Abstract
Multiple myeloma (MM) is a malignant disease based on differentiated plasma cells (PCs) in the bone marrow (BM). Flow cytometry and fluorescence microscopy, used to identify a large combination of clusters of differentiation (CDs), are applied for MM immunophenotyping. However, due to the heterogeneous MM immunophenotypes, more antibody panels are necessary for a preliminary diagnosis and for the monitoring of minimal residual disease (MRD). In this study, we evaluated the use of phage clones as probes for the identification of several PCs immunophenotypes from MM patients. First, A 9-mer M13-pVIII phage display library was screened against an MM.1 cells line to identify peptides that selectively recognize MM.1 cells. Then, the most representative phage clones, with amino acid sequences of foreign peptides closer to the consensus, were labelled with isothiocyanate of fluorescein (FITC) and were used to obtain a fluorescent signal on cells in ex-vivo samples by fluorescence microscopy. Selected phage clones were able to discriminate different MM immunophenotypes from patients related to CD45, CD38, CD56, and CD138. Our results highlight the possibility of using a phage-fluorescence probe for the simultaneous examination of the presence/absence of CDs associated with disease usually detected by combination of anti-CD antibodies. The design of a multi-phage imaging panel could represent a highly sensitive approach for the rapid detection of immunophenotype subtypes and the subsequent characterization of patient disease status.
1. Introduction
Multiple myeloma (MM) is a hematological malignancy characterized by expansion of immunophenotypically heterogeneous plasma cells (PC) in the bone marrow (BM) []. In order to improve the assessment of response to the therapy and prognostication, many researchers focused their goal on the development of systems for preliminary diagnosis and detection of minimal residual disease (MRD) in BM samples. Flow cytometry represents the gold standard in the detection/differentiation of several MM subtypes, thanks to the use of labelled antibodies against cell surface markers [,]. These clusters of differentiation (CDs) permit the discrimination between normal/reactive PCs and aberrant myeloma PCs, including the identification of non-PC normal/reactive BM cell compartments [,]. However, the differentiation of a single marker is not sufficient for a clear discrimination of PC subtypes; diagnosis strategies require a large combination of CDs for the detection and the identification of abnormal PCs from an MM patient []. Assessment of CD38 and CD138 is usually applied to PCs identification. In particular, CD138, also called syndecan-1, is a cell adhesion molecule involved in cell–cell or cell–matrix interactions, and is involved in several cell activities []. Despite its usual expression in normal cells [], CD138 results in being one of the most abundant surface proteins in MM PCs and is associated with cell proliferation and other carcinogenesis activities [,]. Due to that, several research studies are focusing their goal on its use as an antigen receptor for diagnosis and therapy in MM disease [].
Due to very high expression and uniformity on surfaces of MM PCs, CD38 is also being considered as a good target for new diagnostic and therapeutic approaches []. CD38 is a type II transmembrane glycoprotein, which has transmembrane receptor functions in addition to ecto-enzymatic activities []. It is expressed on normal cell subsets, such as T, NK, B, and dendritic cells, but it is also expressed during cell activation and proliferation []. In addition to CD138 and CD38, other markers—namely, CD45, CD56, CD19, CD81, CD27, and CD117—are used to further distinguish between normal/reactive PCs and MM PCs during preliminary diagnosis or detection of MRD [,,]. Among all, a multiparametric flow cytometry evaluation of BM from MM patients reveals two subpopulations of clonal PCs, based on the presence/absence of CD45 expression []. CD45 is a key regulator of antigen-mediated signaling and activation in lymphocytes, initially characterized as a leukocyte-common antigen. Despite its role remaining controversial, CD45 expression allows MM prognostic stratification. Specifically, MM patients with high-grade angiogenesis generally show a lower number of CD45+ PCs [].
Specific biological probes, able to detect and identify specific immunophenotypes of PC, including those of myeloma, could support flow cytometry in the simultaneous evaluation of the presence/absence of molecular targets (surface or intracellular) associated with MM disease. In this context, phage display technology is a powerful tool for the identification of new ligands that specifically recognize targets of interest []. Phage-displayed random peptide libraries consist of billions of M13 bacteriophage (or simply phage) clones that expose random peptides on their capsid protein. From these combinatorial phage libraries, specific peptides able to recognize and selectively bind molecular targets on whole cells can be screened by the affinity selection technique of biopanning [,]. M13 is a filamentous phage that measures 900 nm (length) and 6 nm (width). Its capsid consists of approximately 2700 copies of the major coat protein—namely, pVIII—capped on the two ends by 5 copies of 4 different minor coat proteins (pIX, pVII, pVI, and pIII) []. Consequently, M13 phage clones, engineered in PVIII, were considered advantageous in terms of their nanoscale size, high solubility, and multivalent binding sites compared with the antibodies. In fact, they have been successfully applied to several detection systems with fluorescent dyes of noble metal nanoparticles (such as gold, silver), silica-doped with quantum dots and a surface of micro-materials (such as magnetic beads) [,,,,,,,,].
The aim of this study was to use an engineered phage as a probe for a phage–phenotype identification of plasma cells derived from an MM patient. Specifically, phage clones from the M13-pVIII-9aa phage peptide library were directly labelled with isothiocyanate of fluorescein and used in recognition matrix tests of MM immunophenotype based on CD45, CD 56, CD38, and CD138.
2. Materials and Methods
2.1. Bacteriophage, Bacteria Host, and Eukaryotic Cells Line
Escherichia coli TG1, bacterium strain Lac-Z delete, Kan− and Amp−, was used to propagate phage clones. The bacterium was grown and maintained in Luria–Bertani medium (LB; tryptone 10 g/L, NaCl 10 g/L, and yeast extract 5 g/L) and Luria–Bertani agar (LB with addition of agar bacteriology 20 g/L), respectively. Selected phage clones (Amp+) and insert-less phagemid (namely, vector pC89) were maintained at 4 °C in TBS (Tris-buffered saline; 7.88 g/L Tris hydrochloride and sodium chloride 8.77 g/L; pH 7.5). MM.1 ATCC®CRL-2974™ (American Type Culture Collection; Rockville, MD, USA) cell line was cultured in humidified atmosphere containing 5% CO2 at 37 °C using a RPMI 1640 medium (Sigma-Aldrich, Milan, Italy) supplemented with fetal bovine serum 10%; l-glutamine, 2 mM; penicillin 100 units/mL; and streptomycin 100 μg/mL.
2.2. Phage Selection and Binding Affinity
A 9-mer M13-pVIII random phage display library was constructed in the vector pC89, by cloning a random DNA insert in-frame of encoding segments of the pVIII gene []. The procedure described in Lentini et al. [] was used to isolate specific phage clone against multiple myeloma cell line (MM.1). The MM.1 cell line was used because several cell phenotypes expressing mixed surface markers are present. First, the phage library was used in a pre-selection against plastic materials, used for the selection protocol against MM.1 cells. Non-binding phage were used in four rounds of affinity selection against MM.1 cells (1 × 106 cells/mL in Hank’s buffered salt solution, Sigma-Aldrich; pH 7.6). Each round included the incubation of phage with the target for 1 h at room temperature (RT) with gentle agitation. Cells–phage complex was precipitated by spinning (8000× g, 5 min) and separated from unbound phage by five-times washing and centrifugation steps (8000× g, 5 min) with 1 mL 0.1%TBS/Tween (50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 0.1% Tween-20). Phage were finally eluted from the cells–phage complex by 250 μL of 0.2 M glycine–HCl (pH 2.2) in gentle shaking at RT for 10 min. Then, the solutions were neutralized with 25 μL of 1 M Tris–HCl (pH 9.1).
Phage clones from the last round of affinity selection were identified by plating infected E. coli TG1 cells on LB agar added with X-Gal (5-bromo-4-chloro-3-indolyl-beta-d-galactopyranoside), IPTG (isopropyl thiogalactoside), and ampicillin. The presence of the insert was detected by X-gal, which produces a characteristic blue dye when cleaved by β-galactosidase. Blue bacterial colonies, from the fourth round of selection, were randomly selected, considering that each colony contained a single phage clone. The binding ability of selected phage clones to the target was tested in ELISA using as a negative control vector pC89. Briefly, 2.5 × 104 MM.1 cells in phosphate-buffered saline (PBS) were dispensed in each well of a 96-well plate and incubated overnight at 4 °C. After the incubation, cells were fixed by methanol (10 min at room temperature), washed three times with washing buffer (PBS + 0.05% Tween 20), and blocked with 100 µL of PBS + 5% lactalbumin + 0.05% Tween 20 (2 h at 37 °C in slow shaking 30 rpm). The 1 × 1010/100 µL phage clones, in PBS + 1% lactalbumin + 0.1% Tween 20, were added to each well, and the plate was incubated for 1 h at 37 °C in slow shaking 30 rpm. Then, cells were washed three times with washing buffer and incubated with monoclonal anti-M13 peroxidase conjugate antibody (Amersham Biosciences, Buckinghamshire, UK) at a dilution of 1/5000 in PBS + 1% lactalbumin + 0.1% Tween-20 (100 μL/well) for 1 h at 37 °C. Finally, cells were washed three times with washing buffer and incubated for 45 min at room temperature with 100 µL of 3,3′,5,5′-Tetra-methylbenzidine (TMB) for antibody binding detection. Reaction was stopped with 25 μL of 1 M H2SO4 and optical absorbance recorded at 450 nm using a MultisKan FC ThermoScientific.
2.3. DNA Sequencing and Peptides Analysis
For each phage clone, insert DNA, encoding the foreign peptide, was obtained from colonies of infected bacteria amplified by polymerase chain reaction (PCR) using sequencing primers M13–40 reverse (5′-GTTTTCCCAGTCACGAC-3′) and E24 forward (5′-GCTACCCTCGTTCCGATGCTGTC-3′), obtained from Proligo, Sigma (Milan, Italy). The PCR products were purified by the QIAquick PCR purification kit (Qiagen). Insert DNA sequence was obtained from the DNA sequencing service of CRIBI (University of Padova, Italy) using the M13 primer-40 (5′-GTTTTCCCAGTCACGAC-3′).
The DNA sequences were translated into amino acids by the “translate” program on the proteomics server of the Swiss Institute of Bioinformatics Expert Protein Analysis System (ExPASy http://www.expasy.ch/, accessed on 1 Decembre 2020). The amino acid sequences were aligned according to their similarity using the IDENTITY series matrix in Clustal X 2.1 sequence alignment software (available at http://clustalx.software.informer.com/2.1). Frequency (fxi) of amino acids, within the pool of peptides in the selected phage clones, were calculated using the following formula:
where Xi and N are amino acid frequency at position i and the total number of amino acids in the alignment, respectively. All frequencies were compared with the threshold value, which was conventionally set at 40%. All amino acid with a frequency greater than 40% were included in the “consensus”, a theoretical sequence containing the amino acids most involved in binding with the target. The resulting amino acid sequence of the peptide inserts was converted into FASTA files and analyzed with MEME Suite (motif-based sequence analysis tools v.5.0.5; http://meme-suite.org/, accessed on 1 Decembre 2020); Eukaryotic Linear Motif (ELM, http://elm.eu.org/, accessed on 1 Decembre 2020) resource, for SLiMs (short linear motif), and Domain Motif Interactions (DMIs) identification, fitting data at “Taxonomic Context” Homo Sapiens and restricted to nucleus, cytosol, and plasma membrane; BLAST-p (BLAST suite, https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 1 Decembre 2020) to search regions of similarity with human proteins involved in hematological neoplastic diseases using the following parameters: (i) Protein Data Bank proteins (pdb), as database; (ii) human taxid: 9606, as organism; and (iii) blastp protein–protein BLAST, as algorithm [,,,].
2.4. Labelling of Phage with FITC
Each clone was propagated in E. coli TG1, the bacteria host, and purified by twice precipitation in PEG/NaCl and filtered at 0.22 µm []. After the protocol, a phage titer of about 1012 virions/mL, estimated by measuring the number of transducing units per mL (TU/mL), was obtained. Labelling of phage with FITC fluorochrome was carried out according to the previously described procedure [], and adapted to the experimental condition of the present study. An amount of 5 × 1012 TU (transducing units) were resuspended in 200 μL buffer Na2CO3/NaHCO3 (pH 9.2) with 5 μL of fluorescein isothiocyanate (FITC, 5 mg/mL). Phage were incubated in a dark environment on a rotator for 2 hat RT to allow the reaction with fluorochrome. The sample was then incubated at 4 °C overnight with 200 μL of PEG/NaCl and subsequently centrifuged at 15,300× g (Eppendorf Centrifuge 5417R) at 4 °C for 1 h. The supernatant was discarded and the pellet re-suspended in 100 μL of Tris-buffered saline. Insert-less phagemid (pC89) was also labelled as negative control. Labelled phage was stored in the dark at 4 °C until utilization.
2.5. Sample Collection
Bone marrow (BM) samples were obtained from three MM patients (one with IgA-Lambda gammopathy and two with IgA-Kappa). All patients were admitted at the Hematology Unit of University Hospital Policlinico G. Martino, Messina. The study was approved by the institutional ethics committees, and all participants gave written informed consent according to the Declaration of Helsinki.
2.6. Isolation of Myeloma Plasma Cells from Bone Marrow
Bone marrow (BM) mononuclear cells (BMMCs) were isolated from heparinized BM aspirated samples by Ficoll Hypaque density gradient centrifugation (30 min, 25 °C, 400× g). BMMCs were incubated for 20 min at 4 °C in the dark with the following fluorochrome-conjugated monoclonal antibodies: CD45 (APC-conjugated, clone HI30, BD Bioscience), CD56 (PeCy7-conjugated, clone NCAM16.2, BD Bioscience), CD38 (PerCp5.5-conjugated, clone HIT2, BD Bioscience), CD138 (PE-conjugated, clone M115, BD Bioscience). Cells were washed twice with sterile PBS (Sigma-Aldrich, Milan, Italy) before proceeding to cell purification. PCs were isolated by using fluorescence-activated cell sorting (FACS) on a FACSAria sorter (BD Bioscience) with the following gating strategy: CD45−/CD56−/CD38+ cells, CD45−/CD56+/CD38+ cells, CD45−/CD38+/CD138+ cells, and CD45+/CD38+/138− cells. Dead cells were excluded by staining with LIVE/DEAD FIXABLE Aqua Dead dye (Invitrogen). Cell populations sorted by flow cytometry displayed purity >95% and were immediately used for fluorescence imaging.
2.7. Sample Preparation for Fluorescence Imaging
For each sample (MM.1 cell line or cell from MM patients after cell sorter), 25 µL of sorted cells was placed on poly-l-lysine (PLL)-coated glass slides and incubated for 15 min at room temperature. After the incubation, non-adherent cells were removed by washing three times with PBS, while the remaining ones were fixed by paraformaldehyde 4% (v/v in PBS), applied for 15 min at room temperature. After the incubation, excess of paraformaldehyde was removed by PBS washing (thrice) and cells were permeabilized with Triton X-100 0.1% (v/v in PBS) for 10 min at room temperature. Triton X-100 was removed by PBS washing (thrice), then 15 µL of phage labelled-FITC was added for 1 h at 37 °C. After the incubation, unbound phage were removed by PBS washing (thrice) and cells for fluorescence analysis were prepared by adding glycerol mounting buffer 90% (v/v in PBS) and coverslips to the slides. The samples were analyzed by fluorescence microscope (LeicaDMRE) with 63× magnification.
3. Results and Discussion
Twenty-four phage clones were selected from the 9-mer M13-pVIII phage display library by three rounds of biopanning against the MM.1 cells. Twenty-four phage clones were amplified, and their DNA were sequenced to derive the amino acids sequence of the foreign peptide displayed in their major coat protein. Positively charged amino acids were the most frequently present in all the sequences, immediately adjacent to the N-terminus of the peptide insert. These findings indicate that the amino acids, which do not vary in position, could be likely close to optimal sequence binding of the target. In this position, the most represented amino acids were arginine (R), lysine (K), and the alanine (A). Based on these most representative amino acids in the N-terminus, three families of motifs were deduced, as shown in Table 1.

Table 1.
Sequence consensus of selected phages.
In the families, four phage clones, displaying the foreign peptides ARKMNPPAG, RRANNPSPQ, RRYTHPNNS, and KKAHPGGSP, were chosen because they were the most representative and closest to the consensus sequence; they were then used for the next analysis.
The speculative bioinformatic analysis, aimed at the possibility of identifying motifs in the selected peptides that are present in human proteins with functions that are already known in the literature, are shown in Table S1.
Prior to ex-vivo fluorescence imaging of myeloma plasma cells from heparinized BM aspirated samples, a fluorescent validation test of the phage clones, labelled with FITC fluorochrome, was performed on MM.1, using the insert-less vector pC89 as negative control (Figure 1).
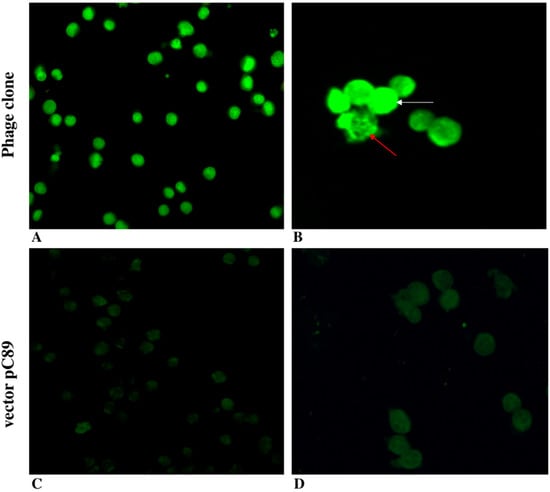
Figure 1.
Validation of phage clones by fluorescence microscopy. Fluorescence imaging of MM.1 cells incubated with ARKMNPPAG FITC-labelled phage (magnification 40×, (A), and 63×, (B)) and insert-less vector pC89 (magnification 40×, (C), and 63×, (D)).
Phage clones were found to specifically bind the MM cells line, as seen by the bright green fluorescence of FITC (Figure 1A,B) compared with the absence of fluorescence when the pC89 vector insert-less was used (Figure 1C,D). These findings indicate that the binding specificity was due to the foreign peptides displayed by phage clones, and the amine-reactive fluorochrome in the major coat protein, pVIII, did not modify the phage’s ability to bind the MM.1 target cells. However, we observed a different phage location, homogeneously distributed (white arrows in Figure 1B) or limited to specific subcellular sites (red arrows in Figure 1B) within the same cell population. These results could depend on the presence of different phenotypes originating within the same cell line. Although still controversial, the immunophenotype of the MM.1 cell line shows CD38+/45−, with the presence of two subpopulations CD138++ (96%) and CD138low (4%) [,]. In addition, CD56 expression was decreased in the MM.1R cell line compared with the sensitive counterpart (MM.1S) [].
Based on the above considerations, to verify the phage clone’s ability to identify specific PC immunophenotypes from MM samples, FITC-labelled phage were used in ex-vivo fluorescence imaging with sorted MM cells from heparinized BM aspirated samples. Figure 2 shows ARKMNPPAG phage clones’ ability to bind/discriminate MM subtype cells, sorted by using the CD45, CD56, and CD38 antibodies (Figure 2).
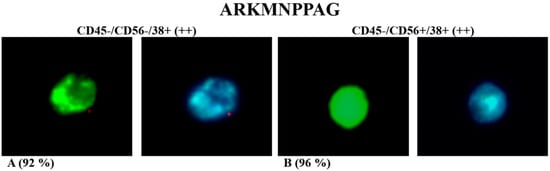
Figure 2.
Ex-vivo fluorescence test of ARKMNPPAG FITC-labelled phage incubated with immunophenotypes CD45−/CD56−/CD38+ (A) and CD45−/CD56+/CD38+ (B). Green fluorescence was due to FITC-labelled phage, while blue fluorescence was due to DAPI nucleus staining (magnification 100×). Percentage in brackets refers to only cells with evident green fluorescence due to phage clone binding.
The ARKMNPPAG phage, previously selected against the MM.1 cell line, was able to bind both the immunophenotypes CD45−/CD56−/CD38+ and CD45−/CD56+/CD38+, but it was differently located (Figure 2A,B and Figure S1). Specifically, the non-specific marking of the nucleus by DAPI staining (blue fluorescence) showed that phage (green fluorescence) was specifically localized in subcellular sites, close or identified as nuclear, in CD56− PCs. Otherwise, in CD56+ PCs, the phage is homogeneously distributed on the whole cell surface. It is known that 50–80% of MM patients express CD56 on PCs [,]. On the other hand, the biological function of CD56 is extremely controversial in MM patients with undefined prognosis [,].
CD45 can be used in MM prognosis, specifically to discriminate MM patients with high-grade angiogenesis []. Therefore, the CD45 immunophenotypic gate was associated with a second surface antigen—namely, CD138—with the aim of testing the phage clones towards myeloma immunophenotypes with different prognostic significance. Specifically, the CD45−/CD38+/CD138+ and CD45+/CD38+/138− PC immunophenotypes were chosen for ex-vivo fluorescence imaging using the most representative selected phage clones (Figure 3).
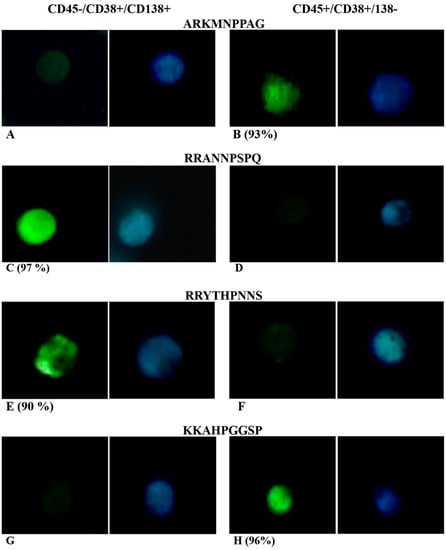
Figure 3.
Ex-vivo fluorescence test of ARKMNPPAG (A,B), RRANNPSPQ (C,D), RRYTHPNNS (E,F), and KKAHPGGSP (G,H) FITC-labelled phage incubated with MM cell sorted using CD45, CD38, CD138 gates. Green fluorescence was due to FITC-labelled phage, while blue fluorescence was due to DAPI nucleus staining. Percentage in brackets refers to only cells with evident green fluorescence due to phage clone binding.
Figure 3 show results from ex-vivo fluorescence test of ARKMNPPAG (Figure 3A,B), RRANNPSPQ (Figure 3C,D), RRYTHPNNS (Figure 3E,F), and KKAHPGGSP (Figure 3G,H) FITC-labelled phage incubated with MM cell sorted using CD45, CD38, CD138 gates. Data showed that recognition/detection of both ARKMNPPAG and KKAHPGGSP phage was directed to CD45+/CD138− phenotypic patterns (Figure 3B,H and Figure S2), while CD138+/CD45− PCs were not recognized, as evidenced by the absence of green fluorescence (Figure 3A,G). These findings suggest that phage were able to recognize/detect the MM immunophenotype with lower expression levels of CD138, related to the alteration of the membrane protein rather than the nucleus. It has been demonstrated that downregulation of CD138 may be linked to a clinically advanced disease; in particular, it can promote dissemination and spread to other bones []. Otherwise, CD38+/CD138+ PCs identify, in MM patients, an abnormal immunophenotype in the proliferation phase, with high capacity to bind and accumulate growth factors [,,]. Consequently, the selective recognition of ARKMNPPAG and KKAHPGGSP phage could direct the diagnosis to an early stage of multiple myeloma (MM) in the patient.
Otherwise, the RRANNPSPQ and RRYTHPNNS phage were able to selectively bind CD45−/CD38+/CD138+ PCs, as indicated by the presence of green fluorescence (Figure 3C,E), while no fluorescence was detected on other PCs (Figure 3D,F). By the comparison with DAPI staining, we observed that FITC-labelled phage was homogeneously distributed on the whole cell surface for the RRANNPSPQ phage (Figure 3C and Figure S3), and it was specifically localized in subcellular sites for the RRYTHPNNS phage (Figure 3E and Figure S3). The low or absence of CD45 expression has been found in MM PCs with high-grade angiogenesis []. Therefore, the two phage clones could constitute a useful prognostic index to be validated in subsequent studies [,].
In summary, phage fluorescence imaging permitted the identification of different immunophenotypes from MM samples. Specifically, ARKMNPPAG phage showed a different recognition trend with respect to CD56, which means recognition of CD56− in the nuclear and perinuclear region and CD56+ in the plasma membrane regions. In PC CD38+, normally co-expressed with CD138, ARKMNPPAG and KKAHPGGSP phage discriminated the myeloma subtype CD45+/CD138− (late-stage disease). Otherwise, RRANNPSPQ and RRYTHPNNS phage were able to bind cells with an immunophenotype pattern CD45−/CD138+ (early-stage disease), albeit with a different fluorescence biodistribution. The described approach allowed an experimental design of a phage array that is able to recognize/bind different MM immunophenotypes based on the CD45/CD56/CD38/CD138 gates, which could be used for the detection and monitoring of disease (Table 2).

Table 2.
Phage array for MM immunophenotype discrimination.
4. Conclusions
In this study, the use of phage clones as fluorescent probes for an immunophenotype identification of MM PCs was described. Specifically, we found that the most representative selected phage were able to discriminate two MM immunophenotype cells—namely, CD45−/CD38+/CD138+ and CD45+/CD38+/CD138−. These results, although preliminary, highlight the possibility of using a phage-fluorescence probe for the simultaneous examination of the presence/absence of CDs associated with disease usually detected by a combination of more anti-CD antibodies. From this perspective, the possibility of using whole phage clones could open the way to the development of low cost, innovative biosensor devices to monitor residual diseases, and also to identify a new stage-associated target disease. The approach described in this study consists of a multi-phage imaging panel of MM PCs and could be easily scaled and tuned for specific cell subsets of MM and other neoplasia that would be critical to the characterization of a patient’s disease status.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/app11177910/s1, Table S1: Significant motifs in foreign peptide, Figure S1: Ex-vivo fluorescence test of ARKMNPPAG FITC-labelled phage incubated with MM cell sorted using CD45, CD56, and CD38 gates. Green fluorescence was due to FITC-labelled phage, while blue fluorescence was due to DAPI nucleus staining (magnification 40×), Figure S2: Ex-vivo fluorescence test of ARKMNPPAG and KKAPGGSP FITC-labelled phage incubated with CD45+/CD38+/CD138− MM cells. Green fluorescence was due to FITC-labelled phage, while blue fluorescence was due to DAPI nucleus staining (magnification 40×). Figure S3: Ex-vivo fluorescence test of RRANNPSPQ and RRYTHPNNS FITC-labelled phage incubated with CD45−/CD38+/CD138+ MM cells. Green fluorescence was due to FITC-labelled phage, while blue fluorescence was due to DAPI nucleus staining (magnification 40×).
Author Contributions
Conceptualization, L.M.D.P., D.F., E.F., A.A., G.F. and S.P.P.G.; methodology, L.M.D.P., D.F., M.B. and R.C.; software, L.M.D.P. and S.T.; validation, L.M.D.P., D.F. and S.P.P.G.; formal analysis, all authors; data curation, all authors; writing—original draft preparation, L.M.D.P., D.F. and S.P.P.G.; writing—review and editing, L.M.D.P., D.F., E.F., A.A., C.M., G.F., F.N. and S.P.P.G.; supervision, L.M.D.P., D.F., E.F., A.A., G.F. and S.P.P.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Ethics Committees.
Informed Consent Statement
The study was approved by the institutional ethics committees, and all participants gave written informed consent according to the Declaration of Helsinki.
Acknowledgments
This work was supported by Associazione pro Bambini e Adulti Leucemici (A.B.A.L.) onlus Messina (Italy) (http://www.abalmessina.it). Authors gratefully acknowledge A.B.A.L. for the use of the XploRA Raman spectrometer and for the scholarship. The authors thank Franco Felici for the kind gift of phage display M13 libraries.
Conflicts of Interest
The authors have no conflict of interest to declare.
References
- Zagozdzon, R.; Golab, J. Cancer stem cells in haematological malignancies. Wspolczesna Onkol. 2015, 19, A1–A6. [Google Scholar] [CrossRef]
- Rawstron, A.C.; Davies, F.E.; Das Gupta, R.; Ashcroft, A.J.; Patmore, R.; Drayson, M.T.; Owen, R.G.; Jack, A.S.; Child, J.A.; Morgan, G.J. Flow cytometric disease monitoring in multiple myeloma: The relationship between normal and neoplastic plasma cells predicts outcome after transplantation. Blood 2002, 100, 3095–3100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- San Miguel, J.F.; Almeida, J.; Mateo, G.; Bladé, J.; López-Berges, C.; Caballero, D.; Hernández, J.; Moro, M.J.; Fernández-Calvo, J.; Díaz-Mediavilla, J.; et al. Immunophenotypic evaluation of the plasma cell compartment in multiple myeloma: A tool for comparing the efficacy of different treatment strategies and predicting outcome. Blood 2002, 99, 1853–1856. [Google Scholar] [CrossRef] [PubMed]
- Flores-Montero, J.; de Tute, R.; Paiva, B.; Perez, J.J.; Bottcher, S.; Wind, H.; Sanoja, L.; Puig, N.; Lecrevisse, Q.; Vidriales, M.B.; et al. Immunophenotype of normal vs. myeloma plasma cells: Toward antibody panel specifications for MRD detection in multiple myeloma. Cytom. B Clin. Cytom. 2016, 90, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Fumey, W.; Koenigsdorf, J.; Kunick, V.; Menzel, S.; Schütze, K.; Unger, M.; Schriewer, L.; Haag, F.; Adam, G.; Oberle, A.; et al. Nanobodies effectively modulate the enzymatic activity of CD38 and allow specific imaging of CD38+ tumors in mouse models in vivo. Sci. Rep. 2017, 7, 14289. [Google Scholar] [CrossRef] [PubMed]
- Akhmetzyanova, I.; McCarron, M.J.; Parekh, S.; Chesi, M.; Bergsagel, P.L.; Fooksman, D.R. Dynamic CD138 surface expression regulates switch between myeloma growth and dissemination. Leukemia 2020, 34, 245–256. [Google Scholar] [CrossRef] [Green Version]
- Palaiologou, M.; Delladetsima, I.; Tiniakos, D. CD138 (syndecan-1) expression in health and disease. Histol. Histopathol. 2014, 29, 177–189. [Google Scholar] [CrossRef]
- Coombe, D.R. Biological implications of glycosaminoglycan interactions with haemopoietic cytokines. Immunol. Cell Biol. 2008, 86, 598–607. [Google Scholar] [CrossRef]
- Casu, B.; Naggi, A.; Torri, G. Heparin-derived heparan sulfate mimics to modulate heparan sulfate-protein interaction in inflammation and cancer. Matrix Biol. 2010, 29, 442–452. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Mahendravada, A.; Ballard, B.; Kale, B.; Ramos, C.; West, J.; Maguire, T.; McKay, K.; Lichtman, E.; Tuchman, S.; et al. Safety and efficacy of targeting CD138 with a chimeric antigen receptor for the treatment of multiple myeloma. Oncotarget 2019, 10, 2369–2383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, F.; Dalla Palma, B.; Giuliani, N. CD38 Expression by Myeloma Cells and Its Role in the Context of Bone Marrow Microenvironment: Modulation by Therapeutic Agents. Cells 2019, 8, 1632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deaglio, S.; Aydin, S.; Vaisitti, T.; Bergui, L.; Malavasi, F. CD38 at the junction between prognostic marker and therapeutic target. Trends Mol. Med. 2008, 14, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Menke, D.M.; Horny, H.P.; Griesser, H.; Atkinson, E.J.; Kaiserling, E.; Kyle, R.A. Immunophenotypic and genotypic characterisation of multiple myelomas with adverse prognosis characterised by immunohistological expression of the T cell related antigen CD45RO (UCHL-1). J. Clin. Pathol 1998, 51, 432–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rheinländer, A.; Schraven, B.; Bommhardt, U. CD45 in human physiology and clinical medicine. Immunol. Lett. 2018, 196, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Dass, J.; Arava, S.; Mishra, P.C.; Dinda, A.K.; Pati, H.P. Role of CD138, CD56, and light chain immunohistochemistry in suspected and diagnosed plasma cell myeloma: A prospective study. South Asian J. Cancer. 2019, 8, 60–64. [Google Scholar] [CrossRef]
- Gonsalves, W.I.; Timm, M.M.; Rajkumar, S.V.; Morice, W.G.; Dispenzieri, A.; Buadi, F.K.; Lacy, M.Q.; Dingli, D.; Leung, N.; Kapoor, P.; et al. The prognostic significance of CD45 expression by clonal bone marrow plasma cells in patients with newly diagnosed multiple myeloma. Leuk. Res. 2016, 44, 32–39. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Rajkumar, S.V.; Kimlinger, T.; Greipp, P.R.; Witzig, T.E. CD45 expression by bone marrow plasma cells in multiple myeloma: Clinical and biological correlations. Leukemia 2015, 19, 1466–1470. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P.; Petrenko, V.A. Phage display. Chem. Rev. 1997, 97, 391–410. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.H.; Liu, I.J.; Lu, R.M.; Wu, H.C. Advancement and applications of peptide phage display technology in biomedical science. J. Biomed. Sci. 2016, 23, 8. [Google Scholar] [CrossRef] [Green Version]
- Gillespie, J.W.; Yang, L.; De Plano, L.M.; Stackhouse, M.A.; Petrenko, V.A. Evolution of a Landscape Phage Library in a Mouse Xenograft Model of Human Breast Cancer. Viruses 2019, 11, 988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morag, O.; Sgourakis, N.G.; Baker, D.; Goldbourt, A. The NMR-Rosetta capsid model of M13 bacteriophage reveals a quadrupled hydrophobic packing epitope. Proc. Natl. Acad. Sci. USA 2015, 112, 971–976. [Google Scholar] [CrossRef] [Green Version]
- Jaye, D.L.; Geigerman, C.M.; Fuller, R.E.; Akyildiz, A.; Parkos, C.A. Direct fluorochrome labeling of phage display library clones for studying binding specificities: Applications in flow cytometry and fluorescence microscopy. J. Immunol. Methods 2004, 295, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Kelly, K.A.; Waterman, P.; Weissleder, R. In vivo imaging of molecularly targeted phage. Neoplasia 2006, 8, 1011–1018. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Chen, Y.; Li, S.; Nguyen, H.G.; Niu, Z.; You, S.; Mello, C.M.; Lu, X.; Wang, Q. Chemical modification of M13 bacteriophage and its application in cancer cell imaging. Bioconjug. Chem. 2010, 21, 1369–1377. [Google Scholar] [CrossRef] [PubMed]
- Torrisi, L.; Guglielmino, S.; Silipigni, L.; De Plano, L.M.; Kovacik, L.; Lavrentiev, V.; Torrisi, A.; Fazio, M.; Fazio, B.; Di Marco, G. Study of gold nanoparticle transport by M13 phages towards disease tissues as targeting procedure for radiotherapy applications. Gold Bull. 2019, 52, 135–144. [Google Scholar] [CrossRef]
- Lee, L.A.; Niu, Z.; Wang, Q. Viruses and virus-like protein assemblies-Chemically programmable nanoscale building blocks. Nano Res. 2009, 2, 349–364. [Google Scholar] [CrossRef] [Green Version]
- De Plano, L.M.; Scibilia, S.; Rizzo, M.G.; Crea, S.; Franco, D.; Mezzasalma, A.M.; Guglielmino, S.P. One-step production of phage–silicon nanoparticles by PLAL as fluorescent nanoprobes for cell identification. Appl. Phys. A 2018, 124, 222. [Google Scholar] [CrossRef]
- Rakonjac, J.; Bennett, N.J.; Spagnuolo, J.; Gagic, D.; Russel, M. Filamentous bacteriophage: Biology, phage display and nanotechnology applications. Curr. Issues Mol. Biol. 2011, 13, 51–76. [Google Scholar] [PubMed]
- De Plano, L.M.; Fazio, E.; Rizzo, M.G.; Franco, D.; Carnazza, S.; Trusso, S.; Neri, F.; Guglielmino, S.P. Phage-based assay for rapid detection of bacterial pathogens in blood by Raman spectroscopy. J. Immunol. Methods 2019, 465, 45–52. [Google Scholar] [CrossRef]
- Franco, D.; De Plano, L.M.; Rizzo, M.G.; Scibilia, S.; Lentini, G.; Fazio, E.; Neri, F.; Guglielmino, S.P.P.; Mezzasalma, A.M. Bio-hybrid gold nanoparticles as SERS probe for rapid bacteria cell identification. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 224, 117394. [Google Scholar] [CrossRef] [PubMed]
- Felici, F.; Castagnoli, L.; Musacchio, A.; Jappelli, R.; Cesareni, G.J. Selection of antibody ligands from a large library of oligopeptides expressed on a multivalent exposition vector. Mol. Biol. 1991, 222, 301–310. [Google Scholar] [CrossRef]
- Lentini, G.; Fazio, E.; Calabrese, F.; De Plano, L.M.; Puliafico, M.; Franco, D.; Nicolò, M.S.; Carnazza, S.; Trusso, S.; Allegra, A.; et al. Phage–AgNPs complex as SERS probe for U937 cell identification. Biosens. Bioelectron. 2015, 74, 398–405. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Gouw, M.; Michael, S.; Sámano-Sánchez, H.; Kumar, M.; Zeke, A.; Lang, B.; Bely, B.; Chemes, L.B.; Davey, N.E.; Deng, Z.; et al. The eukaryotic linear motif resource—2018 update. Nucleic Acids Res. 2018, 46, D428–D434. [Google Scholar] [CrossRef] [PubMed]
- Petrenko, V.A.; Gillespie, J.W.; Xu, H.; O’Dell, T.; De Plano, L.M. Combinatorial Avidity Selection of Mosaic Landscape Phages Targeted at Breast Cancer Cells-An Alternative Mechanism of Directed Molecular Evolution. Viruses 2019, 11, 785. [Google Scholar] [CrossRef] [Green Version]
- De Plano, L.M.; Carnazza, S.; Franco, D.; Rizzo, M.G.; Conoci, S.; Petralia, S.; Nicoletti, A.; Zappia, M.; Campolo, M.; Esposito, E.; et al. Innovative IgG biomarkers based on phage display microbial amyloid mimotope for state and stage diagnosis in Alzheimer’s disease. ACS Chem. Neurosci. 2020, 11, 1013–1026. [Google Scholar] [CrossRef]
- Herman, R.E.; Makienko, E.G.; Prieve, M.G.; Fuller, M.; Houston, M.E., Jr.; Johnson, P.H. Phage display screening of epithelial cell monolayers treated with EGTA: Identification of peptide FDFWITP that modulates tight junction activity. J. Biomol. Screen. 2007, 12, 1092–1101. [Google Scholar] [CrossRef]
- Paíno, T.; Sarasquete, M.E.; Paiva, B.; Krzeminski, P.; San-Segundo, L.; Corchete, L.A.; Redondo, A.; Garayoa, M.; García-Sanz, R.; Gutiérrez, N.C.; et al. Phenotypic, genomic and functional characterization reveals no differences between CD138++ and CD138 low subpopulations in multiple myeloma cell lines. PLoS ONE 2014, 9, e92378. [Google Scholar] [CrossRef] [PubMed]
- Greenstein, S.; Krett, N.L.; Kurosawa, Y.; Ma, C.; Chauhan, D.; Hideshima, T.; Anderson, K.C.; Rosen, S.T. Characterization of the MM.1 human multiple myeloma (MM) cell lines: A model system to elucidate the characteristics, behavior, and signaling of steroid-sensitive and -resistant MM cells. Exp. Hematol. 2003, 31, 271–282. [Google Scholar] [CrossRef]
- Chang, H.; Samiee, S.; Yi, Q.L. Prognostic relevance of CD56 expression in multiple myeloma: A study including 107 cases treated with high-dose melphalan-based chemotherapy and autologous stem cell transplant. Leuk. Lymphoma 2006, 47, 43–47. [Google Scholar] [CrossRef]
- Mateo, G.; Castellanos, M.; Rasillo, A.; Gutierrez, N.C.; Montalban, M.A.; Martin, M.L.; Hernandez, J.M.; Lopez-Berges, M.C.; Montejano, L.; Blade, J.; et al. Genetic abnormalities and patterns of antigenic expression in multiple myeloma. Clin. Cancer Res. 2005, 11, 3661–3667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palumbo, A.; Avet-Loiseau, H.; Oliva, S.; Lokhorst, H.M.; Goldschmidt, H.; Rosinol, L.; Richardson, P.; Caltagirone, S.; Lahuerta, J.J.; Facon, T.; et al. Revised International Staging System for Multiple Myeloma: A Report from International Myeloma Working Group. J. Clin. Oncol. 2015, 33, 2863–2869. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, K.; Suzuki, K. CD56 for Multiple Myeloma: Lack of CD56 May Be Associated with Worse Prognosis. Acta Haematol. 2018, 140, 40–41. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhu, Z.M.; Sun, K.; Lei, P.C.; Liu, Z.W.; Guo, J.M.; Yang, J.; Zang, Y.Z.; Zhang, Y. Expression of CD45 in newly diagnosed multiple myeloma and the relationship with prognosis. Zhonghua Xue Ye Xue Za Zhi 2019, 40, 744–749. (In Chinese) [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).