Abstract
Due to their small size, microorganisms directly experience only a tiny portion of the environmental heterogeneity manifested in the soil. The microscale variations in soil properties constrain the distribution of fungi and bacteria, and the extent to which they can interact with each other, thereby directly influencing their behavior and ecological roles. Thus, to obtain a realistic understanding of bacterial–fungal interactions, the spatiotemporal complexity of their microenvironments must be accounted for. The objective of this review is to further raise awareness of this important aspect and to discuss an overview of possible methodologies, some of easier applicability than others, that can be implemented in the experimental design in this field of research. The experimental design can be rationalized in three different scales, namely reconstructing the physicochemical complexity of the soil matrix, identifying and locating fungi and bacteria to depict their physical interactions, and, lastly, analyzing their molecular environment to describe their activity. In the long term, only relevant experimental data at the cell-to-cell level can provide the base for any solid theory or model that may serve for accurate functional prediction at the ecosystem level. The way to this level of application is still long, but we should all start small.
1. Introduction
Living organisms are always and constantly interacting with their biotic and abiotic environment, irrespective of habitat, trophic level, or biological function. The network of interacting organisms is a mosaic and entwined reality that, depending on the scale of analysis, may be described at different levels of complexity. Cumulative cell-to-cell interactions among organisms belonging to very different taxa levels and origins, such as animals, protists, fungi, bacteria, archaea, and viruses, determine the overall microbial community activity in a given habitat. These interactions have an effect not just on their surrounding environment (i.e., microenvironment), but also influence large-scale fluxes and, thus, impact global ecosystem processes [1]. However, especially for soil biota, there is still a marked gap between studies performed in laboratory conditions and the in vivo reality [2]. Although providing crucially relevant data, this reduction in complexity cannot resolve general principles of microbial interactions at the ecosystem level [3].
In this review, we focus on bacterial and fungal interactions (BFI) in soil, and how the technologies available today can be applied for a better understanding of the behavior and phenotypic diversity of these organisms when closely linked to their micro-habitat. Information on the properties of soil and on how its spatial and chemical heterogeneity affects the dynamics of BFI will be described first, to justify the selection of strategies and methodologies reported in the later sections. This minireview is not meant to be exhaustive on either topic, as the arrays of approaches and studies available is extensive and beyond the means of this review. When possible, relevant examples on BFI are given but, in many cases, it was inevitable to refer to research on interactions within bacteria or other taxa.
2. Fungi, Bacteria and Their Microenvironment
Interactions within the soil biota are an especially challenging and fascinating topic; as a consequence of the microorganisms’ size, the variable metabolic activities of nearby microbes, and the changes in physicochemical conditions over short intervals of time and distance (soil heterogeneity), numerous microenvironments can co-exist in close proximity within a given habitat. Understanding the fine scale heterogeneity of soil environments is a prerequisite for predicting and contextualizing the physiology of the present organisms and metabolic interactions among community members. Their dynamics, in all their shapes and forms, are the consequences of their adaptation in response to the micro-habitats they experience (Figure 1).
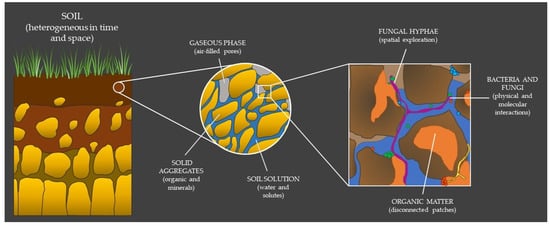
Figure 1.
The complexity of soil depends on the scale of analysis and is ultimately a mosaic of dynamic microenvironments defined at a specific point in time and space. Fungi and bacteria, together with other soil residents, live, grow and interact in this reality to the best of their fitness, perpetually looking for their realized niche.
2.1. The Hidden Properties of Soil
Basic physical laws govern and dictate the properties of each of the microscopic components that compose soils; the surface-to-volume ratio increases with the decreasing dimensions of an object, leading to surface effects, such as surface tension, capillary forces, adhesion, and viscous drag.
Soil is a highly heterogeneous medium, consisting of a mixture of solid material, and of water- or air-filled pores [4] (Figure 1). Thus, soil can be theoretically interpreted, either as the (organized) arrangement of aggregates/particles in the soil (i.e., soil structure) [5,6], or as the connectivity, tortuosity, and heterogeneity of the pore space between these soil components (i.e., soil architecture) [7] (for a complete argumentation on soil structure versus soil architecture, see Baveye et al. [3]). Albeit the hierarchical organization of aggregates highly depends on the amount of energy that is applied to take the soil apart, soils can be classified based on the distribution of their different aggregate species (i.e., soil texture). The aggregates are generally divided depending on their size, i.e., from (very) stable micro-aggregates (<2 µm), which are mainly composed of organic matter and clays, to less stable macro-aggregates, which are commonly composed of silt (2–63 µm) or sand (63 µm to 2 mm) [8]. Physical and chemical processes driven by both biotic and abiotic factors cause changes in aggregate size and continuous particle rearrangement. These constant changes in spatial organization of the solids and voids affect the architecture and connectivity of the pore space, which, in turn, affects the distribution of water and gases [9], as well as the diffusion of substrates (e.g., organic matter) [10,11] and solutes (e.g., elements and ions) [12] in soil.
Water is one of the most important, but at the same time is most variable component of soil. Soil’s water content depends on soil composition, rainfall, drainage, evaporation, temperature, and plant cover. Water is the medium connecting spatially separated areas in the soil matrix and becomes the solvent in which organic matter, microelements, and metabolites of different biological origins are dissolved or suspended (i.e., soil solution). The degree of water retention in soil microenvironments mainly depends on the soil pore neck size [13]. In macro-pores, localized in and between macro-aggregates, water is often well drained, whereas it is fully retained in micro-pores (localized between micro-aggregates) due to capillary action [13,14]. As a consequence, when alternating saturation/desiccation cycles that occur due to changes in precipitation or temperature, water status is more conserved in the micro-pores, generating fine water pockets rich in moisture that remain spatially disconnected from one another [14,15,16,17,18].
Micro-pores are also important for the retention of important biological molecules, such as organic matter, proteins, and nucleic acids [19,20,21,22,23]. As a result of the clay-cation exchange capacity (CEC) [4], they can all be adsorbed and retained by the negative charges of clay in soil micro-aggregates. Indeed, Ranjard and Richaume [14] found that organic matter is not homogenously distributed in soils, and higher concentrations (50–80%) were detected in micro-pores.
Furthermore, at the micro-scale level, micro-elements are to be considered for the growth and survival of microorganisms. Micro-elements are often present in the soil solution, as they are originating from both metabolically catalysed redox reactions as well as from abiotic chemical weathering of rock surfaces. They can diffuse in and out of the smallest pores, including the very narrow 1.8-nm-wide spaces between clay particles [3]. These diffusion processes cause pH and element concentration gradients that are highly dependent on the abiotic properties of the soil (e.g., the mineral composition, morphology, and texture), the geochemistry of the surrounding fluids, as well as the activity of microorganisms secreting highly reactive organic acids (e.g., oxalic acids, citric acid) [24].
Air (gases) resides in between the fractions of the soil pore network that are filled with the soil solution. The soil atmosphere depends on the connectivity of the non-water-filled pores of soil with the atmosphere or with other open pores. It usually consists of varying amounts of gasses such as oxygen, carbon dioxide, nitrogen, and nitrogen dioxide, but also of volatile organic compounds (gasses with biotic origin). The composition of the soil atmosphere highly depends on the production or consumption of a specific gas by local organisms, the solubility and diffusion of the gases in the soil water and, consequently, also on the capacity for water retention of the soil itself, as described before [13,25]. As an example, molecular oxygen has a very low solubility in water and its diffusion rate in water is 10,000-fold lower than in air [25]. Hence, soil micropores filled with water will rapidly turn anoxic upon consumption, limiting the growth and survival of many microbes. On the other hand, micropores can also offer protective microhabitats against “toxic” gases [26].
Independent of the soil component considered, the soil solution present in colloids can eventually diffuse out into wider pores, where it is readily available to microorganisms or is transported with the percolating water. It is also worth mentioning that filamentous fungi and plant roots stabilize the micro-aggregates of soil, and, in turn, together with other organisms living in the soil (e.g., nematodes, worms, larger animals), continuously increase soil solution conductivity through the soil. Nonetheless, the majority of substrates remain isolated and persist in the environment, possibly due to either physical protection or the separation from relevant enzymes [19]. In this regard, given their even smaller size, exoenzymes released by bacteria, archaea, and fungi into the soil solution can diffuse in and out of tiny pores and have significant roles both in the total fitness of the microbial population as well as in geo-chemical nutrient cycles of an ecosystem [27].
Respiration and decomposition rates differ considerably between sandy soils and soils rich in clay. This illustrates impressively how the dynamic interaction between the physical, air, and water components of soil directly influence the access, and thus the activity, of microbial populations to their substrate both in space and time. Sandy soils have lower surface-to-volume ratios than clay soils, meaning less water retention, but also lower segregation between microenvironments. In this context, soil heterogeneity has to be considered. For example, organic matter placed in different regions of the pore network was shown to be decomposed at different rates [28] while a number of other studies reported increased respiration rates only after disruption of the soil structure in soils with high clay content (e.g., [29]). These results taken together suggest that both the local environmental conditions as well as the uneven distribution of microbial populations within the local environment strongly affect the mineralization rates [28,30,31] and the range over which organisms can disperse and interact.
2.2. Bacterial–Fungal Interactions: A Harsh Existence
At the cell-to-cell level, bacteria and fungi interact at many different levels of intimacy that can be considered from two perspectives: in terms of physical associations and in terms of molecular communication [32]. Physical associations can range from seemingly disordered polymicrobial communities (i.e., biofilms) to highly specific symbiotic associations of fungal hyphae and bacterial cells (i.e., ectosymbiotic or endosymbiotic). Molecular interactions, on the other hand, involve a complex and diverse set of chemicals and compounds, and the interaction can be contextualized as antibiosis, signaling, and chemotaxis, metabolite exchange, metabolic conversion, adhesion, protein secretion, genetic exchange, and physicochemical changes. More often than not, multiple mechanism of interaction can be employed by one microorganism. The multitude of interactions and their effect on the partners or surrounding environment is extensive and well reviewed in [1,32,33,34].
Irrespective of the type of interaction, bacteria and fungi need to recognize each other prior to initiating any kind of target-oriented interaction. However, sensing and recognition mechanisms are often hindered by the particularity of soil and by the phenotypical characteristics of the organism (Figure 1).
When considering bacteria, they have their highest diversity in soil micro-aggregates, where they are either “swimming” in the soil solution, or attached to soil particles. This is not surprising, as it is in accordance with the higher concentrations of organic matter found in the micro-pores, the more stable conditions in water content, and a lower predation pressure by protozoan or other predators [35,36,37,38,39]. One of the first studies focusing on the distribution of bacteria in soil revealed that specific bacterial populations are typically residing inside micro-aggregates, with 40–70% of these bacteria being localized in the 2–20 µm and in <2 µm size micro-pores [14]. In some cases, cells can even penetrate pores smaller than themselves [40]. In contrast, more recent studies reported that pores in the 30 to 150 µm size range harbor a greater abundance of specific bacterial groups [41]. However, studies are scarce and only represent a snapshot in time, in a very dynamic environment. In fact, microbes explore a constantly changing environment in search of their realized niche, let it be by either active exploration of soil by fungal hyphae or by chemotactic movement, or passive transportation in bacteria. Any potential in bacterial mobility is limited by surface tension, capillary forces, and viscous drag that increase the energy requirement for their motility, particularly in partially saturated pore networks. Motility was found to cease, virtually completely, when the thickness of the water film was smaller than 1.5 µm [42]. For these reasons, microbial cells are often found as individual cells when associated with highly localized dissolved organic matter, or as patches of dense populations when linked to more conspicuous substrates [43]. In bulk soil, the average distance between neighbouring bacterial cells was found to be around 12.46 µm, with inter-cell distances shorter near the soil surface (10.38 µm) than at depth (>18 µm), due to changes in cell densities [44]. Simplified calculations suggests that, despite the very high number of cells and species in soil (108 cells per gram of soil), the number of neighbours that a single bacterial cell has within an interaction distance of ca. 20 µm is relatively limited (120 cells on average) [44]. Similar crude estimates were also found when the surfaces of soil pores were used to calculate the exclusion zone of cells on soil aggregates (a radius of 178 µm) [45].
A completely different story is depicted when filamentous fungi are considered. Fungi actively explore the soil pore space through hyphal spread, and cope well with heterogeneous distribution of nutrients [46]. They can cross air–water interfaces and nutrient-depleted spots to gain access to distant nutrient resources. The extensive mycelial architecture enables fungi to easily and efficiently re-allocate useful compounds to substrate-limited regions, to the benefit of exploratory colonisation of more unfavourable habitats [47].
A relevant feature of microbial interaction related to mycelial growth is that hyphae serve as dispersal vectors for motile bacteria (i.e., fungal highways) [48] and, hence, allow for bacterial colonization of new micro-habitats [49,50]. Moreover, the mycelia of fungi and oomycetes enhance bacterial activity by nutrient and water transfer (excretion) from the hyphae to the bacterial cells, thus enabling bacterial growth in otherwise too oligotrophic habitats [51], or enhancing microbial activity in dry soils [52]. An important example of such a symbiotic interaction is extraradical mycelia of arbuscular mycorrhizal and other mycorrhizal fungi in the rhizosphere of plants, where a large array of bacteria can directly consume fungal exudates released into the environment [34]. The mutualistic relationship can also be reciprocal: fungi take advantage of their bacterial partners to improve their carbon source pool in a mechanism named bacterial farming [53]. Fungal hyphae can also become an ideal hotspot for horizontal gene transfer [54,55,56] or bacterial prey populations [57], as they facilitate dispersal and preferential contact of bacteria in the hyphosphere. In this regard, it should be mentioned that, apart from fungal hyphae, plant roots and dead organisms also behave as hot spots for highly interactive (e.g., competition, pathogeny, mutualism, predation) microbial communities. Nonetheless, these many physical interactions between bacteria and hyphae-forming fungi may represent short-lived associations, as microscale communities frequently assemble and disassemble by migration, attachment, and detachment from surfaces and cells.
In this quite lonesome and unforgiving scenario, physical contact between cells is often not realizable. Hence, molecular compounds are employed and secreted in the soil solution to sense potential partners in their surroundings. Importantly, the release of molecular signals represent a cost, both energetic and elemental, and hence are regulated to maximize the fitness of the organism [27]. As molecules diffuse freely from any pore and are easily removed from the microenvironments, molecular interactions can only occur at relatively short distances. On leaf surfaces, for example, interactions among bacteria have been found to occur principally in the 5 to 20 µm range [58], whereas in soil, where local patches of cells may reside in a pocket of soil solution, diffusible metabolites can reach neighbouring cells up to 100 µm away [44,59]. The interactive dynamics are very different in soil crusts, biofilms, or mats where cells are well physically constrained and in direct contact with each other (e.g., [60]).
Sensing and recognition of fungi and bacteria includes also solution-independent ways. Volatile organic compounds (VOCs) are one important quorum sending vehicle-enabling communication over longer distances, especially in vision of the spatial separation between soil particles often occurring in unsaturated water conditions [61,62].
Finally, it is worth mentioning that bacteria and fungi can also indirectly interact by modifying their microenvironment in ways that can positively or negatively affect their partners (i.e., niche modulation) [34]. For example, the acidification or the neutralization of pH values influences the solubility of soil nutrients (e.g., phosphorus [24]), thus inhibiting or stimulating overall bacterial growth and metabolism [63,64,65]. Nutrient depletion is one example of the many ways of indirect interaction. Iron depletion by the excretion of bacterial or fungal siderophores can negatively affect the performance of surrounding organisms, including plants [66,67].
In terms of BFI in different scenarios of soil heterogeneity, there is a limited number of studies available. They show both a random distribution of soil microorganisms and a high degree of microbial networking. We will now discuss methodologies and techniques well-suitable for observing microbial interactions and we hope to stimulate the reader to further expand the research in this field.
3. Get Your Hands Dirty: Methods for Studying BFI
Advances in molecular methods and bioinformatics have provided enormous benefits in enlightening BFI from many different perspectives. The availability of genomic methods, such as DNA sequencing, especially with high-throughput sequencing methods, enabled deeper insights into the soil microbial diversity, and contributed enormously to a more comprehensive database build-up, and thus to better network analysis inferences. They surely provide the theoretical background of what to expect from a specific soil sample and undoubtedly increased the knowledge on unculturable microorganisms (some of the relevant literature on the topic include work by [68,69,70,71,72,73]). However, virtually all -omics approaches entirely ignore the geometry of the pore space in soils or the characteristics of microenvironments (e.g., [26,74,75,76]). Even highly confident co-occurrence network analyses cannot distinguish between true ecological interactions and other non-random processes (e.g., cross-feeding versus niche overlap) [69]. Although benchmarking studies are few and urgently needed, their relatively high false-positive interaction rate can probably be decreased by omitting interactions that could physically not have occurred. Thus, our experimental approach should be adapted to make it a better fit for exploring the complexity of BFI in soil (Figure 2).
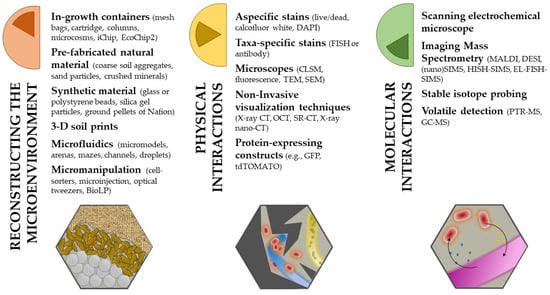
Figure 2.
Overview of methods and technologies discussed in the review. They are divided by the applicability and information they can provide in regards to studies researching bacterial—fungal interactions. Often, by combining more than one together (e.g., microfluidics and FISH-SIP-nanoSIMS), information at different scales and levels are obtained.
As microorganisms interact at micro-scale, micro-scale data from microbiological, physical (e.g., porosity, water, air) and (bio)chemical (soil organic matter, micro-elements, pH) methods, including their new technological advances, should be combined to study BFI. As such, some of the methodologies discussed in the following chapters have also been applied to the investigation of abiotic and biotic aspects of the soil matrix [3], but here they will only be presented in vision of bacterial and fungal interactions. An overview of the abbreviations used to shorten the techniques’ names is reported in Table 1.

Table 1.
Abbreviations of technique’s names discussed in the review.
Although not discussed, the combination of -omics techniques (e.g., metagenomics, transcriptomics, proteomics) alongside other standard protocols remains a very valuable strategy, provided that high-quality raw material is available. Furthermore, the reader should bear in mind other important methodological factors. It is essential to consider the heterogeneity of the soil for sampling strategies [26], the consistency of the experiment for reproducible results, and the applicability of the techniques for eventual high-throughput applications [77].
3.1. Reconstructing the Spatial Heterogeneity of Soil
In an ideal word, one would monitor in real time the dynamics of microbial interactions at the soil microscale, retrieving information about microbial taxonomy, distribution, and behaviour as well as about dependencies of these traits from the physicochemical properties of their micro-niches, directly in their natural microenvironment. Needless to say, this is far from the modern reality as it has proven impossible to quantify and analyse the interaction of microorganisms in actual soil.
Gause’s co-culture experiments [78] are today a standard procedure for experimentally investigating microbial interactions. With inventive approaches (e.g., [79,80,81,82,83,84,85]), agar-based co-culture experiments re-create simplified communities in a controlled environment and thus provide ideal conditions to test ecological concepts concerning community stability and dynamics. However, a clear disadvantage of classical solid nutrient medium is the absence of habitat heterogeneity.
Artificial matrixes have been created that try to simulate as much spatial complexity in which BFI exists as possible but, at the same time, ease downstream analysis. Such soil matrices can often be sterilized, can be buried in soil within in-growth containers under natural conditions for a defined period of time, and then can be retrieved and transported to the laboratory with minimal disturbance. The applied in-growth method enables in situ cultivation of the local microbial communities [86,87] with the assumption that their interaction strategies are retained. Different designs of in-growth containers can be distinguished, such as nylon mesh bags with a mesh size >50 µm [88], or cartridge [87]. Depending on the downstream analysis to follow, they can be filled with different types of pre-fabricated natural material, such as macro-aggregates of soil fragments (5 mm-sized) [41], coarse sand particles [88,89], or mixtures of different crushed minerals [87]. These materials offer good field-representative soil aggregates that can be used also for in vitro experiments (e.g., [90]). However, they have the disadvantage of being opaque, such as real soil. This limits the range of applications to which they can be implemented, and thus are often substituted with artificial materials, as it will be described further down below.
Simon and colleagues [50], and more recently Junier and colleagues [91], developed the fungal highway column in order to detect bacterial dispersal by fungal hyphae in a forest soil. The fungal highway column consists of a small tube in which one medium section is in contact with the soil, while the second one, which is physically separated from the first, can only be colonized by bacteria when they are transported into it via fungal in-growth. Other sophisticated methods for in situ cultivation are iChip [92] and EcoChip2 [93]. They enable parallel cultivation and isolation of up to now uncultivated microbial species, and can elucidate the dependency of some organisms to one another, due to in situ growth factors, that cannot be simulated under standard laboratory conditions.
A common concern for all these types of in situ cultivation is the flow of water. Fungal hyphae are well-stabilized in the soil matrix, but bacteria can easily be transported or washed in and out of the in-growth containers by percolating water. This might ultimately bias BFI analyses. On the other hand, it has been shown that water often follows preferential pre-existing flow lines in the soil [94]. In addition, bacteria can tightly adhere to exogenous material, form biofilms, or be safely sheltered in micro-pores [26]. Ultimately, the real extent to which water can affect the BFI networks is still unknown.
A number of new manufacturing approaches have been developed to simulate the soil architecture, but with the advantages of reproducibility, accessibility, and laboratory manipulation. They are usually inoculated with defined fungal and bacteria isolates.
As it was aforementioned, synthetic materials, such as clean, spherical 500-µm (or bigger) glass beads [95,96,97,98]; crushed silica gel particles (irregularly shaped grains) [99,100]; polystyrene beads [101]; or ground pellets of Nafion [102,103,104,105], have been developed and widely applied as artificial porous media [106] to avoid the limitations given by the opacity of natural materials. Although soil is tremendously more heterogeneous (smaller pore size) and chemically complex, their low reactivity and their optical refractive index, similar to water, allows for the combination of co-cultivation with different visualization techniques (e.g., brightfield and fluorescence imaging, low-field magnetic resonance system, and X-ray CT), as discussed in the next section. Another interesting method for simulating soil microstructure is 3-D printing. Soil-like structures of nylon 12 or resin with parafilm wax [107,108] can be used to study the exploration and interaction strategies of different microbial inoculates, and may give future insight on BFI in these microhabitats. Some general precautions and limitations should be considered when setting up a microcosm experiment with synthetic soil manufacturing. The material chosen should be: biocompatible or at least not toxic to the organisms of interests, congruent with downstream imaging techniques, and an appropriate proxy for the environment of interest [3].
Microfluidic arrays can be deployed when more “personalized” structural constrains and dynamical chemical gradients are needed. The literature on this topic is extensive and more detailed information can be found in several reviews (e.g., [109,110,111,112,113]). Microfluidic platforms allow for the precisely organization and monitoring of small heterogeneous microbial populations [114,115] in a three-dimensional geometry [116]. They can be constructed in different dimensions (in volumes as small as ~100 fl [117]), materials (e.g., transparent polydimethylsiloxane (PDMS) [116,118,119], hydrogels, proteins crosslinked by multiphoton lithography [120], lipid-silica containers [121,122]), or shapes (e.g., soil micromodels [123,124], arenas [125], channels [126], mazes [127], or single droplets [128]). Some of the microfabricated biomaterials used for constructing a microfluidic system can be responsive to external stimuli, hence acting as both physical barriers, as well as an additional function for active control, manipulation, and observation of the microbes in real time. Some examples of dynamic parameters that can be tuned are pH, temperature, osmolarity, light intensity, as well as the geometry and the size of micro-chambers. Furthermore, the permeability of the walls and the rate of mass transport through the system can be manipulated in order to allow controlled diffusion of nutrients, waste products, or other small molecules [120,129]. The experiment can also be run at extensive temporal scales (e.g., miniaturized chemostats [130]), allowing continuous monitoring of the microbial dynamics over time. A series of limitations often highlighted by the authors comprise the sophisticated fabrication processes and complex technical set-up involved [110]. Furthermore, the porosity and complexity obtained with microfluidics are, as to be expected, lower than in real soil, hence it is necessary to compare the created environmental conditions to adequate references [111]. The majority of microfluidic devices have been used to study interactions in model organisms mainly between different bacterial cells (e.g., [117,131]), bacteria and plant [132], or fungi and other organisms [128,133,134]. A number of studies investigating the cell-to-cell interactions between fungi and bacteria are shyly emerging [135,136]. In these exclusive examples, interaction-induced physiological changes and metabolic exchange between fungal hyphae and their associated bacteria were investigated. Clearly, many more possibilities are awaiting.
Micromanipulation of single cells in time and space is also possible [117] that allows either their precise arrangement in a three-dimensional manner or their extraction from a complex heterogenous space. The precise arrangement of single cells can be achieved, for example, by fluorescence-activated droplet- or cell-sorter (FADS, FACS, respectively). These systems are based on the presence or absence of a fluorescent compound (e.g., GFP [137]), resorufin from resazurin [138], FISH-labelling, or stained antibodies [139], and work very well along with the set-up of microfluidic experiment where a defined combination of organisms distributed in specific locations can be achieved. Alternately, Partida-Martinez et al. [140] successfully implemented a microinjection technique based on a laser beam to re-introduce an endohyphal bacterium in its original curated fungal host and to later study their physiological states. Otherwise, to extract specific microorganisms from their microenvironment, micromanipulators of different types have been used [141,142,143,144], including the so-called ‘optical tweezers’ (e.g., [145,146]) and biological laser printing (BioLP) [147].
3.2. Playing Hide and Seek with Bacteria and Fungi: Unravel Their Physical Interactions
In mixed culture of bacteria and fungi growing in a matrix with defined topographical and chemical features providing the microorganisms with the opportunity to spatially organize, a set of methodologies can be applied to visualize, identify, and monitor their interaction.
Basic techniques for visualization of selected microbes in a matrix include non-specific stains, such as live-dead staining [148], stains with calcofluor white M2R [149,150], or DAPI [151]. These allow to spatially visualize and quantify bacteria and fungi in close proximity with minimal effort [44], although taxonomical information is not provided.
Hence, more advanced fluorescence-conjugated techniques are permanently being developed. They can be based on fluorescent probes targeting distinct short nucleotide sequences (fluorescence probe technique (FPT)), or on direct fluorescent antibodies targeting a specific protein domain (direct fluorescent antibody (DFA)) [139]. Fluorescence in situ hybridization (FISH) is a type of FPT that combines the phylogenetic identification of the taxon targeted on a single cell basis (e.g., prokaryotic cells) with the visualization of their distribution in situ (e.g., [152,153,154]). The technique is a pillar in molecular biology, and countless applications and specific developments exists (see for example the reviewing book by Azevedo and colleagues just published [155]). As it will be described in the next section, FISH as well as DFA are widely used in combination with many classical or modern applications in the recognition of taxa involved in fungal and bacterial molecular interplay. When dealing with a very bright background of fluorescing soil constituents, FISH probing can be combined with tyramide signal amplification. This technique is called catalysed reporter deposition (CARD)-FISH, and is very useful for obtaining higher signal intensities and reduced background interference [156,157,158]. The combination of CARD-FISH with confocal laser scanning microscopes (CLSM) allows quantification, localization, and visualization of microbial cells with even more depth-resolution. Transmission and scanning electron microscopes (TEM and SEM, respectively) can also be combined with cellular dyes in order to obtain higher resolutions for both qualitative and quantitative assessment of the targeted organisms. For example, a CARD-based approach using gold nanoparticles (GOLD-FISH) can be used when dealing with very small portions of soil [159].
Fixation techniques for whole soil blocks with special chemicals can be used in order to study the distribution of microorganisms in their specific microenvironments. Blocks of soils are first impregnated with resin. Very thin layers of material can then be serially removed either by a traditional microtome, or by broad or focused ion-beam scanning electron microscopy (BIB- or FIB-SEM) [150]. Alternately, based on the same principle, thin sections of fixed soil can be cut with a diamond-tipped blade and mounted on a microscope slide. If microbes have been previously stained, the soil organisms of interest can be specifically observed [160]. By merging the different layers, three-dimensional images can then be reconstructed from a series of z-dependent two-dimension images with special software and statistical interpolation techniques [160,161,162]. Stains and dyes are not without limitations, however. They often unspecifically bind to soil components (including organic matter), thus increasing the background noise that, together with auto-fluorescent objects naturally occurring in the sample, make the recognition of cells in a soil matrix challenging. This is especially true for bacterial and archaeal cells in respect to fungal hyphae, whose physical arrangement and spatial distribution are relatively more straightforward to identify. In addition, bleaching caused during image acquisition and the impossibility to view “the whole picture” in a single ocular field can be an additional problem. Visualization techniques do generally require long processing times, a lot of handling skills, and result interpretation experience. This probably explains why there is still a relatively low number of articles published on BFI, especially given that these tools have been around for already few decades. Lastly, but most importantly, a fundamental problem remains: when samples are physically altered and chemically fixed (i.e., killing), it is impossible to monitor microbial cell dynamics in situ and in vivo.
With regard to non-invasively studies on soil microbes in their undisturbed, structured environment, several new technical approaches have been developed and proposed during recent years: X-ray computed tomography (CT) [3,111] and its variations (e.g., optical coherence tomography (OCT) [60], synchroton tomography (SR-CT) [163], and X-ray nano-tomography [164]) have been used, in combination with other methods, to study the reciprocal interaction between microbes and their surrounding environment, in both native and artificial soils [41,96,97,98,102,165]. With X-ray CT, for example, the effect of the soil environment on fungal exploration behaviour and gas release was investigated by repeated scanning of microcosm systems over several weeks [90]. This type of approach was also adopted for direct observation of plant–pathogen interactions [166], and to detect the migratory capabilities of Pseudomonas fluorescens [167,168] in situ at high spatial resolution for the first time. However, studies on BFI implementing these techniques have lacked knowledge up to now. A fundamental problem of radiation-based imaging is that it is ionising and penetrative, meaning that it could possibly damage both structures and microorganisms within the substrate [164]. On the other hand, low irradiation doses applied to plant–soil interfaces were reported to neither influence root growth [169], nor microbial cell number, microbial community structure, or their potential activity [165,170]. Nevertheless, even if not lethal, care should be taken for the insurgence of mutations with unpredictable consequences. Lastly, the imaging resolution chosen can limit either the range or the feature of detection [111].
When concerned with the in vivo study of dynamically interacting organisms, simple and reliable applications for the live-cell screening of microbial organisms is the use of fluorescent protein-expressing bacteria (e.g., GFP, [85,142,171]) or fungi (e.g., tdTomato, [172]). A combination of red and green fluorescent proteins allows for a simultaneous detection of multiple organisms, and can be used to follow BFI both in situ and in vivo [172,173]. Although these modern molecular tools only permits to work with isolates or model organisms, if combined with a relevant experimental set-up, such as microfluidics, video microscopy, or other techniques, described in the next section, they can give a great deal of advantages [174].
3.3. What Are They Doing? Investigating the Molecular Interactions in BFI
The ability to confine small numbers of microorganisms and determine their location in space is an important step towards understanding the impact of spatial structure on microbial behaviour. However, the development of techniques for the in situ measurement of small molecule signals, and for the detection of other factors responsible for behavioural modulation, are equally important. These techniques are applicable to simple bacterial and fungal communities and ultimately allow cell-to-cell resolution.
Scanning electrochemical microscopy (SECM) can be considered as the most straightforward technique to quantify and spatially map the concentration of specific redox-active molecules (i.e., one compound at the time) proximal to populations of cells [175], such as the small molecules that are involved in the interaction in multispecies biofilms [176].
For more comprehensive studies, with imaging mass spectrometry (IMS), in all its variations, one can detect and describe multiple molecules at the same time, and superimpose their spatial distribution onto optical or fluorescence images of the sample. To put these advantages in perspective, a key difference between IMS and standard fluorescence microscopy is the capability of IMS to detect up to thousands of unique signals from one biological substrate, compared to only up to eight in the average fluorescence microscopes [177]. The interplay between competing organisms, the metabolic exchange in complex intra- and inter- species signaling, or the detection of new and uncharacterized molecules can be studied with this IMS technique. Furthermore, the direct link between the distribution of detected compounds and the observed phenotypes of the biological sample may give essential information on the function of the molecules themselves. Finally, the endogenous molecules can directly be analysed in their microenvironment, without the indirect biases that artificial substrates may cause (e.g., isotope feeding experiments) [177,178,179].
Matrix-assisted laser desorption-ionization (MALDI) [180] and desorption electrospray ionization (DESI) [181] are two types of IMS techniques that suit particularly well for studying the production of natural products by microorganisms in the laboratory [178]. Whereas the soft and absorbing nature of agar medium limits DESI-IMS analysis, with MALDI-IMS microorganisms can be grown on a 0.5–1.5-mm layer of agar for a defined period of time before being covered with a matrix and subjected to the analyses. Albeit certain media that have a high salt or sugar content may prove difficult to analyse, owing to ion suppression or uneven matrix crystallization, the use of agar-based IMS approach can provide important time-dependent correlations [180]. MALDI- and DESI-IMS have been widely applied for the characterization of microbial monocultures, such as cyanobacteria [182], bacterial colonies [183], or fungal–fungal interaction [184], and their application in BFI studies is slowly increasing, especially for the detection of antifungal compounds in co-culturing experiments [185,186,187]. Some of the limitations of these techniques derive from biases due to sample preparation, the simplification of otherwise complex environments, and signal distortion from background noise [179]. Secondary ion mass spectrometry (SIMS) [188] has the highest resolution (order of nanometres) of all the IMS techniques due to its extremely narrow ion beam. It can be applied to have either a focus on the analytical mass range (static) or to have a greater spatial resolution (dynamic). SIMS platforms can also perform very fine depth-profiling experiments by etching away the sample surface with the ion beam [178]. When an ultra-sensitive characterization of the molecular microenvironment between interacting cells is needed (down to 50 nm of resolution), nanoSIMS can be used. Obviously, with this level of resolution, the sampling area that can be analysed decreases to a few mm2. Sample preparation is also a fundamental step. Samples should be mounted on a conductive surface and be extremely flat, although a notable exception was given by Vaidyanathan et al. [189].
Although experiments with these new modern techniques are on the rise, stable isotopes feed to a biological sample is still the most widespread technique to follow metabolic activities and interactions of bacterial–fungal communities in their natural environments. The literature available on stable isotope probing (SIP) is extensive and detailed descriptions can be found elsewhere (e.g., [141,190,191]). An incredible advantage of isotope labelling is that the substrate of interest can be directly fed in any artificial set-up chosen to simulate the (micro)-habitats of the organisms of interest (e.g., nature, microcosms, artificial matrixes). For example, different 13C-marked carbon sources were supplied in microcosms with soil material to monitor respiration rates and C utilization by bacteria and fungi [192]. Using airtight containers, Pinto-Tomás et al. [193] discovered that the nitrogen enriched material in the fungus garden of soil leaf-cutter ants was not derived by their extensive foraging activity, but was instead fixed by N2-fixing bacteria hosted by the fungus itself. Another neat example that shows the intricate behaviour of fungi and bacteria is the study of Pion et al. [53] where, by tracing 13C-substrate, the researchers found out that the ascomycete fungus Morchella crassipes first farms Pseudomonas putida bacteria and then use them as carbon source. The precision and sensitivity of the instruments for the downstream analysis will eventually limit the experimental design. In fact, a common problematic intrinsic to all labelling experiments is the insufficient incorporation rate for detection. This can be avoided by long incorporation time, but in turn it increases the risk of cross-feeding i.e., contaminations [139]. Independently from the isotopically labelled substrates applied (13C, 15N, 32P, 2H) and the technology used to detect the isotope incorporation (e.g., mass spectrometer, isotope ratio mass spectrometer, Raman microspectrometry, SIMS) [190], the monitoring of uptake and transfer of metabolites has to always be linked to the identification of the involved microorganisms. This can be achieved by a variety of different methods applied after isotope incorporation. Soil microorganisms can be discriminated based on taxa-specific biomolecules by phosphor lipid fatty acid analysis (PFLA); sterol analysis (e.g., using ergosterol as a biomarker for fungi) DNA-, RNA-, or protein-based methods; by FISH-microautoradiography [194,195]; or isotopic rRNA-arrays [196]. Molecular approaches have also been used for tracing back a specific taxon with labelled DNA, including PCR amplification as well as sequencing [197,198,199]. It is also possible to directly clone isotope-labelled DNA and then sequence it [200].
An exciting alternative to these classical taxa-discriminating approaches combines taxa-specific target probes (e.g., FISH, CARD-FISH, DFA) and stable isotope probing (SIP) with the simultaneous detection of atoms produced by nanoSIMS. If based on creative experimental designs, the combination of these techniques can substantially help for linking identity to functions by tracking the uptake and transfer of isotopically labelled compounds in environmental samples where the individual taxa cannot be isolated from one another. A satisfactory overview of these combination methods is given by Musat et al. [201]. Briefly, a microbial community that was grown with metabolic-labelled tracers (e.g., 13C or 15N) is fixed, hybridized with phylogenetic probes, and visualized by fluorescence microscopy to obtain a picture of the distribution of specific taxa in space. Then, the sample is analysed by nanoSIMS [155] which maps the existing molecules in the sample by detecting and identifying them from their specific locations. Finally, a superimposed picture of the partners is obtained, which involved giving both physical and molecular information on the interaction.
The use of SIP-nanoSIMS has great potential as an attractive alternative to stand-alone autoradiography experiments for the study of the molecular micro-environments of microbes, also thanks to the increase in more accessible facilities and the decrease in costs. An increasing number of studies have used the combination of these powerful techniques to link identity and metabolic behaviours within bacterial communities (e.g., [202,203]), between microbe–host interactions (e.g., [204]) or in cyanobacteria mats [205,206]. Less studied are the interaction between bacteria and fungi. An exceptional example in this regard was carried out by Worrich et al. [51] where they investigated the water and nutrient exchange between bacterial cells and fungal hyphae in stress conditions.
Direct label imaging during nanoSIMS analysis is also emerging with the use of EL-FISH nanoSIMS [159] or HISH-SIMS [207], although the studies for microbial interactions using these techniques are still limited.
Finally, but not least importantly, it is worth mentioning that interesting insights in fungal and bacterial communication can be retrieved from instruments that detect VOC (proton transfer reaction (PTR)-MS or GC-MS) [208]. These molecules are very important when considering the porosity of the soil (or similar artificial models) and the connection that air provides among different locations, even between considerable distances.
4. Outlook
Although the list of tools provided in this review may seem extensive, the majority of the available studies carried out on bacterial–fungal interaction used a repetition of a few well-established, often simple, methods such as agar-plating, isotope probing, or fluorescence microscopy, combined in new neat ways to answer research questions never asked before. Surely, there were some exceptions, but negligible if compared to the long-time availability of novel methods such as CARD-FISH, SIP-nanoSIMS, or microfluidics, and the immense number of plausible combinations of these. All methods have strengths and weaknesses, as well as costs, thus the best combination of these applications remains the one that best fits the specific research questions that a researcher is trying to address. Nevertheless, to acquire the best realistic overview of the system, the combination of tools that aims at providing data at different levels of complexity is highly required (Figure 3). Inventiveness and perseverance are the way forward.
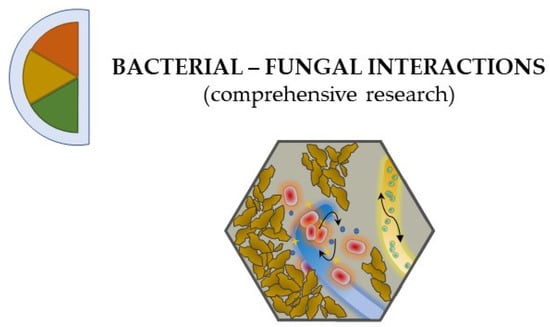
Figure 3.
The more variables and data that an experimental design incorporates or collects at different levels of complexity (i.e., by reconstructing a coherent environmental set-up as well as by investigating the physical and the molecular interactions), the more comprehensive becomes our understanding of bacterial–fungal interactions, setting the field a step closer to accurately depict the reality of the natural environment.
Studies investigating the interaction of fungi and bacteria within each own taxonomical unit are greater than the studies investigating the combination of the two. This, along with the too often bypassed research that tries to untangle the complexity of the physicochemical properties of soils, shows that this field of microbiology often still works in sectorial ways. Just as much there really is a community of microbes living together for their “greater good” that cannot be extracted from their environmental background without seeing it biased, we, as observers, should learn from them and work together, in an interdisciplinary way, to better catch a glimpse of their intricate lives.
Funding
This study was financed by the Open Access Funding by the Austrian Science Fund (FWF) and by the Land Tirol (project Microbial Interactions in Snow-covered soil MICINSNOW-P31038).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
No conflicts of interests to be declared.
References
- Jansson, J.K.; Hofmockel, K.S. The soil microbiome—From metagenomics to metaphenomics. Curr. Opin. Microbiol. 2018, 43, 162–168. [Google Scholar] [CrossRef]
- Baldrian, P. The known and the unknown in soil microbial ecology. FEMS Microbiol. Ecol. 2019, 95, 1–9. [Google Scholar] [CrossRef]
- Baveye, P.C.; Otten, W.; Kravchenko, A.; Balseiro-Romero, M.; Beckers, É.; Chalhoub, M.; Darnault, C.; Eickhorst, T.; Garnier, P.; Hapca, S.; et al. Emergent properties of microbial activity in heterogeneous soil microenvironments: Different research approaches are slowly converging, yet major challenges remain. Front. Microbiol. 2018, 9, 1929. [Google Scholar] [CrossRef]
- Young, I.M. Interactions and self-organization in the soil-microbe complex. Science 2004, 304, 1634–1637. [Google Scholar] [CrossRef]
- Dexter, A. Advances in characterization of soil structure. Soil Tillage Res. 1988, 11, 199–238. [Google Scholar] [CrossRef]
- Hillel, D. Introduction to Environmental Soil Physics; Elsevier: Amsterdam, The Netherlands, 2003. [Google Scholar]
- Young, I.; Crawford, J.; Rappoldt, C. New methods and models for characterising structural heterogeneity of soil. Soil Tillage Res. 2001, 61, 33–45. [Google Scholar] [CrossRef]
- Tisdall, J.M.; Oades, J.M. Organic matter and water-stable aggregates in soils. Eur. J. Soil Sci. 1982, 33, 141–163. [Google Scholar] [CrossRef]
- Horn, R.; Smucker, A. Structure formation and its consequences for gas and water transport in unsaturated arable and forest soils. Soil Tillage Res. 2005, 82, 5–14. [Google Scholar] [CrossRef]
- Mueller, C.W.; Koelbl, A.; Hoeschen, C.; Hillion, F.; Heister, K.; Herrmann, A.M.; Kögel-Knabner, I. Submicron scale imaging of soil organic matter dynamics using NanoSIMS—From single particles to intact aggregates. Org. Geochem. 2012, 42, 1476–1488. [Google Scholar] [CrossRef]
- Rawlins, B.G.; Wragg, J.; Reinhard, C.; Atwood, R.C.; Houston, A.; Lark, R.M.; Rudolph, S. Three-dimensional soil organic matter distribution, accessibility and microbial respiration in macroaggregates using osmium staining and synchrotron X-ray computed tomography. SOIL 2016, 2, 659–671. [Google Scholar] [CrossRef]
- Jasinska, E.; Wetzel, H.; Baumgartl, T.; Horn, R. Heterogeneity of physico-chemical properties in structured soils and its consequences. Pedosphere 2006, 16, 284–296. [Google Scholar] [CrossRef]
- Schurgers, G.; Dörsch, P.; Bakken, L.; Leffelaar, P.; Haugen, L. Modelling soil anaerobiosis from water retention characteristics and soil respiration. Soil Biol. Biochem. 2006, 38, 2637–2644. [Google Scholar] [CrossRef]
- Ranjard, L.; Richaume, A. Quantitative and qualitative microscale distribution of bacteria in soil. Res. Microbiol. 2001, 152, 707–716. [Google Scholar] [CrossRef]
- Otten, W.; Gilligan, C.; Watts, C.; Dexter, A.; Hall, D. Continuity of air-filled pores and invasion thresholds for a soil-borne fungal plant pathogen, Rhizoctonia solani. Soil Biol. Biochem. 1999, 31, 1803–1810. [Google Scholar] [CrossRef]
- Otten, W.; Gilligan, C.A. Soil structure and soil-borne diseases: Using epidemiological concepts to scale from fungal spread to plant epidemics. Eur. J. Soil Sci. 2006, 57, 26–37. [Google Scholar] [CrossRef]
- Kaisermann, A.; Maron, P.; Beaumelle, L.; Lata, J. Fungal communities are more sensitive indicators to non-extreme soil moisture variations than bacterial communities. Appl. Soil Ecol. 2015, 86, 158–164. [Google Scholar] [CrossRef]
- Baveye, P.C.; Ebaveye, J.; Egowdy, J. Soil “ecosystem” services and natural capital: Critical appraisal of research on uncertain ground. Front. Environ. Sci. 2016, 4, 1–49. [Google Scholar] [CrossRef]
- Demanèche, S.; Jocteur-Monrozier, L.; Quiquampoix, H.; Simonet, P. Evaluation of biological and physical protection against nuclease degradation of clay-bound plasmid DNA. Appl. Environ. Microbiol. 2001, 67, 293–299. [Google Scholar] [CrossRef]
- Pietramellara, G.; Ascher, J.; Borgogni, F.; Ceccherini, M.T.; Guerri, G.; Nannipieri, P. Extracellular DNA in soil and sediment: Fate and ecological relevance. Biol. Fertil. Soils 2009, 45, 219–235. [Google Scholar] [CrossRef]
- Romanowski, G.; Lorenz, M.G.; Sayler, G.; Wackernagel, W. Persistence of free plasmid DNA in soil monitored by various methods, including a transformation assay. Appl. Environ. Microbiol. 1992, 58, 3012–3019. [Google Scholar] [CrossRef]
- Stotzky, G. Influence of soil mineral colloids on metabolic processes, growth, adhesion, and ecology of microbes and viruses. Geochem. Soil Radionucl. 2015, 17, 305–428. [Google Scholar] [CrossRef]
- Nielsen, K.M.; Calamai, L.; Pietramellara, G. Stabilization of extracellular DNA and proteins by transient binding to various soil components. Soil Biol. 2006, 8, 141–157. [Google Scholar] [CrossRef]
- Konhauser, K. Introduction to Geomicrobiology, 1st ed.; John Wiley & Sons: Hoboken, NJ, USA; Blackwell Publishing: Hoboken, NJ, USA, 2007. [Google Scholar]
- Rappoldt, C.; Crawford, J. The distribution of anoxic volume in a fractal model of soil. Geoderma 1999, 88, 329–347. [Google Scholar] [CrossRef]
- Lombard, N.; Prestat, E.; van Elsas, J.D.; Simonet, P. Soil-specific limitations for access and analysis of soil microbial communities by metagenomics. FEMS Microbiol. Ecol. 2011, 78, 31–49. [Google Scholar] [CrossRef]
- Nunan, N.; Schmidt, H.; Raynaud, X. The ecology of heterogeneity: Soil bacterial communities and C dynamics. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190249. [Google Scholar] [CrossRef]
- Strong, D.T.; Wever, H.D.E.; Merckx, R.; Recous, S. Spatial location of carbon decomposition in the soil pore system. Eur. J. Soil Sci. 2004, 55, 739–750. [Google Scholar] [CrossRef]
- Salomé, C.; Nunan, N.; Pouteau, V.; Lerch, T.Z.; Chenu, C. Carbon dynamics in topsoil and in subsoil may be controlled by different regulatory mechanisms. Glob. Change Biol. 2010, 16, 416–426. [Google Scholar] [CrossRef]
- Ruamps, L.S.; Nunan, N.; Pouteau, V.; Leloup, J.; Raynaud, X.; Roy, V.; Chenu, C. Regulation of soil organic C mineralisation at the pore scale. FEMS Microbiol. Ecol. 2013, 86, 26–35. [Google Scholar] [CrossRef]
- Ruamps, L.S.; Nunan, N.; Chenu, C. Microbial biogeography at the soil pore scale. Soil Biol. Biochem. 2011, 43, 280–286. [Google Scholar] [CrossRef]
- Frey-Klett, P.; Burlinson, P.; Deveau, A.; Barret, M.; Tarkka, M.; Sarniguet, A. Bacterial–fungal interactions: Hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol. Mol. Biol. Rev. 2011, 75, 583–609. [Google Scholar] [CrossRef]
- Bonfante, P.; Anca, I.-A. Plants, mycorrhizal fungi, and bacteria: A network of interactions. Annu. Rev. Microbiol. 2009, 63, 363–383. [Google Scholar] [CrossRef] [PubMed]
- Deveau, A.; Bonito, G.; Uehling, J.; Paoletti, M.; Becker, M.; Bindschedler, S.; Hacquard, S.; Hervé, V.; Labbé, J.; Lastovetsky, O.A.; et al. Bacterial–fungal interactions: Ecology, mechanisms and challenges. FEMS Microbiol. Rev. 2018, 42, 335–352. [Google Scholar] [CrossRef] [PubMed]
- Clarholm, M. Protozoan grazing of bacteria in soil—Impact and importance. Microb. Ecol. 1981, 7, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Kuikman, P.; van Elsas, J.; Jansen, A.; Burgers, S.; van Veen, J. Population dynamics and activity of bacteria and protozoa in relation to their spatial distribution in soil. Soil Biol. Biochem. 1990, 22, 1063–1073. [Google Scholar] [CrossRef]
- Rutherford, P.M.; Juma, N.G. Influence of texture on habitable pore space and bacterial-protozoan populations in soil. Biol. Fertil. Soils 1992, 12, 221–227. [Google Scholar] [CrossRef]
- Wright, D.A.; Killham, K.; Glover, L.A.; Prosser, J.I. Role of pore size location in determining bacterial activity during predation by protozoa in soil. Appl. Environ. Microbiol. 1995, 61, 3537–3543. [Google Scholar] [CrossRef]
- Wright, D.; Killham, K.; Glover, L.; Prosser, J. The effect of location in soil on protozoal grazing of a genetically modified bacterial inoculum. Geoderma 1993, 56, 633–640. [Google Scholar] [CrossRef]
- Männik, J.; Driessen, R.; Galajda, P.; Keymer, J.E.; Dekker, C. Bacterial growth and motility in sub-micron constrictions. Proc. Natl. Acad. Sci. USA 2009, 106, 14861–14866. [Google Scholar] [CrossRef]
- Kravchenko, A.N.; Negassa, W.C.; Guber, A.K.; Hildebrandt, B.; Marsh, T.; Rivers, M.L. Intra-aggregate pore structure influences phylogenetic composition of bacterial community in macroaggregates. Soil Sci. Soc. Am. J. 2014, 78, 1924–1939. [Google Scholar] [CrossRef]
- Dechesne, A.; Wang, G.; Gulez, G.; Or, D.; Smets, B.F. Hydration-controlled bacterial motility and dispersal on surfaces. Proc. Natl. Acad. Sci. USA 2010, 107, 14369–14372. [Google Scholar] [CrossRef]
- Vos, M.; Wolf, A.B.; Jennings, S.J.; Kowalchuk, G.A. Micro-scale determinants of bacterial diversity in soil. FEMS Microbiol. Rev. 2013, 37, 936–954. [Google Scholar] [CrossRef] [PubMed]
- Raynaud, X.; Nunan, N. Spatial ecology of bacteria at the microscale in soil. PLoS ONE 2014, 9, e87217. [Google Scholar] [CrossRef]
- Pennell, K.D. Specific surface area. In Reference Module in Earth Systems and Environmental Sciences; Elias, S.A., Ed.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 1–8. [Google Scholar]
- Boswell, G.P.; Jacobs, H.; Ritz, K.; Gadd, G.M.; Davidson, F. The development of fungal networks in complex environments. Bull. Math. Biol. 2007, 69, 605–634. [Google Scholar] [CrossRef] [PubMed]
- Fricker, M.D.; Heaton, L.L.M.; Jones, N.S.; Boddy, L. The mycelium as a network. Fungal Kingd. 2017, 5, 335–367. [Google Scholar] [CrossRef]
- Kohlmeier, S.; Smits, T.H.; Ford, R.M.; Keel, C.; Harms, H.; Wick, L.Y. Taking the fungal highway: Mobilization of pollutant-degrading bacteria by fungi. Environ. Sci. Technol. 2005, 39, 4640–4646. [Google Scholar] [CrossRef]
- Warmink, J.A.; Nazir, R.; van Elsas, J.D. Universal and species-specific bacterial ‘fungiphiles’ in the mycospheres of different basidiomycetous fungi. Environ. Microbiol. 2009, 11, 300–312. [Google Scholar] [CrossRef]
- Simon, A.; Hervé, V.; Al-Dourobi, A.; Verrecchia, E.; Junier, P. An in situ inventory of fungi and their associated migrating bacteria in forest soils using fungal highway columns. FEMS Microbiol. Ecol. 2016, 93, fiw217. [Google Scholar] [CrossRef]
- Worrich, A.; Stryhanyuk, H.; Musat, N.; König, S.; Banitz, T.; Centler, F.; Frank, K.; Thullner, M.; Harms, H.; Richnow, H.-H.; et al. Mycelium-mediated transfer of water and nutrients stimulates bacterial activity in dry and oligotrophic environments. Nat. Commun. 2017, 8, 15472. [Google Scholar] [CrossRef]
- Guhr, A.; Borken, W.; Spohn, M.; Matzner, E. Redistribution of soil water by a saprotrophic fungus enhances carbon mineralization. Proc. Natl. Acad. Sci. USA 2015, 112, 14647–14651. [Google Scholar] [CrossRef] [PubMed]
- Pion, M.; Spangenberg, J.E.; Simon, A.; Bindschedler, S.; Flury, C.; Chatelain, A.; Bshary, R.; Job, D.; Junier, P. Bacterial farming by the fungus Morchella crassipes. Proc. R. Soc. B Boil. Sci. 2013, 280, 20132242. [Google Scholar] [CrossRef] [PubMed]
- Haq, I.U.; Zhang, M.; Yang, P.; van Elsas, J.D. The interactions of bacteria with fungi in soil. Adv. Appl. Microbiol. 2014, 89, 185–215. [Google Scholar] [CrossRef]
- Berthold, T.; Centler, F.; Hübschmann, T.; Remer, R.; Thullner, M.; Harms, H.; Wick, L.Y. Mycelia as a focal point for horizontal gene transfer among soil bacteria. Sci. Rep. 2016, 6, 36390. [Google Scholar] [CrossRef] [PubMed]
- Nazir, R.; Mazurier, S.; Yang, P.; Lemanceau, P.; van Elsas, J.D. The ecological role of type three secretion systems in the interaction of bacteria with fungi in soil and related habitats is diverse and context-dependent. Front. Microbiol. 2017, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Otto, S.; Bruni, E.P.; Harms, H.; Wick, L.Y. Catch me if you can: Dispersal and foraging of Bdellovibrio bacteriovorus 109J along mycelia. ISME J. 2016, 11, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Esser, D.S.; Leveau, J.H.; Meyer, K.M.; Wiegand, K. Spatial scales of interactions among bacteria and between bacteria and the leaf surface. FEMS Microbiol. Ecol. 2015, 91, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Gantner, S.; Schmid, M.; Dürr, C.; Schuhegger, R.; Steidle, A.; Hutzler, P.; Langebartels, C.; Eberl, L.; Hartmann, A.; Dazzo, F.B. In situ quantitation of the spatial scale of calling distances and population density-independent N-acylhomoserine lactone-mediated communication by rhizobacteria colonized on plant roots. FEMS Microbiol. Ecol. 2006, 56, 188–194. [Google Scholar] [CrossRef]
- Cai, P.; Sun, X.; Wu, Y.; Gao, C.; Mortimer, M.; Holden, P.A.; Redmile-Gordon, M.; Huang, Q. Soil biofilms: Microbial interactions, challenges, and advanced techniques for ex-situ characterization. Soil Ecol. Lett. 2019, 1, 85–93. [Google Scholar] [CrossRef]
- Schmidt, R.; Cordovez, V.; de Boer, W.; Raaijmakers, J.; Garbeva, P. Volatile affairs in microbial interactions. ISME J. 2015, 9, 2329–2335. [Google Scholar] [CrossRef]
- Effmert, U.; Kalderás, J.; Warnke, R.; Piechulla, B. Volatile mediated interactions between bacteria and fungi in the soil. J. Chem. Ecol. 2012, 38, 665–703. [Google Scholar] [CrossRef]
- Nazir, R.; Warmink, J.A.; Boersma, H.; van Elsas, J.D. Mechanisms that promote bacterial fitness in fungal-affected soil microhabitats. FEMS Microbiol. Ecol. 2009, 71, 169–185. [Google Scholar] [CrossRef]
- Bignell, E. The molecular basis of pH sensing, signaling, and homeostasis in fungi. Adv. Appl. Microbiol. 2012, 79, 1–18. [Google Scholar] [PubMed]
- Braunsdorf, C.; Mailänder-Sánchez, D.; Schaller, M. Fungal sensing of host environment. Cell. Microbiol. 2016, 18, 1188–1200. [Google Scholar] [CrossRef]
- Lurthy, T.; Cantat, C.; Jeudy, C.; Declerck, P.; Gallardo, K.; Barraud, C.; Leroy, F.; Ourry, A.; Lemanceau, P.; Salon, C.; et al. Impact of bacterial siderophores on iron status and ionome in pea. Front. Plant Sci. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Haas, H. Fungal siderophore metabolism with a focus on Aspergillus fumigatus. Nat. Prod. Rep. 2014, 31, 1266–1276. [Google Scholar] [CrossRef] [PubMed]
- Barberán, A.; Bates, S.T.; Casamayor, E.O.; Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2011, 6, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Rev. Genet. 2012, 10, 538–550. [Google Scholar] [CrossRef]
- Jiang, D.; Armour, C.R.; Hu, C.; Mei, M.; Tian, C.; Sharpton, T.J.; Jiang, Y. Microbiome multi-omics network analysis: Statistical considerations, limitations, and opportunities. Front. Genet. 2019, 10, 995. [Google Scholar] [CrossRef]
- Matchado, M.S.; Lauber, M.; Reitmeier, S.; Kacprowski, T.; Baumbach, J.; Haller, D.; List, M. Network analysis methods for studying microbial communities: A mini review. Comput. Struct. Biotechnol. J. 2021, 19, 2687–2698. [Google Scholar] [CrossRef]
- Weiss, S.; van Treuren, W.; Lozupone, C.; Faust, K.; Friedman, J.; Deng, Y.; Xia, L.C.; Xu, Z.Z.; Ursell, L.; Alm, E.J.; et al. Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME J. 2016, 10, 1669–1681. [Google Scholar] [CrossRef]
- Telagathoti, A.; Probst, M.; Peintner, U. Habitat, snow-cover and soil pH, affect the distribution and diversity of mortierellaceae species and their associations to bacteria. Front. Microbiol. 2021, 12, 669784. [Google Scholar] [CrossRef]
- Nannipieri, P.; Ascher, J.; Ceccherini, M.T.; Landi, L.; Pietramellara, G.; Renella, G. Microbial diversity and soil functions. Eur. J. Soil Sci. 2003, 54, 655–670. [Google Scholar] [CrossRef]
- Nannipieri, P.; Ascher, J.; Ceccherini, M.T.; Landi, L.; Pietramellara, G.; Renella, G. Landmark Papers: No. 6. Eur. J. Soil Sci. 2017, 70, 1. [Google Scholar] [CrossRef]
- Nunan, N. The microbial habitat in soil: Scale, heterogeneity and functional consequences. J. Plant Nutr. Soil Sci. 2017, 180, 425–429. [Google Scholar] [CrossRef]
- Cordero, O.X.; Datta, M.S. Microbial interactions and community assembly at microscales. Curr. Opin. Microbiol. 2016, 31, 227–234. [Google Scholar] [CrossRef]
- Gause, G.F. The Struggle for Existence; Williams and Wilkins Co.: Baltimore, MD, USA, 1934. [Google Scholar]
- Bravo, D.; Cailleau, G.; Bindschedler, S.; Simon, A.; Job, D.; Verrecchia, E.; Junier, P. Isolation of oxalotrophic bacteria able to disperse on fungal mycelium. FEMS Microbiol. Lett. 2013, 348, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Ingham, C.J.; Kalisman, O.; Finkelshtein, A.; Ben-Jacob, E. Mutually facilitated dispersal between the nonmotile fungus Aspergillus fumigatus and the swarming bacterium Paenibacillus vortex. Proc. Natl. Acad. Sci. USA 2011, 108, 19731–19736. [Google Scholar] [CrossRef] [PubMed]
- Krug, L.; Erlacher, A.; Markut, K.; Berg, G.; Cernava, T. The microbiome of alpine snow algae shows a specific inter-kingdom connectivity and algae-bacteria interactions with supportive capacities. ISME J. 2020, 14, 2197–2210. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, D.; Vitale, S.; Lima, G.; di Pietro, A.; Turrà, D. A bacterial endophyte exploits chemotropism of a fungal pathogen for plant colonization. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef]
- Schmidt, R.; de Jager, V.; Zühlke, D.; Wolff, C.; Bernhardt, J.; Cankar, K.; Beekwilder, J.; van Ijcken, W.; Sleutels, F.; de Boer, W.; et al. Fungal volatile compounds induce production of the secondary metabolite sodorifen in Serratia plymuthica PRI-2C. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Stopnišek, N.; Zühlke, D.; Carlier, A.; Barberán, A.; Fierer, N.; Becher, D.; Riedel, K.; Eberl, L.; Weisskopf, L. Molecular mechanisms underlying the close association between soil Burkholderia and fungi. ISME J. 2015, 10, 253–264. [Google Scholar] [CrossRef]
- Aspray, T.J.; Jones, E.E.; Davies, M.W.; Shipman, M.; Bending, G.D. Increased hyphal branching and growth of ectomycorrhizal fungus Lactarius rufus by the helper bacterium Paenibacillus sp. Mycorrhiza 2013, 23, 403–410. [Google Scholar] [CrossRef]
- Dresch, P.; Falbesoner, J.; Ennemoser, C.; Hittorf, M.; Kuhnert, R.; Peintner, U. Emerging from the ice-fungal communities are diverse and dynamic in earliest soil developmental stages of a receding glacier. Environ. Microbiol. 2019, 21, 1864–1880. [Google Scholar] [CrossRef]
- Casar, C.P.; Kruger, B.R.; Flynn, T.M.; Masterson, A.L.; Momper, L.M.; Osburn, M.R. Mineral-hosted biofilm communities in the continental deep subsurface, Deep Mine Microbial Observatory, SD, USA. Geobiology 2020, 18, 508–522. [Google Scholar] [CrossRef]
- Wallander, H.; Nilsson, L.O.; Hagerberg, D.; Bååth, E. Estimation of the biomass and seasonal growth of external mycelium of ectomycorrhizal fungi in the field. New Phytol. 2001, 151, 753–760. [Google Scholar] [CrossRef]
- Harvey, H.; Wildman, R.D.; Mooney, S.J.; Avery, S.V. Soil aggregates by design: Manufactured aggregates with defined microbial composition for interrogating microbial activities in soil microhabitats. Soil Biol. Biochem. 2020, 148, 107870. [Google Scholar] [CrossRef]
- Helliwell, J.; Miller, T.; Whalley, R.; Mooney, S.; Sturrock, C. Quantifying the impact of microbes on soil structural development and behaviour in wet soils. Soil Biol. Biochem. 2014, 74, 138–147. [Google Scholar] [CrossRef]
- Junier, P.; Cailleau, G.; Palmieri, I.; Vallotton, C.; Trautschold, O.C.; Junier, T.; Paul, C.; Bregnard, D.; Palmieri, F.; Estoppey, A.; et al. Democratization of fungal highway columns as a tool to investigate bacteria associated with soil fungi. FEMS Microbiol. Ecol. 2021, 97, fiab003. [Google Scholar] [CrossRef] [PubMed]
- Nichols, D.; Cahoon, N.; Trakhtenberg, E.M.; Pham, L.; Mehta, A.; Belanger, A.; Kanigan, T.; Lewis, K.; Epstein, S.S. Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl. Environ. Microbiol. 2010, 76, 2445–2450. [Google Scholar] [CrossRef] [PubMed]
- Das, P.S.; Gagnon-Turcotte, G.; Ouazaa, K.; Bouzid, K.; Hosseini, S.N.; Bharucha, E.; Tremblay, D.; Moineau, S.; Messaddeq, Y.; Corbeil, J.; et al. The EcoChip 2: An autonomous sensor platform for multimodal bio-environmental monitoring of the northern habitat. In Proceedings of the 42nd Annual International Conferences of the IEEE Engineering in Medicine and Biology Society, Montreal, QC, Canada, 20–24 July 2020; pp. 4101–4104. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Z.; Ma, Z.; Chen, J.; Akbar, J.; Zhang, S.; Che, C.; Zhang, M.; Cerdà, A. A review of preferential water flow in soil science. Can. J. Soil Sci. 2018, 98, 604–618. [Google Scholar] [CrossRef]
- Lilje, O.; Lilje, E.; Marano, A.V.; Gleason, F.H. Three dimensional quantification of biological samples using micro-computer aided tomography (microCT). J. Microbiol. Methods 2013, 92, 33–41. [Google Scholar] [CrossRef]
- Davit, Y.; Iltis, G.; Debenest, G.; Veran-Tissoires, S.; Wildenschild, D.; Gerino, M.; Quintard, M. Imaging biofilm in porous media using X-ray computed microtomography. J. Microsc. 2010, 242, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Iltis, G.C.; Armstrong, R.T.; Jansik, D.P.; Wood, B.D.; Wildenschild, D. Imaging biofilm architecture within porous media using synchrotron-based X-ray computed microtomography. Water Resour. Res. 2011, 47, 1–5. [Google Scholar] [CrossRef]
- Peszynska, M.; Trykozko, A.; Iltis, G.; Schlueter, S.; Wildenschild, D. Biofilm growth in porous media: Experiments, computational modeling at the porescale, and upscaling. Adv. Water Resour. 2016, 95, 288–301. [Google Scholar] [CrossRef]
- Sanderlin, A.B.; Vogt, S.; Grunewald, E.; Bergin, B.A.; Codd, S.L. Biofilm detection in natural unconsolidated porous media using a low-field magnetic resonance system. Environ. Sci. Technol. 2012, 47, 987–992. [Google Scholar] [CrossRef]
- Lee, B.H.; Lee, S.K. Probing the water distribution in porous model sands with two immiscible fluids: A nuclear magnetic resonance micro-imaging study. J. Hydrol. 2017, 553, 637–650. [Google Scholar] [CrossRef]
- Vogt, S.J.; Sanderlin, A.B.; Seymour, J.D.; Codd, S.L. Permeability of a growing biofilm in a porous media fluid flow analyzed by magnetic resonance displacement-relaxation correlations. Biotechnol. Bioeng. 2012, 110, 1366–1375. [Google Scholar] [CrossRef]
- Carrel, M.; Beltran, M.A.; Morales, V.L.; Derlon, N.; Morgenroth, E.; Kaufmann, R.; Holzner, M. Biofilm imaging in porous media by laboratory X-ray tomography: Combining a non-destructive contrast agent with propagation-based phase-contrast imaging tools. PLoS ONE 2017, 12, e0180374. [Google Scholar] [CrossRef]
- Downie, H.F.; Valentine, T.; Otten, W.; Spiers, A.; Dupuy, L. Transparent soil microcosms allow 3D spatial quantification of soil microbiological processes in vivo. Plant Signal. Behav. 2014, 9, e970421. [Google Scholar] [CrossRef]
- Downie, H.; Holden, N.; Otten, W.; Spiers, A.; Valentine, T.; Dupuy, L.X. Transparent soil for imaging the rhizosphere. PLoS ONE 2012, 7, e44276. [Google Scholar] [CrossRef] [PubMed]
- O’Callaghan, F.E.; Braga, R.; Neilson, R.; Macfarlane, S.A.; Dupuy, L.X. New live screening of plant-nematode interactions in the rhizosphere. Sci. Rep. 2018, 8, 1–17. [Google Scholar] [CrossRef]
- Foster, R.C. Microenvironments of soil microorganisms. Biol. Fertil. Soils 1988, 6, 189–203. [Google Scholar] [CrossRef]
- Lamandé, M.; Schjønning, P.; Ferro, N.D.; Morari, F. Soil pore system evaluated from gas measurements and CT images: A conceptual study using artificial, natural and 3D-printed soil cores. Eur. J. Soil Sci. 2021, 72, 769–781. [Google Scholar] [CrossRef]
- Otten, W.; Pajor, R.; Schmidt, S.; Baveye, P.; Hague, R.; Falconer, R.E. Combining X-ray CT and 3D printing technology to produce microcosms with replicable, complex pore geometries. Soil Biol. Biochem. 2012, 51, 53–55. [Google Scholar] [CrossRef]
- Aleklett, K.; Kiers, E.T.; Ohlsson, P.; Shimizu, T.S.; Caldas, V.E.; Hammer, E.C. Build your own soil: Exploring microfluidics to create microbial habitat structures. ISME J. 2018, 12, 312–319. [Google Scholar] [CrossRef]
- Burmeister, A.; Grünberger, A. Microfluidic cultivation and analysis tools for interaction studies of microbial co-cultures. Curr. Opin. Biotechnol. 2020, 62, 106–115. [Google Scholar] [CrossRef]
- Harvey, H.J.; Wildman, R.D.; Mooney, S.J.; Avery, S.V. Challenges and approaches in assessing the interplay between microorganisms and their physical micro-environments. Comput. Struct. Biotechnol. J. 2020, 18, 2860–2866. [Google Scholar] [CrossRef]
- Wessel, A.K.; Hmelo, L.; Parsek, M.R.; Whiteley, M. Going local: Technologies for exploring bacterial microenvironments. Nat. Rev. Genet. 2013, 11, 337–348. [Google Scholar] [CrossRef]
- Zhou, W.; Le, J.; Chen, Y.; Cai, Y.; Hong, Z.; Chai, Y. Recent advances in microfluidic devices for bacteria and fungus research. TrAC Trends Anal. Chem. 2019, 112, 175–195. [Google Scholar] [CrossRef]
- Weibel, D.B.; DiLuzio, W.R.; Whitesides, G.M. Microfabrication meets microbiology. Nat. Rev. Microbiol. 2007, 5, 209–218. [Google Scholar] [CrossRef]
- Seymour, J.R.; Ahmed, T.; Stocker, R. A microfluidic chemotaxis assay to study microbial behavior in diffusing nutrient patches. Limnol. Oceanogr. Methods 2008, 6, 477–488. [Google Scholar] [CrossRef]
- Kim, H.J.; Boedicker, J.Q.; Choi, J.W.; Ismagilov, R.F. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc. Natl. Acad. Sci. USA 2008, 105, 18188–18193. [Google Scholar] [CrossRef]
- Boedicker, J.Q.; Vincent, M.E.; Ismagilov, R.F. Microfluidic confinement of single cells of bacteria in small volumes initiates high-density behavior of quorum sensing and growth and reveals its variability. Angew. Chem. Int. Ed. 2009, 48, 5908–5911. [Google Scholar] [CrossRef]
- Mukhopadhyay, R. When PDMS isn’t the best. What are its weaknesses, and which other polymers can researchers add to their toolboxes? Anal. Chem. 2007, 79, 3248–3253. [Google Scholar] [CrossRef] [PubMed]
- Mcdonald, J.C.; Duffy, D.C.; Anderson, J.R.; Chiu, D.T. Review general fabrication of microfluidic systems in poly (dimethylsiloxane). Electrophoresis 2000, 21, 27–40. [Google Scholar] [CrossRef]
- Connell, J.L.; Wessel, A.K.; Parsek, M.R.; Ellington, A.D.; Whiteley, M.; Shear, J.B. Probing prokaryotic social behaviors with bacterial “lobster traps”. mBio 2010, 1, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Baca, H.K.; Ashley, C.; Carnes, E.; Lopez, D.; Flemming, J.; Dunphy, D.; Singh, S.; Chen, Z.; Liu, N.; Fan, H.; et al. Cell-directed assembly of lipid-silica nanostructures providing extended cell viability. Science 2006, 313, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Carnes, E.C.; Lopez, D.M.; Donegan, N.; Cheung, A.; Gresham, H.; Timmins, G.; Brinker, C.J. Confinement-induced quorum sensing of individual Staphylococcus aureus bacteria. Nat. Chem. Biol. 2009, 6, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Orner, E.; Chau, J.F.; Anderson, E.M.; Kadilak, A.L.; Rubinstein, R.; Bouchillon, G.M.; Goodwin, R.; Gage, D.; Shor, L.M. Synergistic effects of soil microstructure and bacterial EPS on drying rate in emulated soil micromodels. Soil Biol. Biochem. 2015, 83, 116–124. [Google Scholar] [CrossRef]
- Rubinstein, R.; Kadilak, A.L.; Cousens, V.C.; Gage, D.; Shor, L.M. Protist-facilitated particle transport using emulated soil micromodels. Environ. Sci. Technol. 2015, 49, 1384–1391. [Google Scholar] [CrossRef] [PubMed]
- Wondraczek, L.; Pohnert, G.; Schacher, F.H.; Köhler, A.; Gottschaldt, M.; Schubert, U.S.; Küsel, K.; Brakhage, A.A. Artificial microbial arenas: Materials for observing and manipulating microbial consortia. Adv. Mater. 2019, 31, e1900284. [Google Scholar] [CrossRef]
- Gimeno, A.; Stanley, C.E.; Ngamenie, Z.; Hsung, M.-H.; Walder, F.; Schmieder, S.S.; Bindschedler, S.; Junier, P.; Keller, B.; Vogelgsang, S. A versatile microfluidic platform measures hyphal interactions between Fusarium graminearum and Clonostachys rosea in real-time. Commun. Biol. 2021, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hanson, K.L.; Nicolau, D.V.; Filipponi, L.; Wang, L.; Lee, A.P. Fungi use efficient algorithms for the exploration of microfluidic networks. Small 2006, 2, 1212–1220. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.Y.; Wang, S.; Dick, G.J.; Young, V.B.; Sherman, D.H.; Burns, M.A.; Lin, X.N. Co-cultivation of microbial sub-communities in microfluidic droplets facilitates high-resolution genomic dissection of microbial ‘dark matter’. Integr. Biol. 2020, 12, 263–274. [Google Scholar] [CrossRef]
- Kaehr, B.; Shear, J.B. Multiphoton fabrication of chemically responsive protein hydrogels for microactuation. Proc. Natl. Acad. Sci. USA 2008, 105, 8850–8854. [Google Scholar] [CrossRef]
- Zhang, Z.; Boccazzi, P.; Choi, H.-G.; Perozziello, G.; Sinskey, A.J.; Jensen, K. Microchemostat—Microbial continuous culture in a polymer-based, instrumented microbioreactor. Lab Chip 2006, 6, 906–913. [Google Scholar] [CrossRef]
- Pamp, S.J.; Sternberg, C.; Tolker-Nielsen, T. Insight into the microbial multicellular lifestyle via flow-cell technology and confocal microscopy. Cytom. Part A 2008, 75, 90–103. [Google Scholar] [CrossRef]
- Massalha, H.; Korenblum, E.; Malitsky, S.; Shapiro, O.H.; Aharoni, A. Live imaging of root–bacteria interactions in a microfluidics setup. Proc. Natl. Acad. Sci. USA 2017, 114, 4549–4554. [Google Scholar] [CrossRef]
- Schmieder, S.S.; Stanley, C.; Rzepiela, A.; van Swaay, D.; Sabotič, J.; Nørrelykke, S.; Demello, A.J.; Aebi, M.; Künzler, M. Bidirectional propagation of signals and nutrients in fungal networks via specialized hyphae. Curr. Biol. 2019, 29, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Tayyrov, A.; Stanley, C.E.; Azevedo, S.; Künzler, M. Combining microfluidics and RNA-sequencing to assess the inducible defensome of a mushroom against nematodes. BMC Genom. 2019, 20, 1–13. [Google Scholar] [CrossRef]
- Uehling, J.K.; Entler, M.R.; Meredith, H.R.; Millet, L.J.; Timm, C.M.; Aufrecht, J.A.; Bonito, G.M.; Engle, N.L.; Labbé, J.L.; Doktycz, M.J.; et al. Microfluidics and metabolomics reveal symbiotic bacterial–fungal interactions between Mortierella elongata and Burkholderia include metabolite exchange. Front. Microbiol. 2019, 10, 2163. [Google Scholar] [CrossRef] [PubMed]
- Stanley, C.; Stöckli, M.; van Swaay, D.; Sabotič, J.; Kallio, P.T.; Künzler, M.; Demello, A.J.; Aebi, M. Probing bacterial–fungal interactions at the single cell level. Integr. Biol. 2014, 6, 935–945. [Google Scholar] [CrossRef]
- Eun, Y.J.; Utada, A.S.; Copeland, M.F.; Takeuchi, S.; Weibel, D.B. Encapsulating bacteria in agarose microparticles using microfluidics for high-throughput cell analysis and isolation. ACS Chem. Biol. 2010, 6, 260–266. [Google Scholar] [CrossRef]
- Churski, K.; Kaminski, T.S.; Jakiela, S.; Kamysz, W.; Barańska-Rybak, W.; Weibel, D.B.; Garstecki, P. Rapid screening of antibiotic toxicity in an automated microdroplet system. Lab Chip 2012, 12, 1629–1637. [Google Scholar] [CrossRef]
- Abraham, W.-R. Applications and impacts of stable isotope probing for analysis of microbial interactions. Appl. Microbiol. Biotechnol. 2014, 98, 4817–4828. [Google Scholar] [CrossRef]
- Partida-Martinez, L.P.; Groth, I.; Schmitt, I.; Richter, W.; Roth, M.; Hertweck, C. Burkholderia rhizoxinica sp. nov. and Burkholderia endofungorum sp. nov., bacterial endosymbionts of the plant-pathogenic fungus Rhizopus microsporus. Int. J. Syst. Evol. Microbiol. 2007, 57, 2583–2590. [Google Scholar] [CrossRef]
- Söderström, B.; Erland, S. Isolation of fluorescein diacetate stained hyphae from soil by micromanipulation. Trans. Br. Mycol. Soc. 1986, 86, 465–468. [Google Scholar] [CrossRef]
- Dennis, P.G.; Miller, A.J.; Clark, I.M.; Taylor, R.G.; Valsami-Jones, E.; Hirsch, P.R. A novel method for sampling bacteria on plant root and soil surfaces at the microhabitat scale. J. Microbiol. Methods 2008, 75, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Ashida, N.; Ishii, S.; Hayano, S.; Tago, K.; Tsuji, T.; Yoshimura, Y.; Otsuka, S.; Senoo, K. Isolation of functional single cells from environments using a micromanipulator: Application to study denitrifying bacteria. Appl. Microbiol. Biotechnol. 2009, 85, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, T.; Tago, K.; Uei, Y.; Ishii, S.; Isobe, K.; Otsuka, S.; Senoo, K. Advantages of functional single-cell isolation method over standard agar plate dilution method as a tool for studying denitrifying bacteria in rice paddy soil. AMB Express 2012, 2, 50. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, J.; König, H. New techniques for isolation of single prokaryotic cells. FEMS Microbiol. Rev. 2000, 24, 567–572. [Google Scholar] [CrossRef]
- Whitley, K.D.; Comstock, M.J.; Chemla, Y.R. High-resolution “fleezers”: Dual-trap optical tweezers combined with single-molecule fluorescence detection. In Optical Tweezers. Methods in Molecular Biology; Gennerich, A., Ed.; Humana Press: New York, NY, USA, 2017; Volume 1486, pp. 183–256. [Google Scholar] [CrossRef]
- Ringeisen, B.R.; Rincon, K.; Fitzgerald, L.A.; Fulmer, P.A.; Wu, P.K. Printing soil: A single-step, high-throughput method to isolate micro-organisms and near-neighbour microbial consortia from a complex environmental sample. Methods Ecol. Evol. 2014, 6, 209–217. [Google Scholar] [CrossRef]
- Sato, Y.; Narisawa, K.; Tsuruta, K.; Umezu, M.; Nishizawa, T.; Tanaka, K.; Yamaguchi, K.; Komatsuzaki, M.; Ohta, H. Detection of betaproteobacteria inside the mycelium of the fungus Mortierella elongata. Microbes Environ. 2010, 25, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Ghodsalavi, B.; Svenningsen, N.B.; Hao, X.; Olsson, S.; Nicolaisen, M.H.; Abu Al-Soud, W.; Sørensen, S.J.; Nybroe, O. A novel baiting microcosm approach used to identify the bacterial community associated with Penicillium bilaii hyphae in soil. PLoS ONE 2017, 12, e0187116. [Google Scholar] [CrossRef] [PubMed]
- Postma, J.; Altemüller, H.-J. Bacteria in thin soil sections stained with the fluorescent brightener calcofluor white M2R. Soil Biol. Biochem. 1990, 22, 89–96. [Google Scholar] [CrossRef]
- Hoffman, M.T.; Gunatilaka, M.K.; Wijeratne, K.; Gunatilaka, L.; Arnold, A.E. Endohyphal bacterium enhances production of indole-3-acetic acid by a foliar fungal endophyte. PLoS ONE 2013, 8, e73132. [Google Scholar] [CrossRef] [PubMed]
- Bertaux, J.; Schmid, M.; Prevost-Boure, N.C.; Churin, J.L.; Hartmann, A.; Garbaye, J.; Frey-Klett, P. In situ identification of intracellular bacteria related to Paenibacillus spp. in the mycelium of the ectomycorrhizal fungus Laccaria bicolor S238N. Appl. Environ. Microbiol. 2003, 69, 4243–4248. [Google Scholar] [CrossRef]
- Hoffman, M.T.; Arnold, A.E. Diverse bacteria inhabit living hyphae of phylogenetically diverse fungal endophytes. Appl. Environ. Microbiol. 2010, 76, 4063–4075. [Google Scholar] [CrossRef]
- Naumann, M.; Schüssler, A.; Bonfante, P. The obligate endobacteria of arbuscular mycorrhizal fungi are ancient heritable components related to the mollicutes. ISME J. 2010, 4, 862–871. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.Y.-Y.; Chen, A. Fluorescence in situ hybridization. J. Investig. Dermatol. 2013, 133, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.; Eickhorst, T.; Mußmann, M. Gold-FISH: A new approach for the in situ detection of single microbial cells combining fluorescence and scanning electron microscopy. Syst. Appl. Microbiol. 2012, 35, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.; Eickhorst, T.; Tippkötter, R. Evaluation of tyramide solutions for an improved detection and enumeration of single microbial cells in soil by CARD-FISH. J. Microbiol. Methods 2012, 91, 399–405. [Google Scholar] [CrossRef]
- Schmidt, H.; Eickhorst, T. Detection and quantification of native microbial populations on soil-grown rice roots by catalyzed reporter deposition-fluorescencein situhybridization. FEMS Microbiol. Ecol. 2014, 87, 390–402. [Google Scholar] [CrossRef]
- Behrens, S.; Lösekann, T.; Pett-Ridge, J.; Weber, P.K.; Ng, W.-O.; Stevenson, B.; Hutcheon, I.D.; Relman, D.A.; Spormann, A.M. Linking microbial phylogeny to metabolic activity at the single-cell level by using enhanced element labeling-catalyzed reporter deposition fluorescence in situ hybridization (EL-FISH) and NanoSIMS. Appl. Environ. Microbiol. 2008, 74, 3143–3150. [Google Scholar] [CrossRef] [PubMed]
- Nunan, N.; Ritz, K.; Crabb, D.; Harris, K.; Wu, K.; Crawford, J.W.; Young, I.M. Quantification of the in situ distribution of soil bacteria by large-scale imaging of thin sections of undisturbed soil. FEMS Microbiol. Ecol. 2001, 37, 67–77. [Google Scholar] [CrossRef]
- Hapca, S.; Baveye, P.; Wilson, C.; Lark, R.; Otten, W. Three-dimensional mapping of soil chemical characteristics at micrometric scale by combining 2D SEM-EDX data and 3D X-ray CT images. PLoS ONE 2015, 10, e0137205. [Google Scholar] [CrossRef] [PubMed]
- Nunan, N.; Wu, K.; Young, I.M.; Crawford, J.W.; Ritz, K. Spatial distribution of bacterial communities and their relationships with the micro-architecture of soil. FEMS Microbiol. Ecol. 2003, 44, 203–215. [Google Scholar] [CrossRef]
- Oburger, E.; Schmidt, H. New methods to unravel rhizosphere processes. Trends Plant Sci. 2016, 21, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Bradley, R.S.; Robinson, I.; Yusuf, M. 3D X-ray nanotomography of cells grown on electrospun scaffolds. Macromol. Biosci. 2016, 17, 1–8. [Google Scholar] [CrossRef]
- Bouckaert, L.; van Loo, D.; Ameloot, N.; Buchan, D.; van Hoorebeke, L.; Sleutel, S. Compatibility of X-ray micro-computed tomography with soil biological experiments. Soil Biol. Biochem. 2013, 56, 10–12. [Google Scholar] [CrossRef][Green Version]
- Sturrock, C.; Woodhall, J.; Brown, M.; Walker, C.; Mooney, S.; Ray, R.V. Effects of damping-off caused by Rhizoctonia solani anastomosis group 2-1 on roots of wheat and oil seed rape quantified using X-ray computed tomography and real-time PCR. Front. Plant Sci. 2015, 6, 461. [Google Scholar] [CrossRef]
- Hingley-Wilson, S.M.; Ma, N.; Hu, Y.; Casey, R.; Bramming, A.; Curry, R.J.; Tang, H.L.; Wu, H.; Butler, R.E.; Jacobs, W.R.; et al. Loss of phenotypic inheritance associated with ydcI mutation leads to increased frequency of small, slow persisters in Escherichia coli. Proc. Natl. Acad. Sci. USA 2020, 117, 4152–4157. [Google Scholar] [CrossRef] [PubMed]
- Juyal, A.; Otten, W.; Baveye, P.C.; Eickhorst, T. Influence of soil structure on the spread of Pseudomonas fluorescens in soil at microscale. Eur. J. Soil Sci. 2021, 72, 141–153. [Google Scholar] [CrossRef]
- Zappala, S.; Helliwell, J.R.; Tracy, S.; Mairhofer, S.; Sturrock, C.; Pridmore, T.; Bennett, M.; Mooney, S. Effects of X-ray dose on rhizosphere studies using X-ray computed tomography. PLoS ONE 2013, 8, e67250. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.; Vetterlein, D.; Köhne, J.M.; Eickhorst, T. Negligible effect of X-ray μ-CT scanning on archaea and bacteria in an agricultural soil. Soil Biol. Biochem. 2015, 84, 21–27. [Google Scholar] [CrossRef]
- Arendt, K.R.; Hockett, K.; Araldi-Brondolo, S.J.; Baltrus, D.A.; Arnold, A.E. Isolation of Endohyphal bacteria from foliar ascomycota and in vitro establishment of their symbiotic associations. Appl. Environ. Microbiol. 2016, 82, 2943–2949. [Google Scholar] [CrossRef]
- Benoit, I.; van den Esker, M.H.; Patyshakuliyeva, A.; Mattern, D.J.; Blei, F.; Zhou, M.; Dijksterhuis, J.; Brakhage, A.A.; Kuipers, O.P.; de Vries, R.P.; et al. Bacillus subtilis attachment to Aspergillus nigerhyphae results in mutually altered metabolism. Environ. Microbiol. 2014, 17, 2099–2113. [Google Scholar] [CrossRef]
- Schuster, M.; Kilaru, S.; Guo, M.; Sommerauer, M.; Lin, C.; Steinberg, G. Red fluorescent proteins for imaging Zymoseptoria tritici during invasion of wheat. Fungal Genet. Biol. 2015, 79, 132–140. [Google Scholar] [CrossRef][Green Version]
- Lichius, A. Live-cell imaging in Trichoderma. In New and Future Developments in Microbial Biotechnology and Bioengineering: Recent Developments in Trichoderma Research; Gupta, V.K., Zeilinger, S., Singh, H.B., Druzhinina, I., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 75–108. [Google Scholar]
- Bard, A.J.; Mirkin, M.V. (Eds.) Scanning Electrochemical Microscopy; Marcel Dekker: New York, NY, USA, 2001. [Google Scholar]
- Liu, X.; Ramsey, M.M.; Chen, X.; Koley, D.; Whiteley, M.; Bard, A.J. Real-time mapping of a hydrogen peroxide concentration profile across a polymicrobial bacterial biofilm using scanning electrochemical microscopy. Proc. Natl. Acad. Sci. USA 2011, 108, 2668–2673. [Google Scholar] [CrossRef]
- Buchberger, A.R.; Delaney, K.; Johnson, J.; Jillian, J. Mass spectrometry imaging: A review of emerging advancements and future insights. Anal. Chem. 2018, 90, 240–265. [Google Scholar] [CrossRef]
- Watrous, J.D.; Dorrestein, P.C. Imaging mass spectrometry in microbiology. Nat. Rev. Genet. 2011, 9, 683–694. [Google Scholar] [CrossRef] [PubMed]
- Esquenazi, E.; Yang, Y.-L.; Watrous, J.; Gerwick, W.H.; Dorrestein, P.C. Imaging mass spectrometry of natural products. Nat. Prod. Rep. 2009, 26, 1521–1534. [Google Scholar] [CrossRef]
- Yang, Y.-L.; Xu, Y.; Straight, P.; Dorrestein, P.C. Translating metabolic exchange with imaging mass spectrometry. Nat. Chem. Biol. 2009, 5, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Watrous, J.; Hendricks, N.; Meehan, M.; Dorrestein, P.C. capturing bacterial metabolic exchange using thin film desorption electrospray ionization-imaging mass spectrometry. Anal. Chem. 2010, 82, 1598–1600. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Esquenazi, E.; Coates, C.; Simmons, L.; Gonzalez, D.; Gerwick, W.H.; Dorrestein, P.C. Visualizing the spatial distribution of secondary metabolites produced by marine cyanobacteria and sponges via MALDI-TOF imaging. Mol. BioSyst. 2008, 4, 562–570. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-T.; Yang, Y.-L.; Xu, Y.; Lamsa, A.; Haste, N.M.; Yang, J.Y.; Ng, J.; Gonzalez, D.; Ellermeier, C.; Straight, P.; et al. Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2010, 107, 16286–16290. [Google Scholar] [CrossRef] [PubMed]
- Holzlechner, M.; Reitschmidt, S.; Gruber, S.; Zeilinger, S.; Marchetti-Deschmann, M. Visualizing fungal metabolites during mycoparasitic interaction by MALDI mass spectrometry imaging. Proteomics 2016, 16, 1742–1746. [Google Scholar] [CrossRef]
- Moree, W.J.; Yang, J.Y.; Zhao, X.; Liu, W.-T.; Aparicio, M.; Atencio, L.; Ballesteros, J.; Sanchez, J.; Gavilan, R.; Gutiérrez, M.; et al. Imaging mass spectrometry of a coral microbe interaction with fungi. J. Chem. Ecol. 2013, 39, 1045–1054. [Google Scholar] [CrossRef]
- Boya, P.C.A.; Fernández-Marín, H.; Mejia, L.C.; Spadafora, C.; Dorrestein, P.C.; Gutiérrez, M. Imaging mass spectrometry and MS/MS molecular networking reveals chemical interactions among cuticular bacteria and pathogenic fungi associated with fungus-growing ants. Sci. Rep. 2017, 7, 5604. [Google Scholar] [CrossRef]
- Lane, A.L.; Nyadong, L.; Galhena, A.S.; Shearer, T.L.; Stout, E.P.; Parry, R.M.; Kwasnik, M.; Wang, M.D.; Hay, M.E.; Fernandez, F.M.; et al. Desorption electrospray ionization mass spectrometry reveals surface-mediated antifungal chemical defense of a tropical seaweed. Proc. Natl. Acad. Sci. USA 2009, 106, 7314–7319. [Google Scholar] [CrossRef]
- Pett-Ridge, J.; Weber, P.K. NanoSIP: NanoSIMS applications for microbial biology. In Microbial Systems Biology. Methods in Molecular Biology (Methods and Protocols); Navid, A., Ed.; Humana Press: Totowa, NJ, USA, 2012; Volume 881, pp. 375–408. [Google Scholar] [CrossRef]
- Vaidyanathan, S.; Fletcher, J.; Goodacre, R.; Lockyer, N.P.; Micklefield, J.; Vickerman, J.C. Subsurface biomolecular imaging of Streptomyces coelicolor using secondary ion mass spectrometry. Anal. Chem. 2008, 80, 1942–1951. [Google Scholar] [CrossRef]
- Murrell, J.C.; Whiteley, A.S. Stable Isotope Probing and Related Technologies; American Society for Microbiology Press: Washington, DC, USA, 2010. [Google Scholar] [CrossRef]
- Dumont, M.G.; Murrell, J.C. Stable isotope probing—Linking microbial identity to function. Nat. Rev. Genet. 2005, 3, 499–504. [Google Scholar] [CrossRef]
- Rime, T.; Hartmann, M.; Frey, B. Potential sources of microbial colonizers in an initial soil ecosystem after retreat of an alpine glacier. ISME J. 2016, 10, 1625–1641. [Google Scholar] [CrossRef]
- Pinto-Tomás, A.A.; Anderson, M.A.; Suen, G.; Stevenson, D.M.; Chu, F.S.T.; Cleland, W.W.; Weimer, P.J.; Currie, C.R. Symbiotic nitrogen fixation in the fungus gardens of leaf-cutter ants. Science 2009, 326, 1120–1123. [Google Scholar] [CrossRef]
- Lee, N.; Nielsen, P.H.; Andreasen, K.H.; Juretschko, S.; Nielsen, J.L.; Schleifer, K.-H.; Wagner, M. Combination of fluorescent in situ hybridization and microautoradiography—A new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 1999, 65, 1289–1297. [Google Scholar] [CrossRef]
- Ouverney, C.C.; Fuhrman, J.A. Combined microautoradiography–16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ. Microbiol. 1999, 65, 1746–1752. [Google Scholar] [CrossRef]
- Adamczyk, J.; Hesselsoe, M.; Iversen, N.; Horn, M.; Lehner, A.; Nielsen, P.H.; Schloter, M.; Roslev, P.; Wagner, M. The isotope array, a new tool that employs substrate-mediated labeling of rRNA for determination of microbial community structure and function. Appl. Environ. Microbiol. 2003, 69, 6875–6887. [Google Scholar] [CrossRef] [PubMed]
- Pilloni, G.; von Netzer, F.; Engel, M.; Lueders, T. Electron acceptor-dependent identification of key anaerobic toluene degraders at a tar-oil-contaminated aquifer by Pyro-SIP. FEMS Microbiol. Ecol. 2011, 78, 165–175. [Google Scholar] [CrossRef]
- Eichorst, S.A.; Kuske, C.R. Identification of cellulose-responsive bacterial and fungal communities in geographically and edaphically different soils by using stable isotope probing. Appl. Environ. Microbiol. 2012, 78, 2316–2327. [Google Scholar] [CrossRef] [PubMed]
- López-Mondéjar, R.; Brabcova, V.; Štursová, M.; Davidová, A.; Jansa, J.; Cajthaml, T.; Baldrian, P. Decomposer food web in a deciduous forest shows high share of generalist microorganisms and importance of microbial biomass recycling. ISME J. 2018, 12, 1768–1778. [Google Scholar] [CrossRef] [PubMed]
- Sheik, A.R.; Brussaard, C.P.D.; Lavik, G.; Lam, P.; Musat, N.; Krupke, A.; Littmann, S.; Strous, M.; Kuypers, M.M.M. Responses of the coastal bacterial community to viral infection of the algae Phaeocystis globosa. ISME J. 2014, 8, 212–225. [Google Scholar] [CrossRef]
- Musat, N.; Musat, F.; Weber, P.K.; Pett-Ridge, J. Tracking microbial interactions with NanoSIMS. Curr. Opin. Biotechnol. 2016, 41, 114–121. [Google Scholar] [CrossRef]
- Dekas, A.E.; Poretsky, R.S.; Orphan, V.J. Deep-sea archaea fix and share nitrogen in methane-consuming microbial consortia. Science 2009, 326, 422–426. [Google Scholar] [CrossRef]
- Orphan, V.J.; House, C.H.; Hinrichs, K.-U.; McKeegan, K.D.; DeLong, E.F. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 2001, 293, 484–487. [Google Scholar] [CrossRef]
- Tai, V.; Carpenter, K.J.; Weber, P.K.; Nalepa, C.A.; Perlman, S.J.; Keeling, P.J. Genome evolution and nitrogen fixation in bacterial ectosymbionts of a protist inhabiting wood-feeding cockroaches. Appl. Environ. Microbiol. 2016, 82, 4682–4695. [Google Scholar] [CrossRef] [PubMed]
- Woebken, D.; Burow, L.C.; Prufert-Bebout, L.; Bebout, B.M.; Hoehler, T.M.; Pett-Ridge, J.; Spormann, A.M.; Weber, P.K.; Singer, S.W. Identification of a novel cyanobacterial group as active diazotrophs in a coastal microbial mat using NanoSIMS analysis. ISME J. 2012, 6, 1427–1439. [Google Scholar] [CrossRef] [PubMed]
- Fike, D.A.; Gammon, C.L.; Ziebis, W.; Orphan, V.J. Micron-scale mapping of sulfur cycling across the oxycline of a cyanobacterial mat: A paired nanoSIMS and CARD-FISH approach. ISME J. 2008, 2, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Alonso, C.; Musat, N.; Adam, B.; Kuypers, M.; Amann, R. HISH–SIMS analysis of bacterial uptake of algal-derived carbon in the Río de la Plata estuary. Syst. Appl. Microbiol. 2012, 35, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Majchrzak, T.; Wojnowski, W.; Lubinska-Szczygeł, M.; Różańska, A.K.; Namieśnik, J.; Dymerski, T. PTR-MS and GC-MS as complementary techniques for analysis of volatiles: A tutorial review. Anal. Chim. Acta 2018, 1035, 1–13. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).