Decellularized Skeletal Muscles Support the Generation of In Vitro Neuromuscular Tissue Models
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Tissue Decellularization
2.3. Cell Culture
2.4. Repopulation of the Scaffold and Co-Culture with oSpC
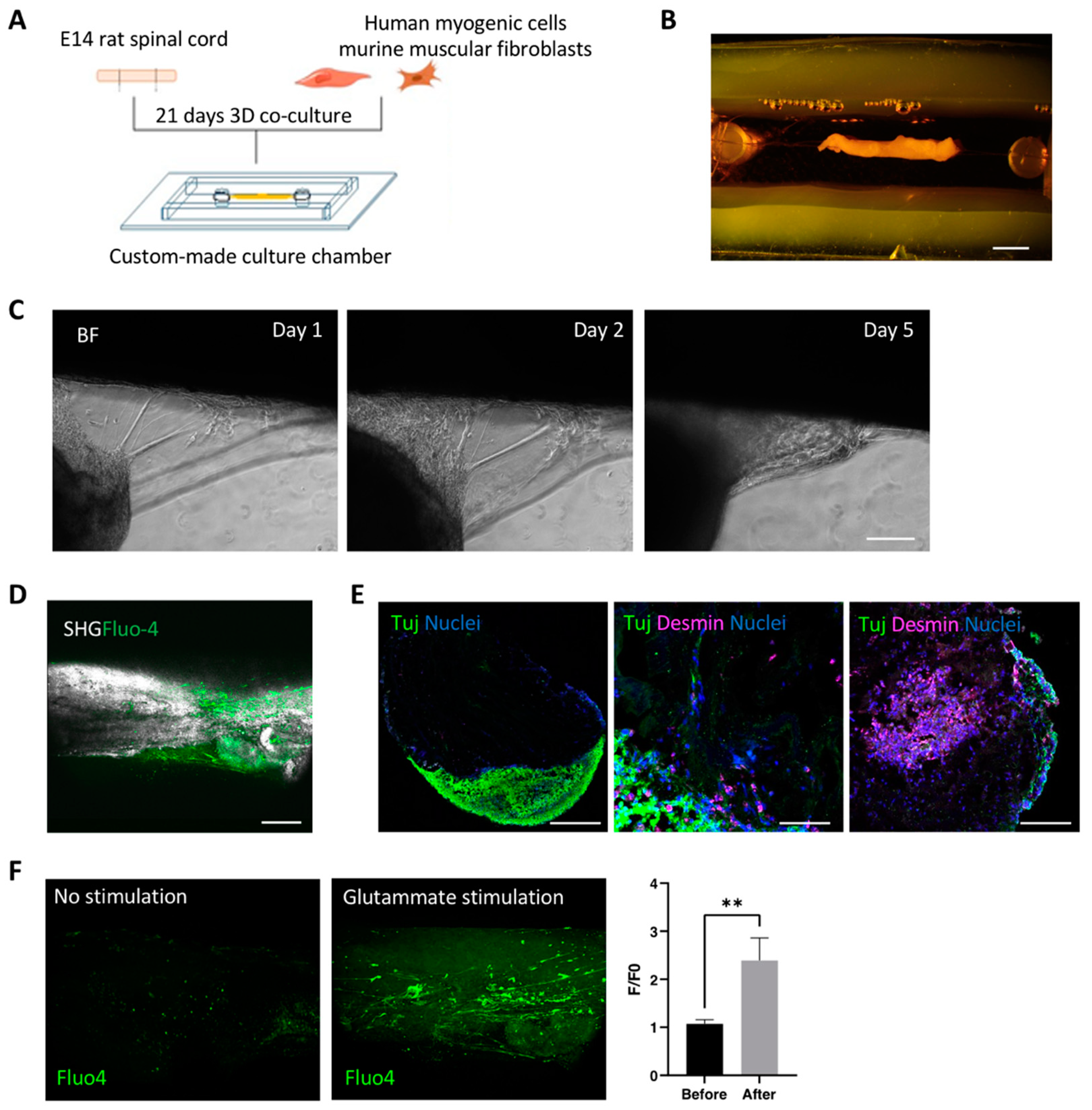
2.5. Custom-Made Culture Chamber
2.6. Cell Viability Assay
2.7. Immunofluorescence Characterization
2.8. Calcium Imaging
2.9. Image Acquisition and Analysis
2.10. Statistical Analysis
3. Results
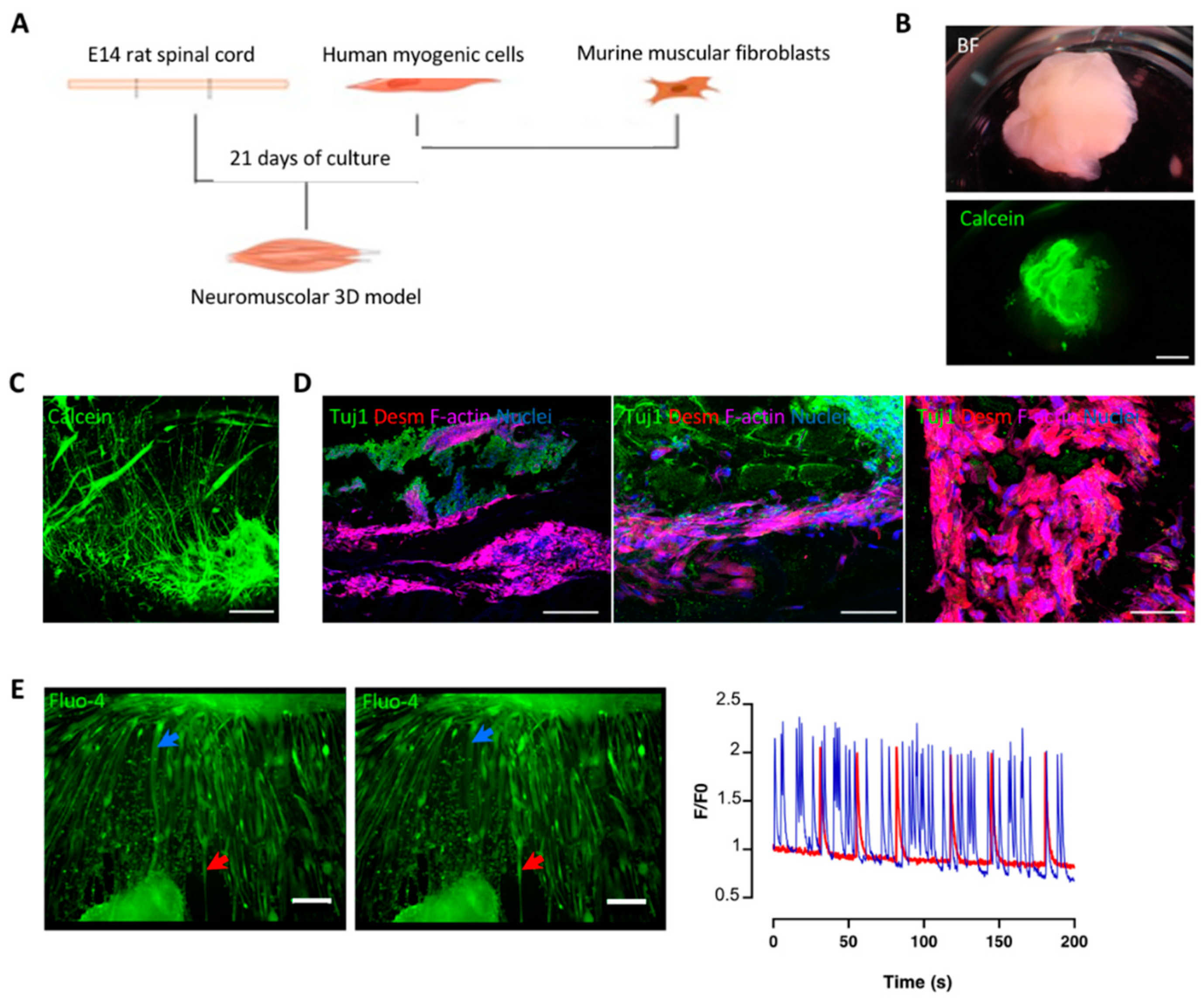
3.1. Decellularized Muscles Allow 3D Co-Culture of Human Myogenic Cells and oSpC Sections
3.2. Optimization and Characterization of 3D Neuromuscular Cultures
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Urciuolo, A.; De Coppi, P. Decellularized Tissue for Muscle Regeneration. Int. J. Mol. Sci. 2018, 19, 2392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Costa, J.M.; Fernández-Garibay, X.; Velasco-Mallorquí, F.; Ramón-Azcón, J. Bioengineered in vitro skeletal muscles as new tools for muscular dystrophies preclinical studies. J. Tissue Eng. 2021, 12, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Quigley, A.; Ngan, C.; Firipis, K.; O’Connell, C.D.; Pirogova, E.; Moulton, S.E.; Williams, R.J.; Kapsa, R.M. Towards bioengineered skeletal muscle: Recent developments in vitro and in vivo. Essays Biochem. 2021, 65, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Mendibil, U.; Ruiz-Hernandez, R.; Retegi-Carrion, S.; Garcia-Urquia, N.; Olalde-Graells, B.; Abarrategi, A. Tissue-Specific Decellularization Methods: Rationale and Strategies to Achieve Regenerative Compounds. Int. J. Mol. Sci. 2020, 21, 5447. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Gordián-Vélez, W.J.; Ledebur, H.C.; Mourkioti, F.; Rompolas, P.; Chen, H.I.; Serruya, M.D.; Cullen, D.K. Innervation: The missing link for biofabricated tissues and organs. NPJ Regen. Med. 2020, 5, 11. [Google Scholar] [CrossRef]
- Fishman, J.M.; Lowdell, M.W.; Urbani, L.; Ansari, T.; Burns, A.J.; Turmaine, M.; North, J.; Sibbons, P.; Seifalian, A.M.; Wood, K.J.; et al. Immunomodulatory effect of a decellularized skeletal muscle scaffold in a discordant xenotransplantation model. Proc. Natl. Acad. Sci. USA 2013, 110, 14360–14365. [Google Scholar] [CrossRef] [Green Version]
- Sun, P.; Wang, Z.; Liu, W.; Li, M.; Wei, S.; Xu, Y.; Qiao, Z.; Wang, W.; Fu, Y.; Bai, H.; et al. Programmed death-1 mediates venous neointimal hyperplasia in humans and rats. Aging 2021, 13, 16656–16666. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Sun, P.; Wei, S.; Xie, B.; Li, M.; Xu, Y.; Wang, W.; Liu, Y.; Zhang, L.; Wu, H.; et al. A novel intramural TGF β 1 hydrogel delivery method to decrease murine abdominal aortic aneurysm and rat aortic pseudoaneurysm formation and progression. Biomed. Pharmacother. 2021, 137, 111296. [Google Scholar] [CrossRef]
- Yao, Q.; Zheng, Y.-W.; Lan, Q.-H.; Kou, L.; Xu, H.-L.; Zhao, Y.-Z. Recent development and biomedical applications of decellularized extracellular matrix biomaterials. Mater. Sci. Eng. C 2019, 104, 109942. [Google Scholar] [CrossRef]
- Kim, J.H.; Seol, Y.-J.; Ko, I.K.; Kang, H.-W.; Lee, Y.K.; Yoo, J.J.; Atala, A.; Lee, S.J. 3D Bioprinted Human Skeletal Muscle Constructs for Muscle Function Restoration. Sci. Rep. 2018, 8, 12307. [Google Scholar] [CrossRef] [Green Version]
- Gresham, R.C.; Bahney, C.S.; Leach, J.K. Growth factor delivery using extracellular matrix-mimicking substrates for musculoskeletal tissue engineering and repair. Bioact. Mater. 2020, 6, 1945–1956. [Google Scholar] [CrossRef]
- Lee, H.; Ju, Y.M.; Kim, I.; Elsangeedy, E.; Lee, J.H.; Yoo, J.J.; Atala, A.; Lee, S.J. A novel decellularized skeletal muscle-derived ECM scaffolding system for in situ muscle regeneration. Methods 2020, 171, 77–85. [Google Scholar] [CrossRef]
- Shapiro, L.; Elsangeedy, E.; Lee, H.; Atala, A.; Yoo, J.J.; Lee, S.J.J.; Ju, Y.M. In vitro evaluation of functionalized decellularized muscle scaffold for in situ skeletal muscle regeneration. Biomed. Mater. 2019, 14, 045015. [Google Scholar] [CrossRef]
- Wassenaar, J.W.; Boss, G.R.; Christman, K.L. Decellularized skeletal muscle as an invitro model for studying drug-extracellular matrix interactions. Biomaterials 2015, 64, 108–114. [Google Scholar] [CrossRef] [Green Version]
- Rossi, C.A.; Flaibani, M.; Blaauw, B.; Pozzobon, M.; Figallo, E.; Reggiani, C.; Vitiello, L.; Elvassore, N.; De Coppi, P. In vivo tissue engineering of functional skeletal muscle by freshly isolated satellite cells embedded in a photopolymerizable hydrogel. FASEB J. 2011, 25, 2296–2304. [Google Scholar] [CrossRef]
- Boso, D.; Carraro, E.; Maghin, E.; Todros, S.; Dedja, A.; Giomo, M.; Elvassore, N.; De Coppi, P.; Pavan, P.; Piccoli, M. Porcine Decellularized Diaphragm Hydrogel: A New Option for Skeletal Muscle Malformations. Biomedicines 2021, 9, 709. [Google Scholar] [CrossRef]
- Raffa, P.; Scattolini, V.; Gerli, M.F.M.; Perin, S.; Cui, M.; De Coppi, P.; Elvassore, N.; Caccin, P.; Luni, C.; Urciuolo, A. Decellularized skeletal muscles display neurotrophic effects in three-dimensional organotypic cultures. STEM CELLS Transl. Med. 2020, 9, 1233–1243. [Google Scholar] [CrossRef] [PubMed]
- Carlson, B.M. The biology of long-term denervated skeletal muscle. Eur. J. Transl. Myol. 2014, 24, 5–11. [Google Scholar] [CrossRef]
- Gilbert-Honick, J.; Grayson, W. Vascularized and Innervated Skeletal Muscle Tissue Engineering. Adv. Health Mater. 2019, 9, e1900626. [Google Scholar] [CrossRef]
- Bakooshli, M.A.; Lippmann, E.S.; Mulcahy, B.; Iyer, N.; Nguyen, C.T.; Tung, K.A.; Stewart, B.; Dorpel, H.V.D.; Fuehrmann, T.; Shoichet, M.; et al. A 3D culture model of innervated human skeletal muscle enables studies of the adult neuromuscular junction. eLife 2019, 8, e44530. [Google Scholar] [CrossRef] [PubMed]
- Gilbert-Honick, J.; Iyer, S.R.; Somers, S.M.; Takasuka, H.; Lovering, R.M.; Wagner, K.R.; Mao, H.-Q.; Grayson, W.L. Engineering 3D skeletal muscle primed for neuromuscular regeneration following volumetric muscle loss. Biomaterials 2020, 255, 120154. [Google Scholar] [CrossRef] [PubMed]
- Urciuolo, A.; Urbani, L.; Perin, S.; Maghsoudlou, P.; Scottoni, F.; Gjinovci, A.; Collins-Hooper, H.; Loukogeorgakis, S.; Tyraskis, A.; Torelli, S.; et al. Decellularised skeletal muscles allow functional muscle regeneration by promoting host cell migration. Sci. Rep. 2018, 8, 8398. [Google Scholar] [CrossRef] [Green Version]
- Urciuolo, A.; Quarta, M.; Morbidoni, V.; Gattazzo, F.; Molon, S.; Grumati, P.; Montemurro, F.; Tedesco, F.S.; Blaauw, B.; Cossu, G.; et al. Collagen VI regulates satellite cell self-renewal and muscle regeneration. Nat. Commun. 2013, 4, 1964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rotundo, R.L. The NMJ as a model synapse: New perspectives on formation, synaptic transmission and maintenance: Acetylcholinesterase at the neuromuscular junction. Neurosci. Lett. 2020, 735, 135157. [Google Scholar] [CrossRef]
- Urbani, L.; Camilli, C.; Phylactopoulos, D.-E.; Crowley, C.; Natarajan, D.; Scottoni, F.; Maghsoudlou, P.; McCann, C.J.; Pellegata, A.F.; Urciuolo, A.; et al. Multi-stage bioengineering of a layered oesophagus with in vitro expanded muscle and epithelial adult progenitors. Nat. Commun. 2018, 9, 4286. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raffa, P.; Easler, M.; Cecchinato, F.; Auletta, B.; Scattolini, V.; Perin, S.; Gerli, M.F.M.; Caccin, P.; Elvassore, N.; De Coppi, P.; et al. Decellularized Skeletal Muscles Support the Generation of In Vitro Neuromuscular Tissue Models. Appl. Sci. 2021, 11, 9485. https://doi.org/10.3390/app11209485
Raffa P, Easler M, Cecchinato F, Auletta B, Scattolini V, Perin S, Gerli MFM, Caccin P, Elvassore N, De Coppi P, et al. Decellularized Skeletal Muscles Support the Generation of In Vitro Neuromuscular Tissue Models. Applied Sciences. 2021; 11(20):9485. https://doi.org/10.3390/app11209485
Chicago/Turabian StyleRaffa, Paolo, Maria Easler, Francesca Cecchinato, Beatrice Auletta, Valentina Scattolini, Silvia Perin, Mattia Francesco Maria Gerli, Paola Caccin, Nicola Elvassore, Paolo De Coppi, and et al. 2021. "Decellularized Skeletal Muscles Support the Generation of In Vitro Neuromuscular Tissue Models" Applied Sciences 11, no. 20: 9485. https://doi.org/10.3390/app11209485
APA StyleRaffa, P., Easler, M., Cecchinato, F., Auletta, B., Scattolini, V., Perin, S., Gerli, M. F. M., Caccin, P., Elvassore, N., De Coppi, P., & Urciuolo, A. (2021). Decellularized Skeletal Muscles Support the Generation of In Vitro Neuromuscular Tissue Models. Applied Sciences, 11(20), 9485. https://doi.org/10.3390/app11209485






