Cytochrome cM Is Probably a Membrane Protein Similar to the C Subunit of the Bacterial Nitric Oxide Reductase
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Roncel, M.; Kirilovsky, D.; Guerrero, F.; Serrano, A.; Ortega, J.M. Photosynthetic cytochrome c550. Biochim. Biophys. Acta 2012, 1817, 1152–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malakhov, M.P.; Wada, H.; Los, D.A.; Semenenko, V.E.; Murata, N. A New Type of Cytochrome c from Synechocystis PCC6803. J. Plant Physiol. 1994, 144, 259–264. [Google Scholar] [CrossRef]
- Bernroitner, M.; Tangl, D.; Lucini, C.; Furtmüller, P.G.; Peschek, G.A.; Obinger, C. Cyanobacterial cytochrome cM: Probing its role as electron donor for CuA of cytochrome c oxidase. Biochim. Biophys. Acta Bioenerg. 2009, 1787, 135–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bialek, W.; Szczepaniak, A.; Kolesinski, P.; Kallas, T. Cryptic c6-like and cM cytochromes of cyanobacteria. In Cytochrome Complexes: Evolution, Structures, Energy Transduction, and Signaling; Kramer, W.A., T. Kallas, T., Eds.; Springer: Dordrecht, The Netherlamnds, 2016; pp. 713–734. [Google Scholar]
- Cho, Y.S.; Pakrasi, H.B.; Whitmarsh, J. Cytochrome cM from Synechocystis 6803. Detection in cells, expression in Escherichia coli, purification and physical characterization. Eur. J. Biochem. 2000, 267, 1068–1074. [Google Scholar] [CrossRef]
- Molina-Heredia, F.P.; Balme, A.; Hervás, M.; Navarro, J.A.; De la Rosa, M.A. A comparative structural and functional analysis of cytochrome cM, cytochrome c6 and plastocyanin from the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 2002, 517, 50–54. [Google Scholar] [CrossRef]
- Solymosi, D.; Nikkanen, L.; Muth-Pawlak, D.; Fitzpatrick, D.; Vasudevan, R.; Howe, C.J.; Lea-Smith, D.J.; Allahverdiyeva, Y. Cytochrome cM decreases photosynthesis under photomixotrophy in Synechocystis sp. PCC 6803. Plant Physiol. 2020, 183, 700–716. [Google Scholar] [CrossRef] [Green Version]
- Baers, L.L.; Breckels, L.M.; Mills, L.A.; Gatto, L.; Deery, M.J.; Stevens, T.J.; Howe, C.J.; Lilley, K.S.; Lea-Smith, D.J. Proteome mapping of a cyanobacterium reveals distinct compartment organization and cell-dispersed metabolism. Plant Physiol. 2019, 181, 1721–1738. [Google Scholar] [CrossRef] [Green Version]
- Malakhov, M.P.; Malakhova, O.A.; Murata, N. Balanced regulation of expression of the gene for cytochrome c(M) and that of genes for plastocyanin and cytochrome c6 in Synechocystis. FEBS Lett. 1999, 444, 281–284. [Google Scholar] [CrossRef] [Green Version]
- Pils, D.; Gregor, W.; Schmetterer, G. Evidence for in vivo activity of three distinct respiratory terminal oxidases in the cyanobacterium Synechocystis sp. strain PCC6803. FEMS Microbiol. Lett. 1997, 152, 83–88. [Google Scholar] [CrossRef]
- Manna, P.; Vermaas, W. Lumenal proteins involved in respiratory electron transport in the cyanobacterium Synechocystis sp. PCC6803. Plant. Mol. Biol 1997, 35, 407–416. [Google Scholar] [CrossRef]
- Shuvalov, V.A.; Allakhverdiev, A.I.; Sakamoto, A.; Malakhov, M.; Murata, N. Optical study of cytochrome cM formation in Synechocystis. IUBMB Life 2001, 51, 93–97. [Google Scholar] [CrossRef]
- Hiraide, Y.; Oshima, K.; Fujisawa, T.; Uesaka, K.; Hirose, Y.; Tsujimoto, R.; Yamamoto, H.; Okamoto, S.; Nakamura, Y.; Terauchi, K.; et al. Loss of cytochrome cM stimulates cyanobacterial heterotrophic growth in the dark. Plant Cell Physiol. 2015, 56, 334–345. [Google Scholar] [CrossRef]
- Furumichi, M.; Sato, Y.; Omata, T.; Ikeuchi, M.; Kanehisa, M. CYORF: Community Annotation of Cyanobacteria Genes. Genome Inf. 2002, 13, 402–403. [Google Scholar]
- Nakao, M.; Okamoto, S.; Kohara, M.; Fujishiro, T.; Fujisawa, T.; Sato, S.; Tabata, S.; Kaneko, T.; Nakamura, Y. CyanoBase: The cyanobacteria genome database update 2010. Nucleic Acids Res. 2009, 38, D379–D381. [Google Scholar] [CrossRef] [Green Version]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinf. 2008, 9, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hino, T.; Matsumoto, Y.; Nagano, S.; Sugimoto, H.; Fukumori, Y.; Murata, T.; Iwata, S.; Shiro, Y. Structural basis of biological N2O generation by bacterial nitric oxide reductase. Science 2010, 330, 1666–1670. [Google Scholar] [CrossRef] [PubMed]
- Molina-Heredia, F.P.; Hervás, M.; Navarro, J.A.; De la Rosa, M.A. Cloning and correct expression in Escherichia coli of the petE and petJ genes respectively encoding plastocyanin and cytochrome c6 from the cyanobacterium Anabaena sp. PCC 7119. Biochem. Biophys. Res. Commun. 1998, 243, 302–306. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Sosa, F.M.; Gil-Martínez, J.; Molina-Heredia, F.P. Cytochrome c6-like protein as a putative donor of electrons to photosystem I in the cyanobacterium Nostoc sp. PCC 7119. Photosynth. Res. 2011, 110, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Torrado, A.; Valladares, A.; Puerto-Galán, L.; Hervás, M.; Navarro, J.A.; Molina-Heredia, F.P. Cyt c6-3: A New Isoform of Photosynthetic Cyt c6 Exclusive to Heterocyst-Forming Cyanobacteria. Plant Cell Physiol. 2017, 58, 256–265. [Google Scholar] [PubMed] [Green Version]
- Büsch, A.; Friedrich, B.; Cramm, R. Characterization of the norB gene, encoding nitric oxide reductase, in the nondenitrifying cyanobacterium Synechocystis sp. strain PCC6803. Appl. Environ. Microbiol. 2002, 68, 668–672. [Google Scholar] [CrossRef] [Green Version]
- Murphy, M.P. Nitric oxide and cell death. Biochim. Biophys. Acta Bioenerg. 1999, 1411, 401–414. [Google Scholar] [CrossRef] [Green Version]
- Oubrie, A.; Gemeinhardt, S.; Field, S.; Marritt, S.; Thomson, A.J.; Saraste, M.; Richardson, D.J. Properties of a soluble domain of subunit c of a bacterial nitric oxide reductase. Biochemistry 2002, 41, 10858–10865. [Google Scholar] [CrossRef] [PubMed]
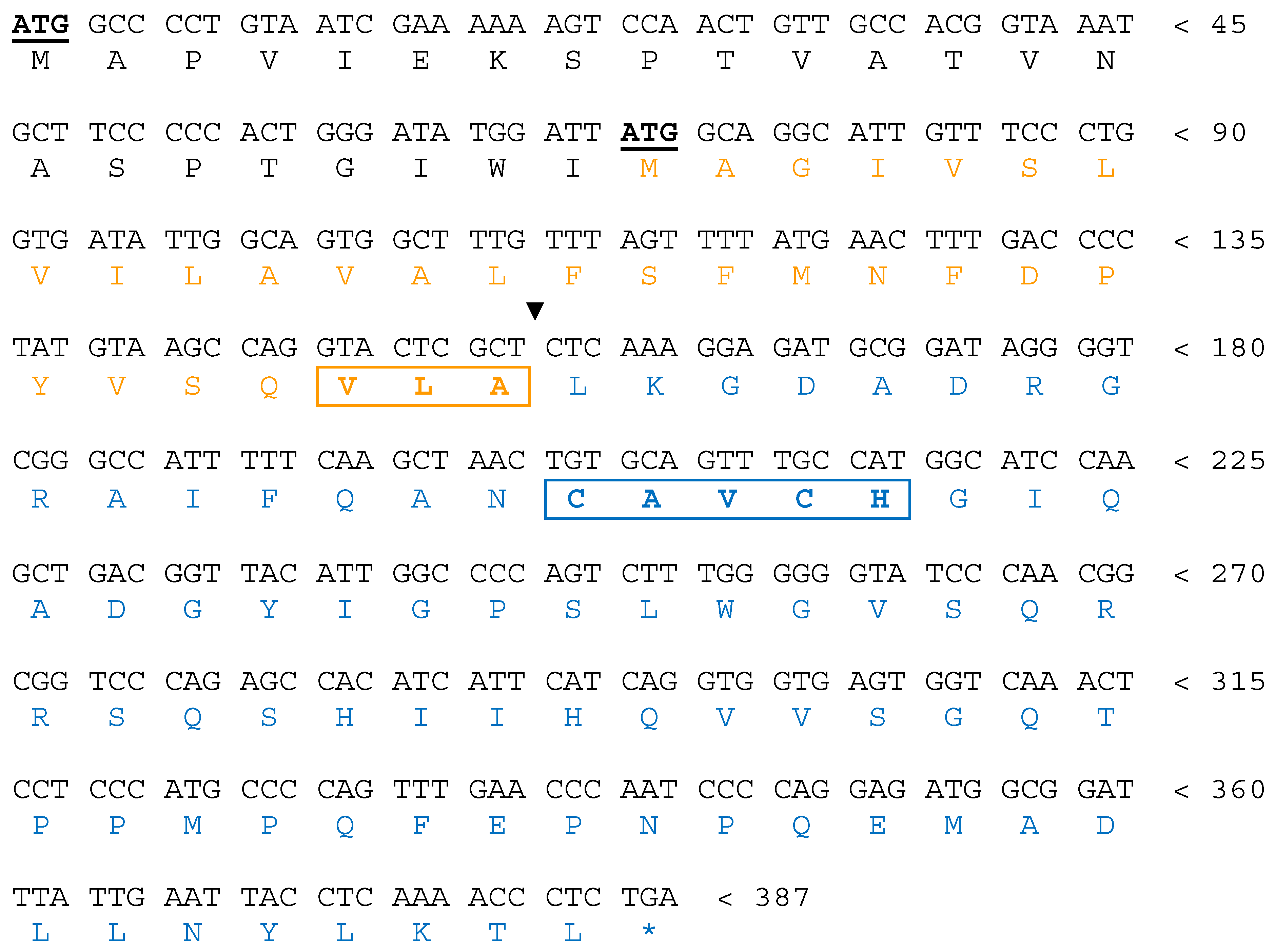
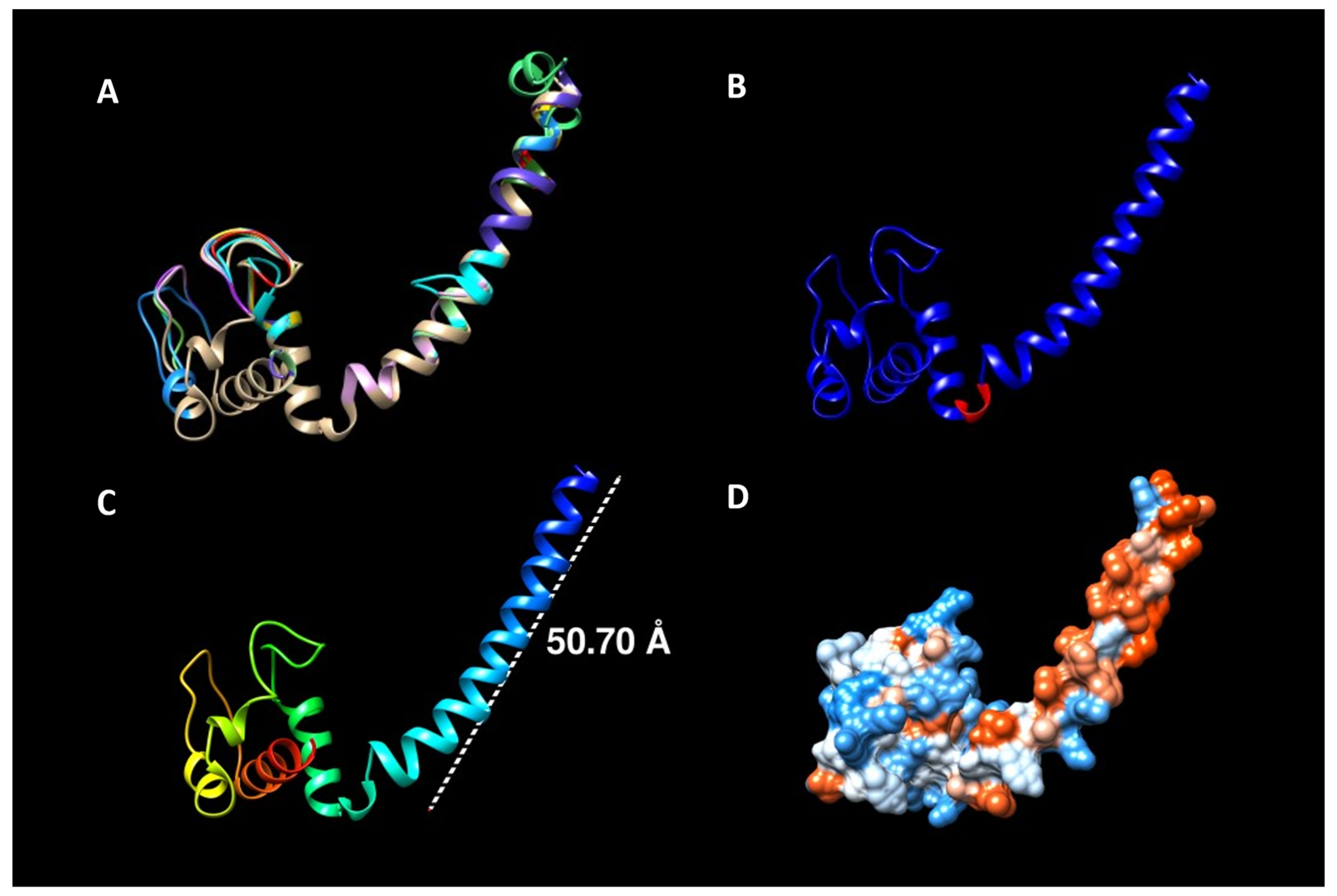
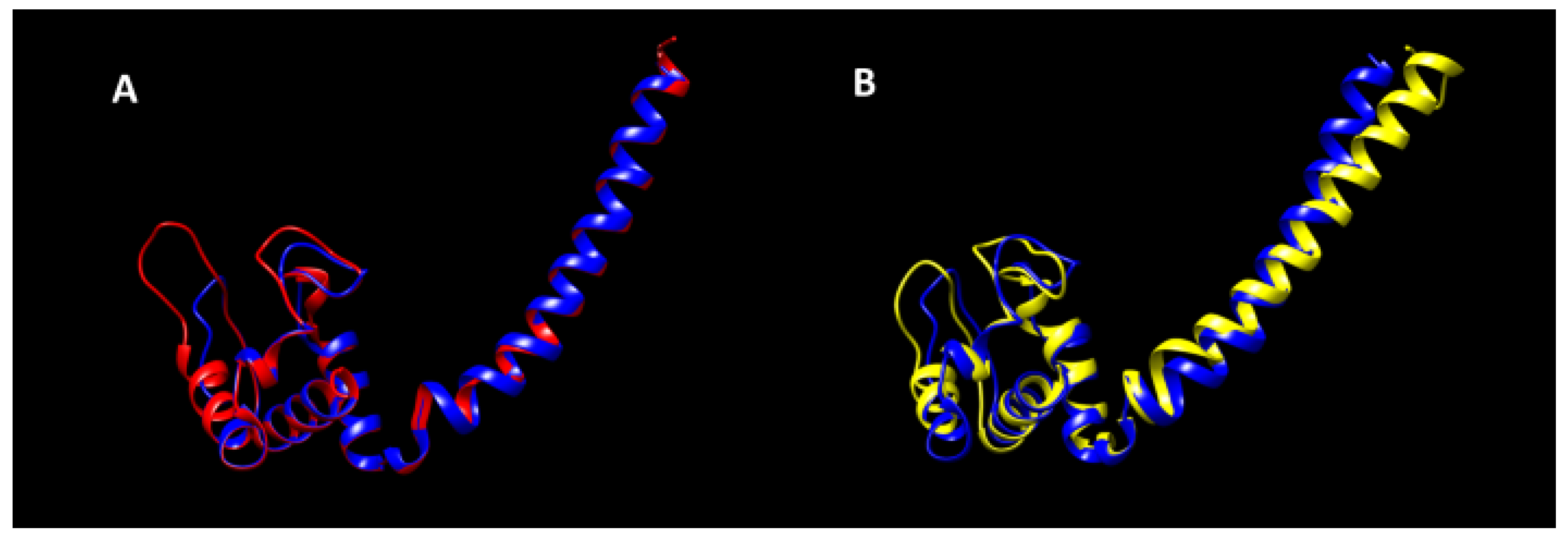
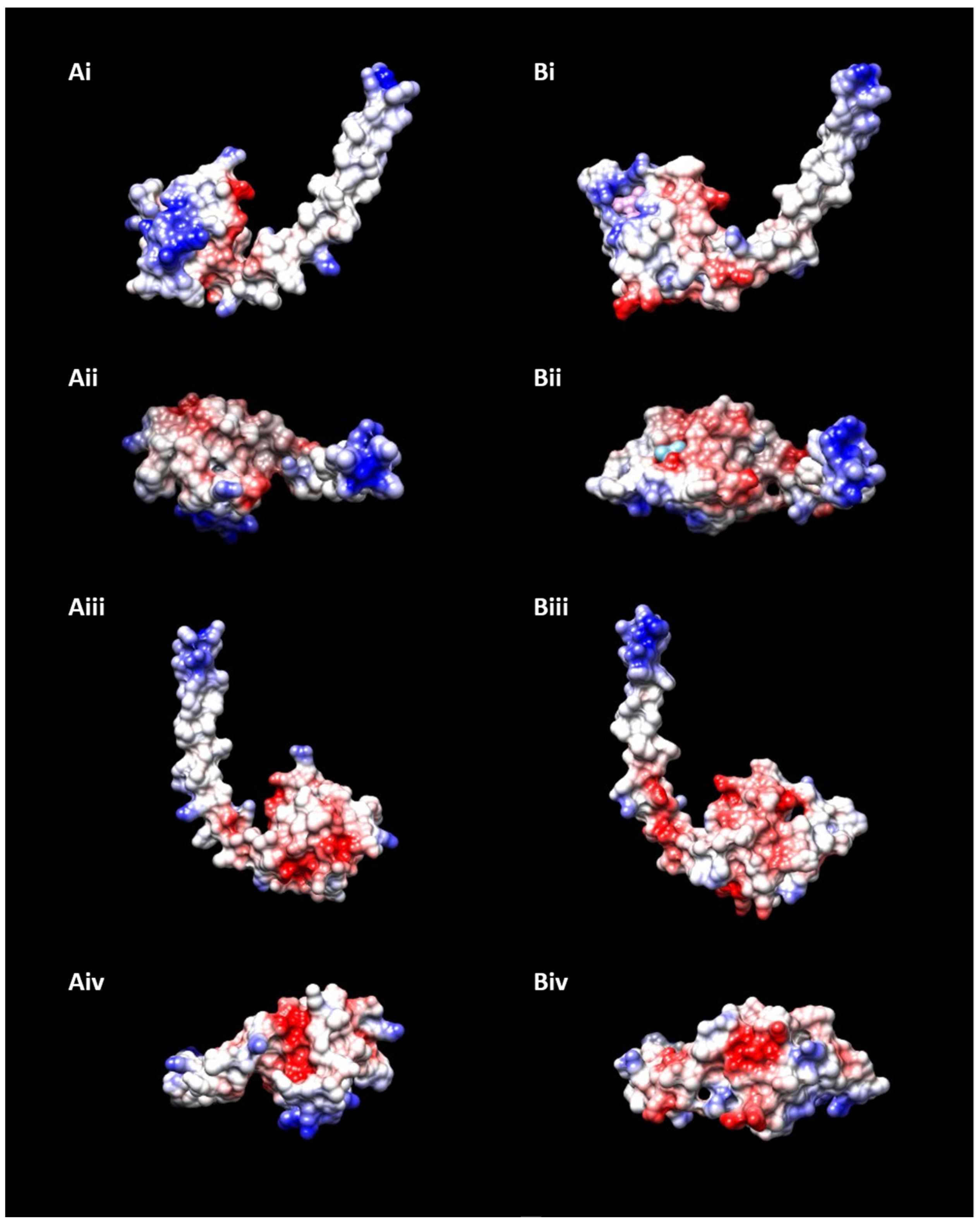
| Organism | Strain | Phyre2 Structure Matching | DB of Retrieval |
|---|---|---|---|
| Gloeobacter violaceus | PCC7421 | Yes | CyanoBase |
| Prochlorococcus marinus | MIT9211 | Yes | CyanoBase |
| Synechococcus elongatus | PCC6301 | No * | CyanoBase & CYORF |
| Synechococcus sp. | WH7803 | No * | CyanoBase & CYORF |
| Synechocystis sp. | PCC6803 | Yes | CyanoBase & CYORF |
| Nostoc sp. | PCC7120 | Yes | CyanoBase & CYORF |
| Prochlorococcus marinus | NATL1A | Yes | CyanoBase & CYORF |
| Prochlorococcus marinus subsp. pastoris | CCMP1986 | Yes | CyanoBase & CYORF |
| Synechococcus elongatus | PCC7942 | No ** | CyanoBase |
| Prochlorococcus marinus | MIT9303 | Yes | CyanoBase & CYORF |
| Synechococcus sp. | RCC307 | Yes | CyanoBase & CYORF |
| Synechococcus sp. | WH8102 | Yes | CyanoBase & CYORF |
| Acaryochloris marina | MBIC11017 | Yes | CyanoBase & CYORF |
| Cyanothece sp. | ATCC51142 | Yes | CyanoBase & CYORF |
| Synechococcus sp. | PCC7002 | Yes | CyanoBase & CYORF |
| Thermosynechococcus elongatus | BP-1 | Yes | CyanoBase & CYORF |
| Microcystis aeruginosa | NIES-843 | Yes | CYORF |
| Prochlorococcus marinus | MIT9313 | Yes | CYORF |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Gil, T.; Torrado, A.; Iniesta-Pallarés, M.; Álvarez, C.; Mariscal, V.; Molina-Heredia, F.P. Cytochrome cM Is Probably a Membrane Protein Similar to the C Subunit of the Bacterial Nitric Oxide Reductase. Appl. Sci. 2021, 11, 9396. https://doi.org/10.3390/app11209396
Rodríguez-Gil T, Torrado A, Iniesta-Pallarés M, Álvarez C, Mariscal V, Molina-Heredia FP. Cytochrome cM Is Probably a Membrane Protein Similar to the C Subunit of the Bacterial Nitric Oxide Reductase. Applied Sciences. 2021; 11(20):9396. https://doi.org/10.3390/app11209396
Chicago/Turabian StyleRodríguez-Gil, Tomás, Alejandro Torrado, Macarena Iniesta-Pallarés, Consolación Álvarez, Vicente Mariscal, and Fernando P. Molina-Heredia. 2021. "Cytochrome cM Is Probably a Membrane Protein Similar to the C Subunit of the Bacterial Nitric Oxide Reductase" Applied Sciences 11, no. 20: 9396. https://doi.org/10.3390/app11209396
APA StyleRodríguez-Gil, T., Torrado, A., Iniesta-Pallarés, M., Álvarez, C., Mariscal, V., & Molina-Heredia, F. P. (2021). Cytochrome cM Is Probably a Membrane Protein Similar to the C Subunit of the Bacterial Nitric Oxide Reductase. Applied Sciences, 11(20), 9396. https://doi.org/10.3390/app11209396









