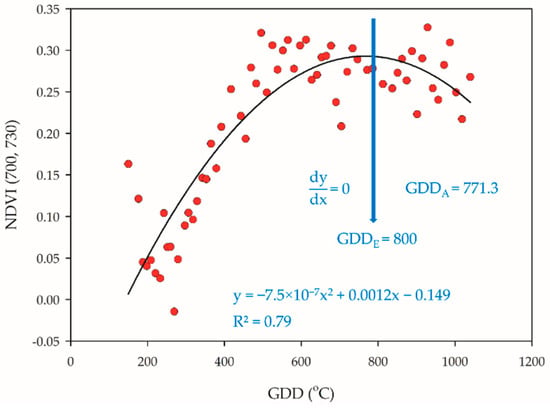
Figure 2.
Estimation of PI occurrence. When the quadratic slope is equal to zero, the actual determination of PI was GDDA = 771.3 through dissection, and the estimation of PI was GDDE = 800.
2.6. Temperature and Growing Degree-Days (GDD)
Temperature data were obtained from Chishan meteorological station (22°35′ N, 120°36′ E), which is located approximately 7 km away from experimental site. The GDD of “Method 1” as stated by preceding study was selected as a substitution to represent the cultivation timing, according to which [35]:
where the Tmax and Tmin are the daily maximum and minimum air temperature, and the Tbase is the base temperature that was set to 10 °C [8].
2.7. Statistical Analysis
The statistical analysis of the data was done by using Microsoft Excel 2013 (Microsoft Corporation, Redmond, WA, USA). The quadratic polynomial regression analysis was performed in SigmaPlot 10.0 (Systat Software Inc., San Jose, CA, USA).
The model was validated by leave-one-out (LOO) cross validation. During the LOO process, each group was progressively and alternately held out for model validation, while the remaining groups were used for model construction. The appropriateness of the estimation of PI occurrence and prediction of the NDVI-based VIs was tested based on relative error (RE) values, where:
in which the Ai and Pi are the actual and predicted value of the ith data point and n is the number of data points. Estimations were considered excellent if RE is <10%, good between 10% and 20%, fair between 20% and 30%, and poor if it is >30% [34,36].
3. Results and Discussion
3.1. PI Determination through Manual Dissection
The PI of each group was observed through manual dissection. The actual determination of PI for group 1 was 60 DAT (GDD = 771.3 °C), group 2 was 56 DAT (GDD = 713.9 °C), group 3 was 57 DAT (GDD = 783.4 °C), and group 4 was 55 DAT (GDD = 791.1 °C) (Table 2). Group 1 encountered low temperature during the tillering stage. As a result, the required DAT of group 1 for PI was five days longer than group 4, which was thoroughly grown under warm conditions. Besides this, group 2 encountered low temperature before the tillering stage. Consequently, the required GDD of group 2 for PI was approximately 80°C lower than group 4. These biases indicated that the DAT and GDD are both largely affected by weather variability.

Table 2.
Observation of PI in each group.
3.2. NDVI-Based VIs Selection
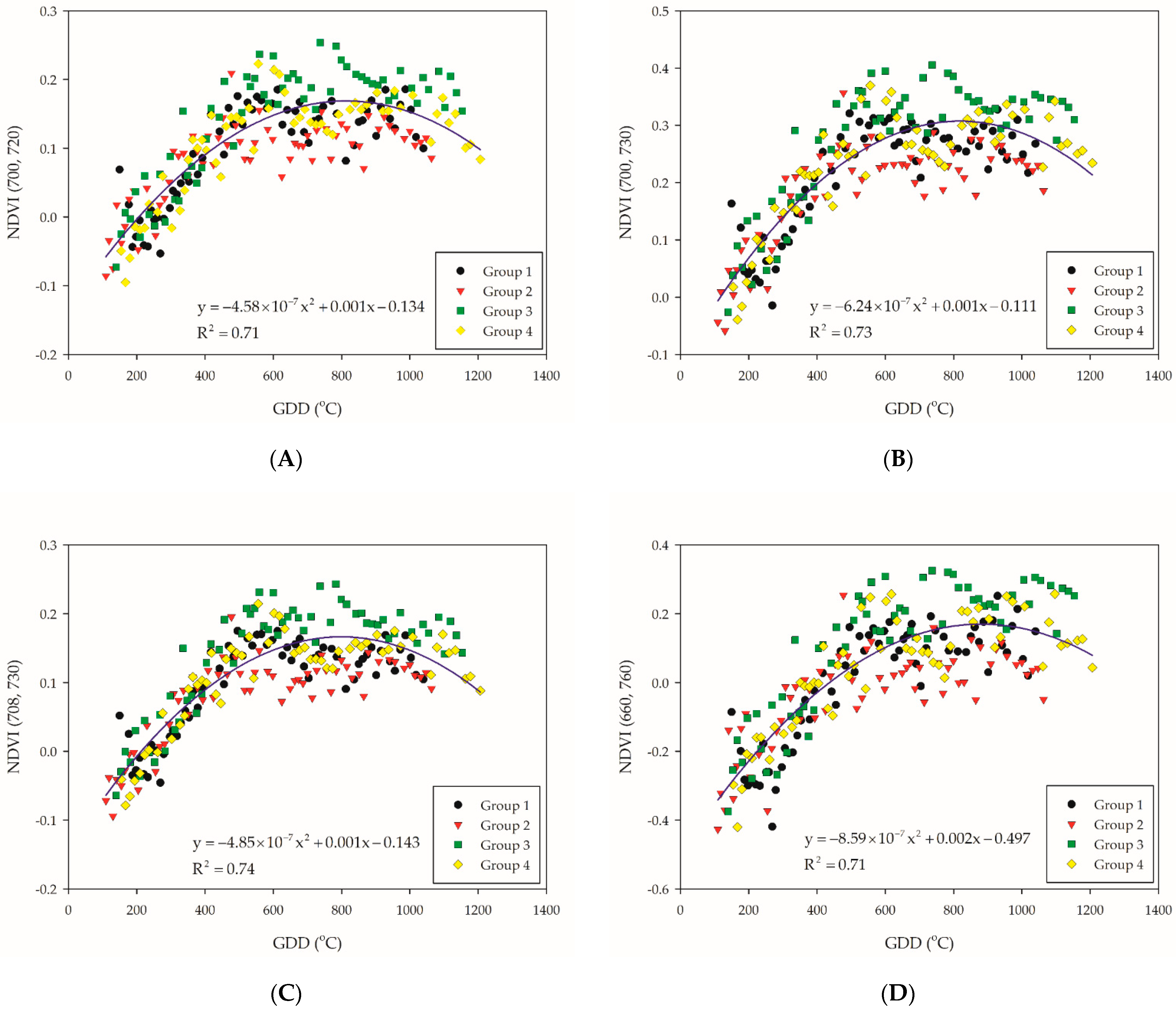
All samples were regressed with quadratic polynomial model, as we assumed that the trend of this model (when the slope is equal to zero) might be correlated with the entrance of the PI stage. Considering the ability of explanation, NDVI (700, 720), NDVI (700, 730), NDVI (708, 730), NDVI (660, 760), NDVI (700, 760), NDVI (708, 760), and NDVI (708, 800) were selected by the R2 value that are above 0.7 of quadratic polynomial regression model (Table 3). On the other hand, all the NDVI-based VIs of groups were stratified, where the distribution of groups 1 and 2 were lower than groups 3 and 4 (Figure 3A–G). The reason could be that group 1 encountered low temperature during the tillering stage, while group 2 encountered low temperature before the tillering stage. A preceding study indicated that more uniquely expressed proteins were found at 20/12 °C (day/night) and frequently alternating stress/non-stress temperature changes, leading the rice plant to complex stress conditions [37]. This also indicated that cold stress at an early stage would have an irreversible impact to rice plants, since the NDVI-based VIs of groups 1 and 2 remained lower than that of groups 3 and 4.

Table 3.
Recombination and selection of NDVI-based VIs from reference wavelengths. The calculation was [(λa − λb)/(λa + λb)], and the R2 values of quadratic polynomial regression models were recorded.
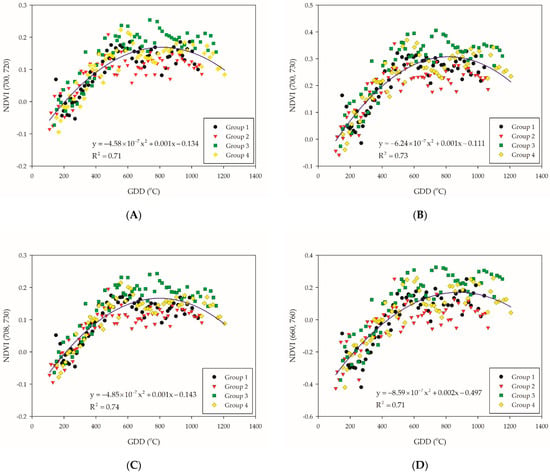
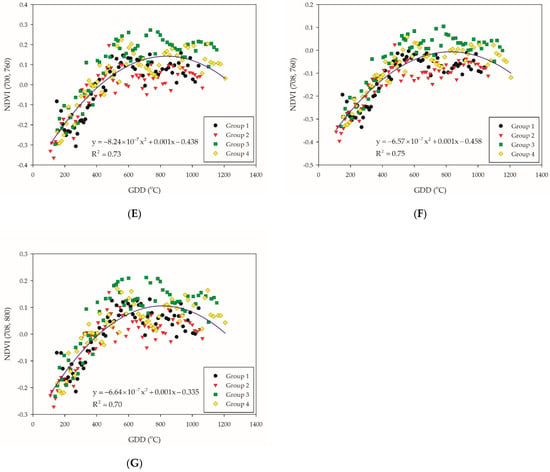
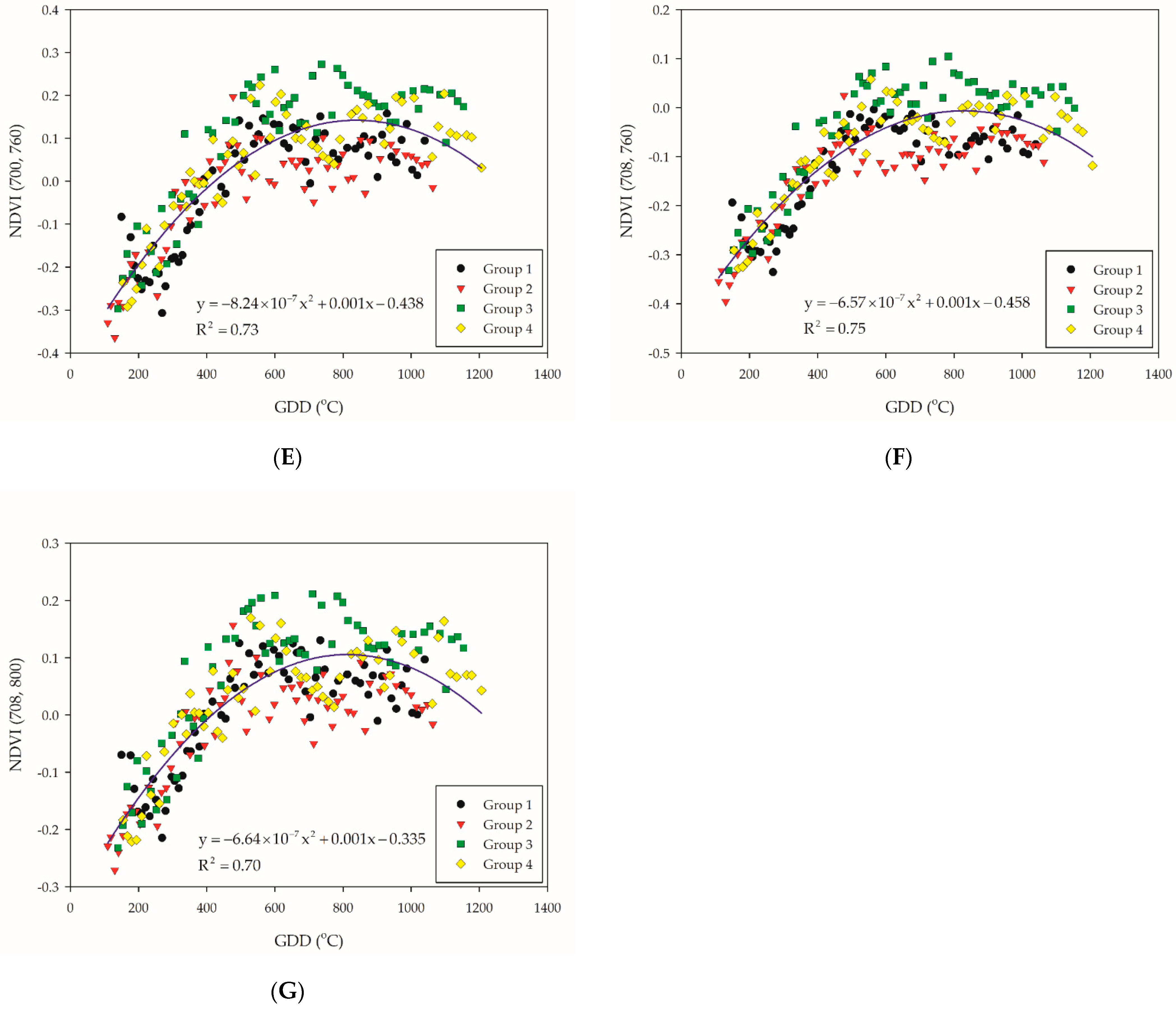
Figure 3.
Illustration for the selected NDVI-based VI combination: (A) NDVI (700, 720); (B) NDVI (700, 730); (C) NDVI (708, 730); (D) NDVI (660, 760); (E) NDVI (700, 760); (F) NDVI (708, 760); and (G) NDVI (708, 800). The blue solid line indicate the quadratic polynomial regression line. The black, red, green, and yellow symbols indicate the NDVI-based VIs data of each group.
3.3. Estimation of PI Occurrence through First-Order Differentiation
The seven NDVI-based VIs were then tested with first-order differentiation in each group to compare the estimated GDD with the actual GDD of PI occurrence (Table 4). Due to the excellent estimation of GDDs for PI occurrence (RE < 10%), NDVI (700, 720), NDVI (700, 730), NDVI (708, 730), NDVI (700, 760), NDVI (708, 760), and NDVI (708, 800) were considered as the suitable NDVI-based VIs for PI determination. Among these combinations, NDVI (708, 760), which was proposed by Tan et al. (2018), coincidentally had the lowest RE value (RE = 5.63%) [18]. This indicated that the entrance of the reproductive phase might have influenced the photosynthetic capacity of the rice plant, such as a lower rate of crop growth and absorption of photosynthetically active radiation (PAR). The other suitable combinations of NDVI-based VIs have not been researched in previous studies.

Table 4.
Seven selected NDVI-based VIs were tested by first-order differentiation in each group.
3.4. Leave-One-Out (LOO) Cross-Validation
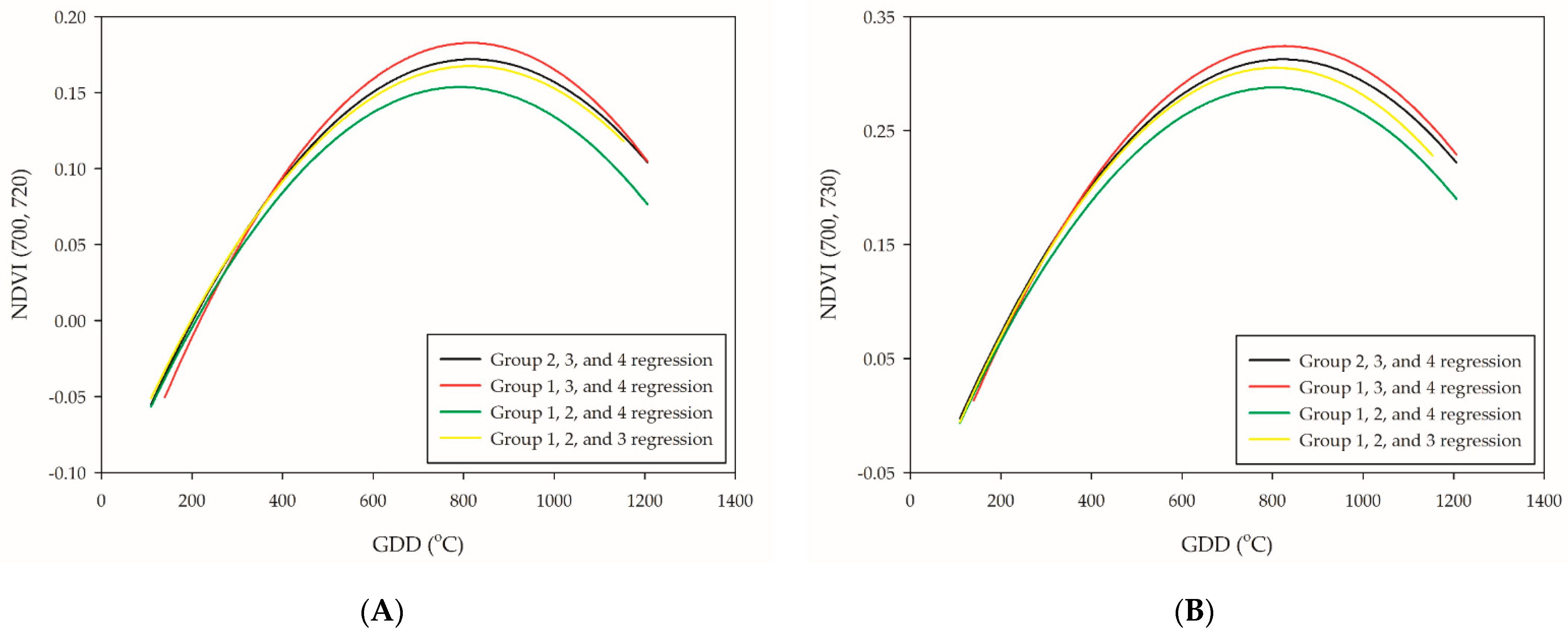
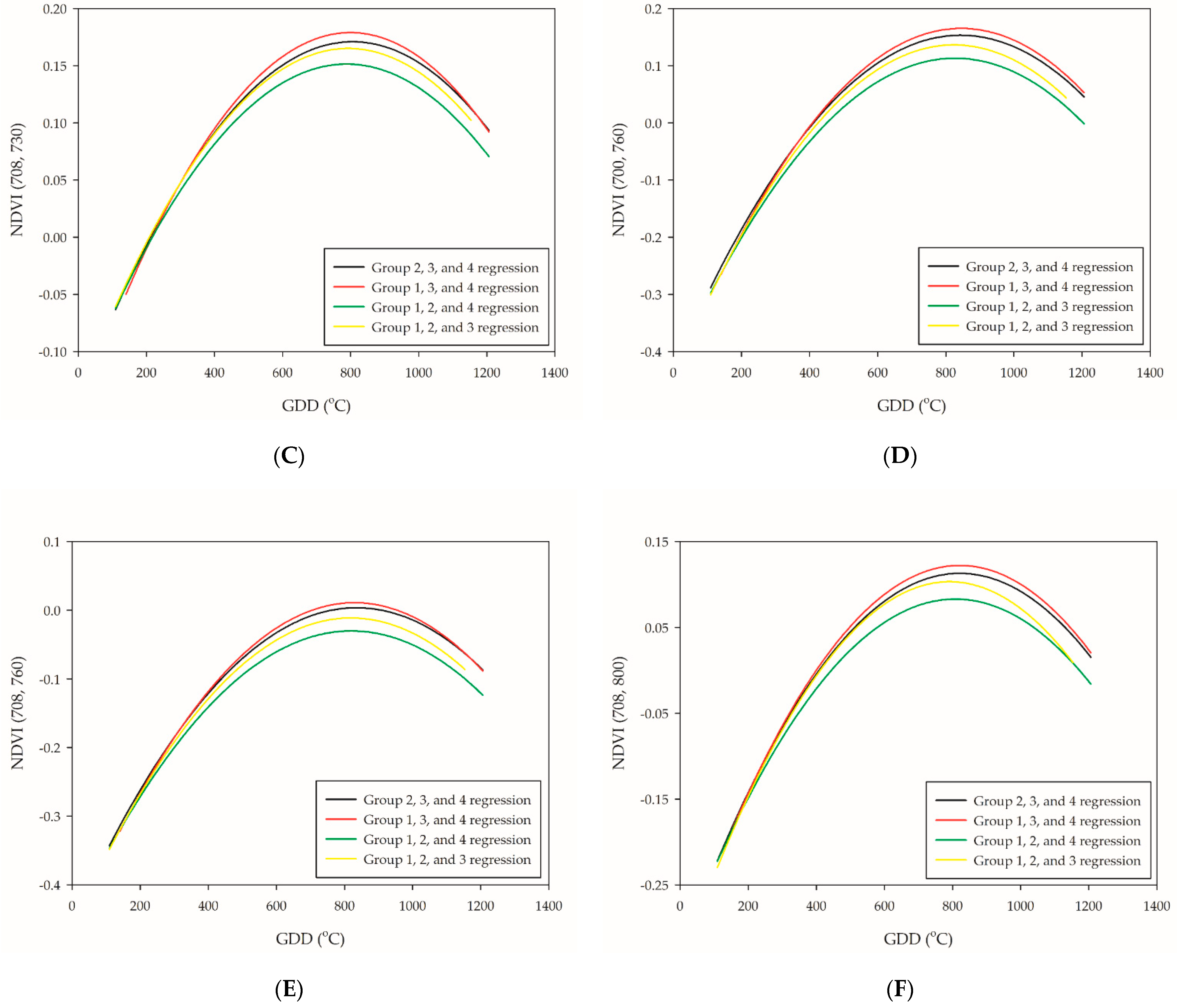
The leave-one-out cross-validation procedure was subsequently performed to test the accuracy of the quadratic polynomial regression of NDVI (700, 720), NDVI (700, 730), NDVI (708, 730), NDVI (700, 760), NDVI (708, 760), and NDVI (708, 800). The LOO regression line of groups 1, 3, and 4 showed the highest plateau, while groups 1, 2, and 4 the lowest (Figure 4A–F). Due to the great variabilities of the plateau distribution, neither setting up an absolute threshold value nor extended the range for the PI occurrence determination seems to be an ideal method. Although the plateau of regression curve between groups was different, the trend of the slopes was relatively similar. This was in agreement with preceding research [38]; however, the plateau of the curve in this study was different, which was at the PI stage rather than the booting-heading stage. The stability between the trend of the slopes and PI occurrence were tested through first-order differentiation in the leave-one-out cross-validation model. The results showed that NDVI (700, 720) was the most reliable NDVI-based VIs for PI determination since it had the lowest RE value (RE = 4.9%). A previous study represented NDVI application on wheat phenology monitoring and obtained satisfying results that achieved the lowest error of 4.61 days during the jointing stage [39]. Our study performed a slightly better approach with the application of NDVI (700, 720) with a quadratic polynomial regression model, which represented a maximum error of 68.9 GDDs (approximately three days) (Table 5). The others, NDVI (700, 730), NDVI (708, 730), and NDVI (708, 800), are potential candidates that are competent for PI assessment (Table 5). At this point, the relationship between NDVI-based VIs and PI occurrence could be confirmed through the first-order differentiation of quadratic polynomial regression model.
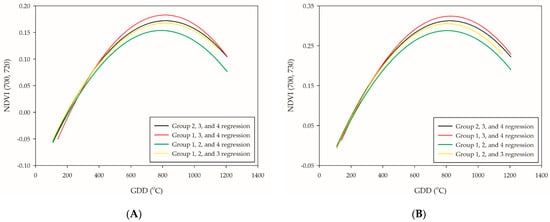
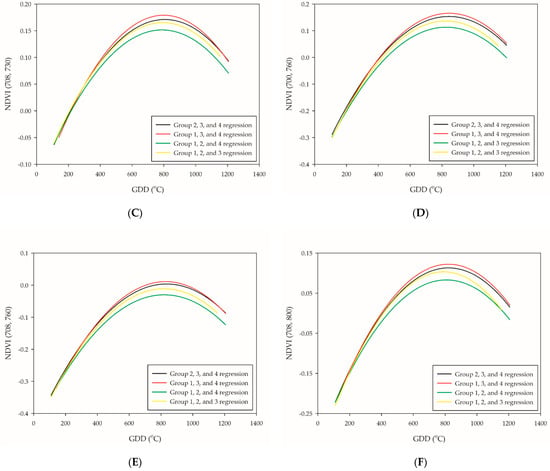
Figure 4.
Leave-one-out cross-validation of the six selected PI-related NDVI-based VIs: (A) NDVI (700, 720); (B) NDVI (700, 730); (C) NDVI (708, 730); (D) NDVI (700, 760); (E) NDVI (708, 760); and (F) NDVI (708, 800). The black line is the validation of group 1, the red line is the validation of group 2, the green line is the validation of group 3, and the yellow line is the validation of group 4.

Table 5.
Leave-one-out cross-validation of the six selected PI-related NDVI-based VIs by first-order differentiation.
4. Conclusions
This study revealed the relationship between rice panicle initiation and NDVI-based VIs by the first-order differentiation of quadratic polynomial regression. The results showed that NDVI (700, 720), NDVI (700, 730), NDVI (708, 730), NDVI (700, 760), NDVI (708, 760), and NDVI (708, 800) are potential candidates to determine PI stage. Although the observed values had great variabilities between groups, the trend is stable—when the population of the target reached their plateau of NDVI-based VIs distribution, PI occurred. Due to the synchronistic correlation between PI and plateau of the trend, the PI estimation might be achieved through determining the increasing proportional of saturated values of NDVI-based VIs. Cooperating with the UAV-mounted multispectral or hyperspectral camera and pixel-based analysis of the region of interest, NDVI-based VIs could be a useful indicator to assess rice plant PI stage, thus optimizing the management of fertilization practice.
Author Contributions
Conceptualization, J.-K.L. and W.-S.L.; Investigation, J.-K.L.; Methodology, J.-K.L.; Resources, W.-S.L.; Writing—original draft preparation, J.-K.L.; Writing—review and editing, J.-K.L. and W.-S.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially funded by the Ministry of Science and Technology (MOST), Taiwan (grant number MOST 110-2637-B-020-005).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors wish to thank the collaborators for their help on the study-site maintenance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ghaley, B.B. Uptake and utilization of 5-split nitrogen topdressing in an improved and a traditional rice cultivar in the Bhutan Highlands. Exp. Agric. 2012, 48, 536–550. [Google Scholar] [CrossRef]
- Inamura, T.; Hamada, H.; Iida, K.; Umeda, M. Correlation of the amount of nitrogen accumulated in the aboveground biomass at panicle initiation and nitrogen content of soil with the nitrogen uptake by lowland rice during the period from panicle initiation to heading. Plant Prod. Sci. 2003, 6, 302–308. [Google Scholar] [CrossRef][Green Version]
- Turner, F.T.; Jund, M.F. Chlorophyll meter to predict nitrogen topdress requirement for semidwarf rice. Agron. J. 1991, 83, 926–928. [Google Scholar] [CrossRef]
- Counce, P.A.; Keisling, T.C.; Mitchell, A.J. A Uniform, Objective, and Adaptive System for Expressing Rice Development. Crop Sci. 2000, 40, 436–443. [Google Scholar] [CrossRef]
- Moldenhauer, K.; Counce, P.; Hardke, J. Rice growth and development. In Rice Production Handbook; Hardke, J., Ed.; University of Arkansas Division of Agriculture: Little Rock, AR, USA, 2001; Volume 192, pp. 7–14. [Google Scholar]
- Ellis, R.H.; Qi, A.; Summerfield, R.J.; Roberts, E.H. Rates of leaf appearance and panicle development in rice (Oryza sativa L.): A comparison at three temperatures. Agric. For. Meteorol. 1993, 66, 129–138. [Google Scholar] [CrossRef]
- Gilmore, E.C.; Rogers, J.S. Heat units as a method of measuring maturity in corn. Agron. J. 1958, 50, 611–615. [Google Scholar] [CrossRef]
- Nielsen, D.; Hinkle, S. Field evaluation of basal crop coefficients for corn based on growing degree days, growth stage, or time. Trans. ASAE 1996, 39, 97–103. [Google Scholar] [CrossRef]
- Snyder, R.L.; Spano, D.; Cesaraccio, C.; Duce, P. Determining degree-day thresholds from field observations. Int. J. Biometeorol. 1999, 42, 177–182. [Google Scholar] [CrossRef]
- Angel, J.R.; Widhalm, M.; Todey, D.; Massey, R.; Biehl, L. The U2U corn growing degree day tool: Tracking corn growth across the US corn belt. Clim. Risk Manag. 2017, 15, 73–81. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring vegetation systems in the Great Plains with ERTS. NASA [Spec. Publ.] SP 1974, 351, 309. [Google Scholar]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Myneni, R.B.; Williams, D.L. On the relationship between FAPAR and NDVI. Remote Sens. Environ. 1994, 49, 200–211. [Google Scholar] [CrossRef]
- Gamon, J.A.; Field, C.B.; Goulden, M.L.; Griffin, K.L.; Hartley, A.E.; Joel, G.; Penuelas, J.; Valentini, R. Relationships between NDVI, canopy structure, and photosynthesis in three Californian vegetation types. Ecol. Appl. 1995, 5, 28–41. [Google Scholar] [CrossRef]
- Carlson, T.N.; Ripley, D.A. On the relation between NDVI, fractional vegetation cover, and leaf area index. Remote Sens. Environ. 1997, 62, 241–252. [Google Scholar] [CrossRef]
- Sims, D.A.; Gamon, J.A. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Zhou, X.; Zheng, H.B.; Xu, X.Q.; He, J.Y.; Ge, X.K.; Yao, X.; Cheng, T.; Zhu, Y.; Cao, W.X.; Tian, Y.C. Predicting grain yield in rice using multi-temporal vegetation indices from UAV-based multispectral and digital imagery. ISPRS J. Photogramm. Remote Sens. 2017, 130, 246–255. [Google Scholar] [CrossRef]
- Tan, C.; Wang, D.; Zhou, J.; Du, Y.; Luo, M.; Zhang, Y.; Guo, W. Remotely assessing fraction of photosynthetically active radiation (FPAR) for wheat canopies based on hyperspectral vegetation indexes. Front. Plant Sci. 2018, 9, 776. [Google Scholar] [CrossRef] [PubMed]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Tanaka, S.; Kawamura, K.; Maki, M.; Muramoto, Y.; Yoshida, K.; Akiyama, T. Spectral index for quantifying leaf area index of winter wheat by field hyperspectral measurements: A case study in Gifu Prefecture, Central Japan. Remote Sens. 2015, 7, 5329–5346. [Google Scholar] [CrossRef]
- Yang, G.; Liu, J.; Zhao, C.; Li, Z.; Huang, Y.; Yu, H.; Xu, B.; Yang, X.; Zhu, D.; Zhang, X.; et al. Unmanned aerial vehicle remote sensing for field-based crop phenotyping: Current status and perspectives. Front. Plant Sci. 2017, 8, 1111. [Google Scholar] [CrossRef]
- Peng, S.; Buresh, R.J.; Huang, J.; Zhong, X.; Zou, Y.; Yang, J.; Wang, G.; Liu, Y.; Hu, R.; Tang, Q.; et al. Improving nitrogen fertilization in rice by sitespecific N management. A review. Agron. Sustain. Dev. 2010, 30, 649–656. [Google Scholar] [CrossRef]
- Xue, L.; Li, G.; Qin, X.; Yang, L.; Zhang, H. Topdressing nitrogen recommendation for early rice with an active sensor in south China. Precis. Agric. 2014, 15, 95–110. [Google Scholar] [CrossRef]
- Ali, A.M.; Thind, H.S.; Varinderpal, S.; Bijay, S. A framework for refining nitrogen management in dry direct-seeded rice using GreenSeeker™ optical sensor. Comput. Electron. Agric. 2015, 110, 114–120. [Google Scholar] [CrossRef]
- Onoyama, H.; Ryu, C.; Suguri, M.; Iida, M. Nitrogen prediction model of rice plant at panicle initiation stage using ground-based hyperspectral imaging: Growing degree-days integrated model. Precis. Agric. 2015, 16, 558–570. [Google Scholar] [CrossRef]
- Cao, Q.; Miao, Y.; Shen, J.; Yu, W.; Yuan, F.; Cheng, S.; Huang, S.; Wang, H.; Yang, W.; Liu, F. Improving in-season estimation of rice yield potential and responsiveness to topdressing nitrogen application with Crop Circle active crop canopy sensor. Precis. Agric. 2016, 17, 136–154. [Google Scholar] [CrossRef]
- Jiang, R.; Sanchez-Azofeifa, A.; Laakso, K.; Wang, P.; Xu, Y.; Zhou, Z.; Luo, X.; Lan, Y.; Zhao, G.; Chen, X. UAV-based partially sampling system for rapid NDVI mapping in the evaluation of rice nitrogen use efficiency. J. Cleaner Prod. 2021, 289, 125705. [Google Scholar] [CrossRef]
- Phyu, P.; Islam, M.R.; Sta Cruz, P.C.; Collard, B.C.Y.; Kato, Y. Use of NDVI for indirect selection of high yield in tropical rice breeding. Euphytica 2020, 216, 74. [Google Scholar] [CrossRef]
- Son, N.-T.; Chen, C.-F.; Chen, C.-R.; Guo, H.-Y.; Cheng, Y.-S.; Chen, S.-L.; Lin, H.-S.; Chen, S.-H. Machine learning approaches for rice crop yield predictions using time-series satellite data in Taiwan. Int. J. Remote Sens. 2020, 41, 7868–7888. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, F.-C.; Jing, Y.-S.; Jiang, X.-D.; Yang, S.-B.; Han, X.-M. Multi-temporal detection of rice phenological stages using canopy spectrum. Rice Sci. 2014, 21, 108–115. [Google Scholar] [CrossRef]
- Yu, F.; Xu, T.; Cao, Y.; Yang, G.; Du, W.; Wang, S. Models for estimating the leaf NDVI of japonica rice on a canopy scale by combining canopy NDVI and multisource environmental data in Northeast China. Int. J. Agric. Biol. Eng. 2016, 9, 132–142. [Google Scholar]
- Zheng, H.; Cheng, T.; Li, D.; Yao, X.; Tian, Y.; Cao, W.; Zhu, Y. Combining unmanned aerial vehicle (UAV)-based multispectral imagery and ground-based hyperspectral data for plant nitrogen concentration estimation in rice. Front. Plant Sci. 2018, 9, 936. [Google Scholar] [CrossRef]
- Lu, J.; Miao, Y.; Shi, W.; Li, J.; Yuan, F. Evaluating different approaches to non-destructive nitrogen status diagnosis of rice using portable RapidSCAN active canopy sensor. Sci. Rep. 2017, 7, 14073. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, G.R.; Pandey, R.N.; Sahoo, R.N.; Gupta, V.K.; Datta, S.C.; Kumar, D. Monitoring nitrogen, phosphorus and sulphur in hybrid rice (Oryza sativa L.) using hyperspectral remote sensing. Precis. Agric. 2017, 18, 736–761. [Google Scholar] [CrossRef]
- McMaster, G.S.; Wilhelm, W.W. Growing degree-days: One equation, two interpretations. Agric. For. Meteorol. 1997, 87, 291–300. [Google Scholar] [CrossRef]
- Jamieson, P.D.; Porter, J.R.; Wilson, D.R. A test of the computer simulation model ARCWHEAT1 on wheat crops grown in New Zealand. Field Crops Res. 1991, 27, 337–350. [Google Scholar] [CrossRef]
- Gammulla, C.G.; Pascovici, D.; Atwell, B.J.; Haynes, P.A. Differential proteomic response of rice (Oryza sativa) leaves exposed to high-and low-temperature stress. Proteomics 2011, 11, 2839–2850. [Google Scholar] [CrossRef]
- Liu, X.; Ferguson, R.B.; Zheng, H.; Cao, Q.; Tian, Y.; Cao, W.; Zhu, Y. Using an active-optical sensor to develop an optimal NDVI dynamic model for high-yield rice production (Yangtze, China). Sensors 2017, 17, 672. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Ma, X.; Wang, K.; Cheng, T.; Tian, Y.; Wang, J.; Zhu, Y.; Hu, Y.; Niu, Q.; Gui, L.; et al. Detection of phenology using an improved shape model on time-series vegetation index in wheat. Comput. Electron. Agric. 2020, 173, 105398. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).