1. Introduction
In Wuhan, China, following an unknown form of pneumonia outbreak, a new coronavirus emerged, dubbed the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), with the current outbreak being dubbed COVID-19 and promptly declared a “Public Health Emergency of International Concern”. As of the time of writing, there are 200 M coronavirus cases worldwide, with the United States alone being responsible for around 33.6 M cases. The worldwide recorded deaths have surpassed 3.85 M. There are now several different vaccines, and a singular therapeutic drug labeled GS-5734 has passed FDA trials, however its efficacy is contested [
1,
2,
3,
4].
In the last decades, the cost of drug discovery has increased exponentially, with the median cost of finding an FDA-approved drug at around USD 19 million when writing this article. Due to this, it is prohibitively expensive to evaluate the potential for the treatment of even a proactively curated set of drugs like the one provided by Recursion Pharmaceuticals [
5]. Standard preclinical trials involve manual assessment of drug efficacy, which is hugely time and labor-intensive, while also subject to human error. Due to the time constraints of such a process, the number of drugs that may be analyzed is limited. As such, for a situation such as COVID-19, which both spreads rapidly and has a risk of mutation, any method of streamlining drug discovery can serve to save countless lives. A viable approach in such a case is to use a deep learning model to identify promising compounds and to validate these compounds through clinical trials [
6].
Drug discovery using machine learning methods such as support vector machines and neural networks has been proven effective in attempting to lower the prohibitive cost and speed up the process, as opposed to the extensive process of human experimentation [
7,
8,
9,
10,
11]. In this research, we attempt to demonstrate the potential of deep learning [
12,
13], a neural network-based machine learning technique, to determine promising drug compounds, using COVID-19 as a proof of concept in order to produce a response for future outbreaks [
6,
11,
12].
Currently, the drug selection process involves target identification and validation, which aims to identify which molecules vary based on the viral status of the cells in order to identify possible gauges for the change caused by drugs [
9,
14]. Target identification also aims to predict the druggability of a target molecule, or the extent to which the activity of the target compound may be changed through therapeutics. The drug then faces compound screening and lead discovery, which aims to identify promising compounds and drugs through determining which compounds result in the most significant variability of the resultant cells and identifying the most promising compounds as leads, which then proceed to the preclinical and clinical development trials. In this paper, we explore the ability of neural networks to accelerate the first two stages of target identification and validation, as well as compound screening and lead verification. Machine learning has various applications in medicine, with one of the most significant benefits being drug discovery. Biological systems are incredibly complex and contain an immense amount of densely stored information, with the new forms of data allow for even more data for pharmaceutical companies to access and organize. The decision process of the neural network relies on its ability to identify specific characteristics of viral cells and healthy control cells, known as “features,” allowing it to analyze new data and classify the data as “healthy” or “viral,” while relative values of these features associated with each class allow the model to provide a probability score indicating its “confidence” in its classification [
5,
6,
7,
12].
In addition, the broader availability of more powerful computers and deep learning algorithms helps increase the accessibility of deep neural networks (DNNs) [
12,
13,
15,
16], and allows for an increasingly robust variety of applications for deep learning within the pharmaceutical industry. There are various effective methods of machine learning, which can be divided into supervised and unsupervised learning techniques. Supervised learning is valuable in its ability to predict labels of data samples, while unsupervised learning is valuable in its ability to categorize and cluster data in ways that the creator may not be aware of. Our model is based on supervised learning, specifically a type of artificial neural network known as a convolutional neural network (CNN). It is well known that different forms of neural networks are suitable in modeling and analyzing different types of data. For example, recurrent neural networks are more capable of analyzing time-variable neural networks, while deep CNNs [
3] excel at image processing. Deep learning is useful in its ability to identify biomarkers and build predictive models on the possible effectiveness of a drug, mainly through its ability to identify which drugs lead to the most significant changes.
Despite their apparent value in rapidly accelerating the drug discovery process, it must be noted that neural networks still face challenges in selecting suitable architectures and in data preparation. In order to effectively identify and test targets and compounds, data must be standardized, comprehensive, and of sufficient quantity. Even then, the model is subject to overfitting, where it begins to recognize the noise of the training data, resulting in reduced performance. Overfitting can be combated with techniques such as regularization, which places a penalty on more complex models, and may also be helped by further processing and cleaning of data [
6].
Our focus in this paper is to identify or validate promising compounds and leads for combating viral cells, particularly SARS-CoV-2 viral cells. In the literature, researchers have presented a wide selection of methods for the analysis of Recursion’s dataset, with most methods arriving at the same conclusion on the most effective drugs, some of which have also been under clinical trials. For the RxRx19a dataset, there have been several research studies that utilized said dataset. In Recursion’s in-house analysis, they concluded that GS-441524 was the clear front runner in terms of preventing cell damage, with Remdesivir, Cx-4945, and Alimitrine all being relatively close to each other in terms of efficacy score [
5]. Another study by researchers from the Simon Frasier University and the University of British Columbia found the leading candidates for viral suppression were Remdesivir, GS-441524, and Aloxistatin, as well as the drugs Mebendazole, Oxibendazole, and Albendazole [
17]. This group of researchers proposed to use support vector machines to identify potential lead compounds, while our model relies on transfer learning. Our analysis, Recursion’s analysis, and analysis from the researchers at Simon Frasier University arrive at similar compounds identified as the most promising, though our approach uses probability as an equivalent efficacy score for each candidate compound.
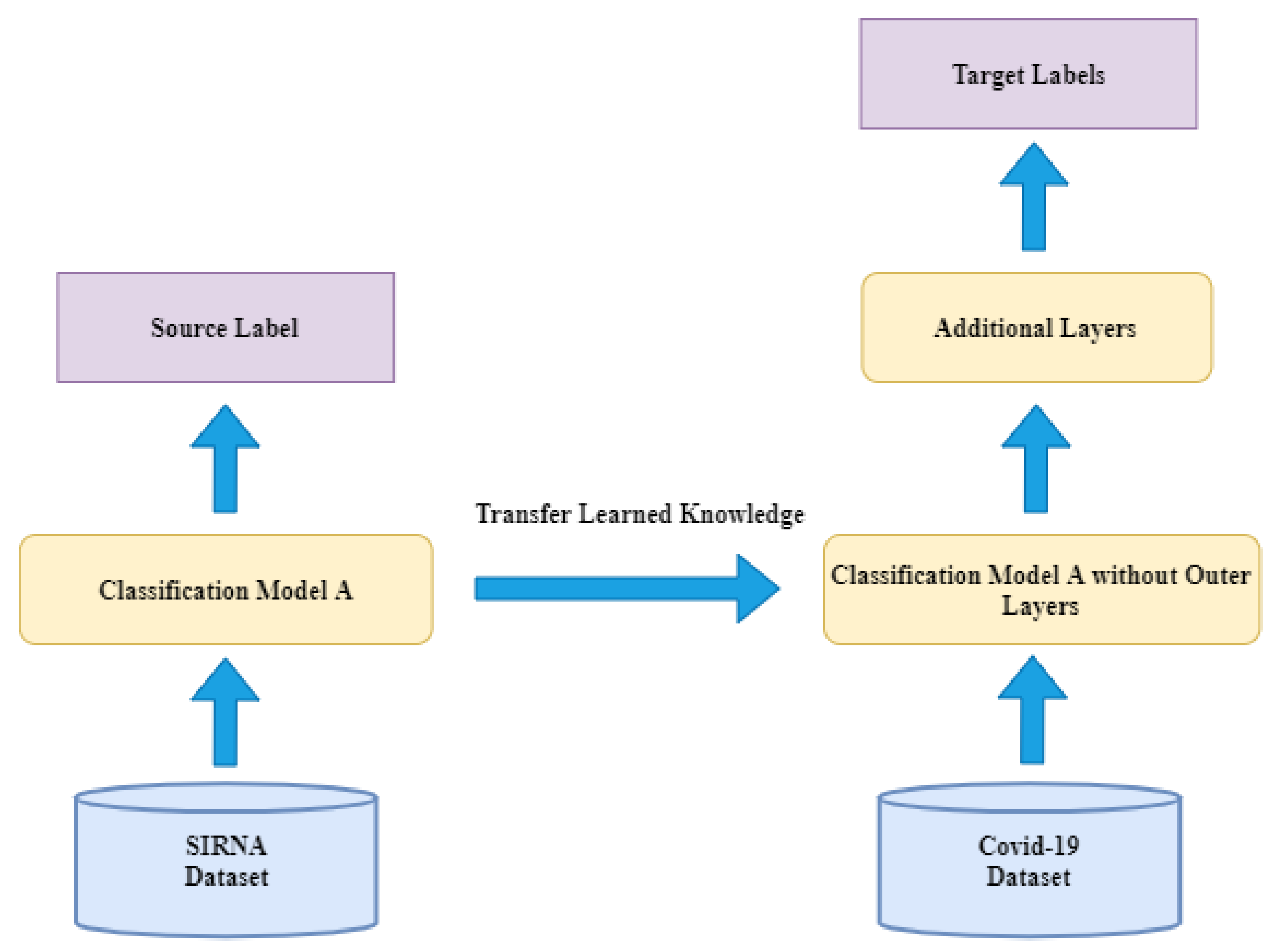
In this study, we applied transfer learning, a technique that refines a deep learning model pre-trained in one domain for a specific application in another domain, to identify promising compounds and leads for COVID-19. A deep learning model, DenseNet, was first trained with the siRNA image dataset with the transfer learning strategy [
18,
19,
20,
21]. The trained DenseNet was then refined, retrained, and experimented on the SARS-CoV-2 dataset with 1752 different compounds, also provided by Recursion. In the process, transfer learning is applied twice; therefore, we call the approach “cascade transfer learning.” The results obtained by the deep learning model were then compared with those obtained by other models and with those approved by FDA and under clinical trials. The experimental study shows that the cascade transfer learning approach is promising in identifying promising compounds and leads. The remainder of the paper is organized in the following manner.
Section 2 introduces the SARS-CoV-2 dataset from Recursion.
Section 3 details the deep learning strategy adopted in this research.
Section 4 presents the experimental results with discussions. The paper ends with concluding remarks.
2. Dataset
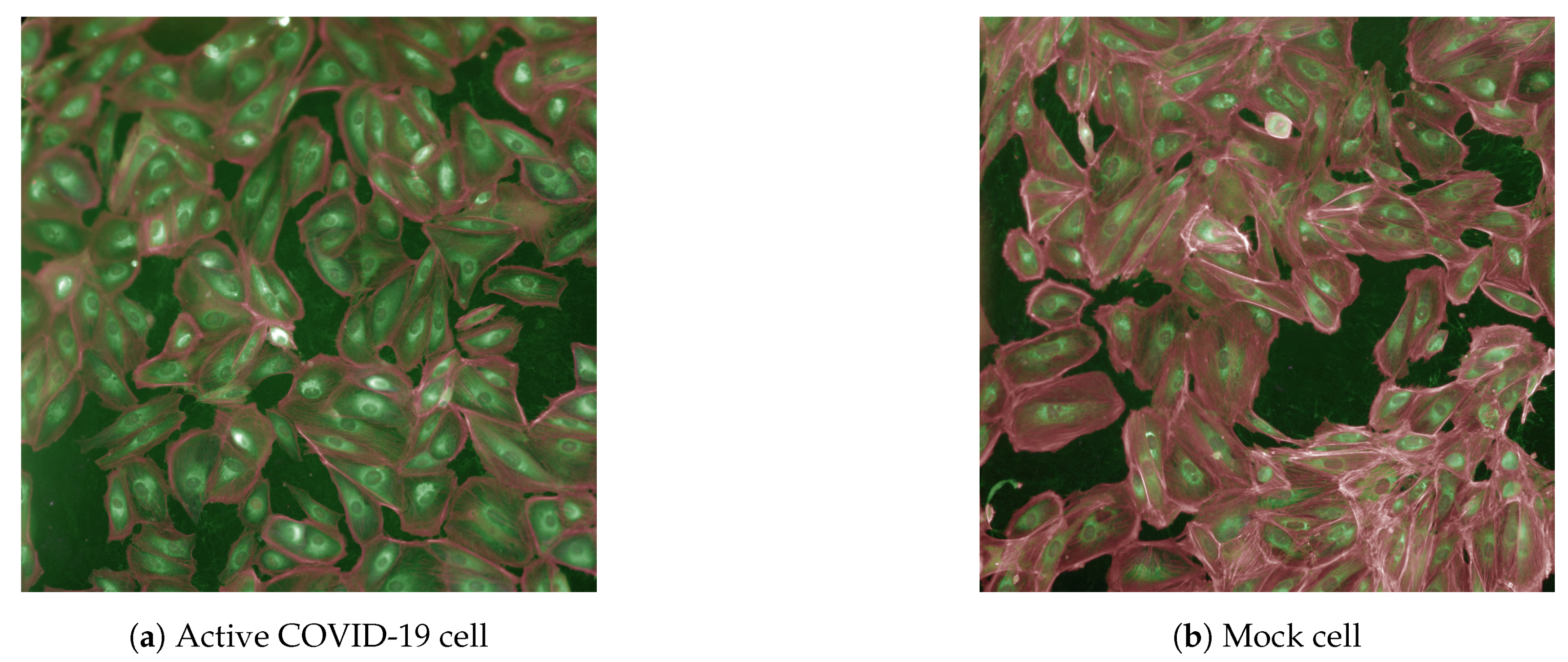
In order to work as efficiently as possible, we utilized a morphological imaging dataset from Recursion Pharmaceuticals with 305,520, 5-channel images. In April of 2020, Recursion developed the dataset, where cells were treated in 1536 various microplates, with fixation through 5% paraformaldehyde, permeabilization through 0.25% Triton, and staining with a variety of different dyes. Three different cell treatments were involved, with mock cells, which served as a control, cells with irradiated and, therefore, deactivated SARS-CoV-2 virons, and cells infected with active SARS-CoV-2 at a ratio of 4 virons to 10 cells. These cells were then treated with a large number of candidate compounds, which came from a library of drugs including FDA approved drugs, European Medicines Agency (EMA) approved drugs, and compounds in clinical trials for SARS, which were compiled into the RxRx19a dataset [
17]. Each compound was given in concentrations, varying in 6 different concentrations, each separated by half-log increments, with each concentration of each compound being repeated 6 times. The SARS-CoV-2 virus targets human kidney cells, with EM observations indicating the presence of virus-like particles within the kidney cells of coronavirus infected patients. The cytopathic nature of the SARS-CoV-2 virus suggests direct tubular damage through cytotoxicity. This knowledge is applied through the usage of continuous renal replacement therapies on COVID-19 patients due to the observation of acute kidney injury in patients, but was also crucial in the creation of the RxRx19a dataset, as tubule damage meant that the infected kidney cells of a COVID-19 patient possess characteristics that would distinguish them from mock cells. Recursion used human renal cortical epithelial cells, or HRCE cells from proximal and distal tubules, as well as VERO cells from the kidney cells of the African green monkey as a control in the dataset. An example of the images described above can be found in
Figure 1 [
5].
Recursion also released another set of data with the same treatment conditions named RxRx1, but with a 384-well plate density, as opposed to the 1536 plate density of RxRx19, an image resolution of 512 × 512 with 6 channels instead of the image resolution of 1024 × 1024 with 5 channels of RxRx19, and 1138 classes of siRNA as opposed to the 1672 compounds of RxRx19. The cells used in the previous experiment were treated with siRNA instead of target compounds. The siRNA dataset shared similar features to the SARS-CoV-2 dataset, but with 1139 classes of siRNA applications instead of the 1672 molecules and three viral conditions of the RxRx19a dataset. This allows us to use the siRNA dataset, a relatively larger dataset, to train the classification model first, and then use the viral/mock cells in the SARS-CoV-2 dataset to retrain the model [
5]. Details on this will be given in
Section 3.3 and
Section 4.
4. Experimental Study
The experiments were implemented using the MATLAB software on a computer server Nvidia DGX Workstation with 256 GB of RAM. First, we created a model to classify siRNA images based on the DenseNet161, and we then compared this model with different pre-trained models, such as VGG10, AlexNet, and GoogleNet.
Table 1 shows the classification results of different models using siRNA image datasets. It is clear from the table that the DenseNet produced the best accuracy performance.
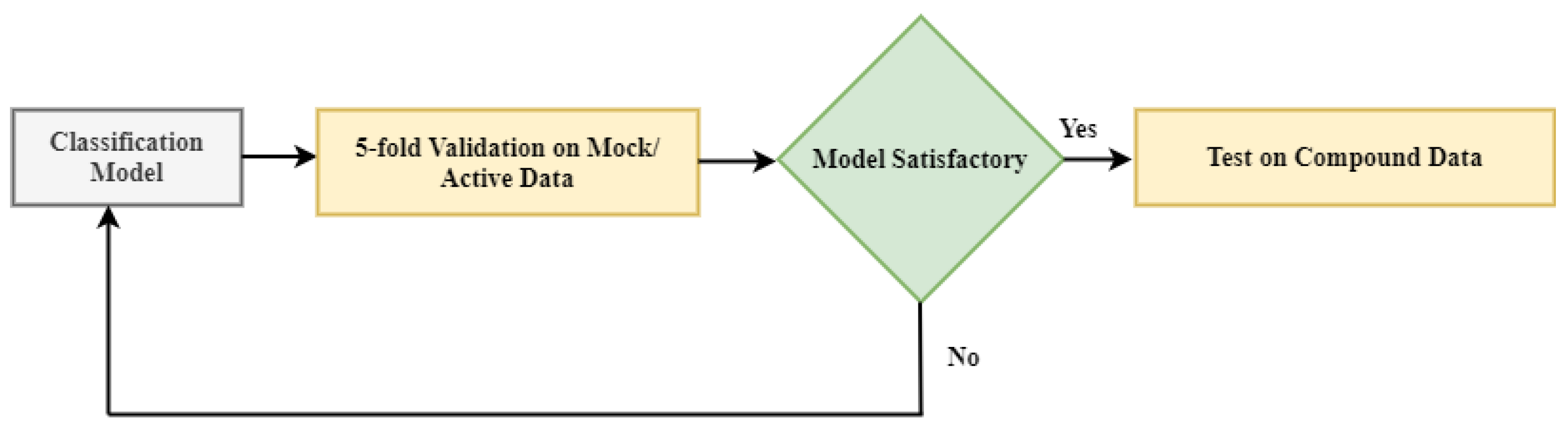
We applied a 5-fold validation procedure to check the performance of the model, as shown in
Figure 5. In this procedure, 80% of the data were randomly chosen for training and the remaining 20% for validation. This step was repeated five times until all mock/active viral cell images in the dataset were validated once. The results from these five tests were averaged. It must be stressed that in the training dataset, there were only active viral cells and mock cells. There were viral cells treated with different compounds in the test dataset, such as Remdesivir and GS-44152. We compare the effectiveness of our model with other well-known pre-trained models in classifying mock and active viral cells.
Table 2 shows the results produced by these models in classifying mock/active viral cells.
Sensitivity serves as a benchmark for the accuracy of a test’s positive prediction, while specificity measures the accuracy of a test’s null prediction [
25]. In the context of our study, sensitivity is the ability of a particular model to determine active viral cells, while specificity is its ability to determine mock/control cells. The F1 score statistically measures the balance of sensitivity and specificity, indicating the ability of the model in preventing both false positives and false negatives [
26]. Finally, the Kappa score is a way of statistically measuring the agreement throughout all the tested cases on a scale of less than and equal to 1, with less than 0 implying little agreement while 1 indicates perfect agreement [
27]. Referencing
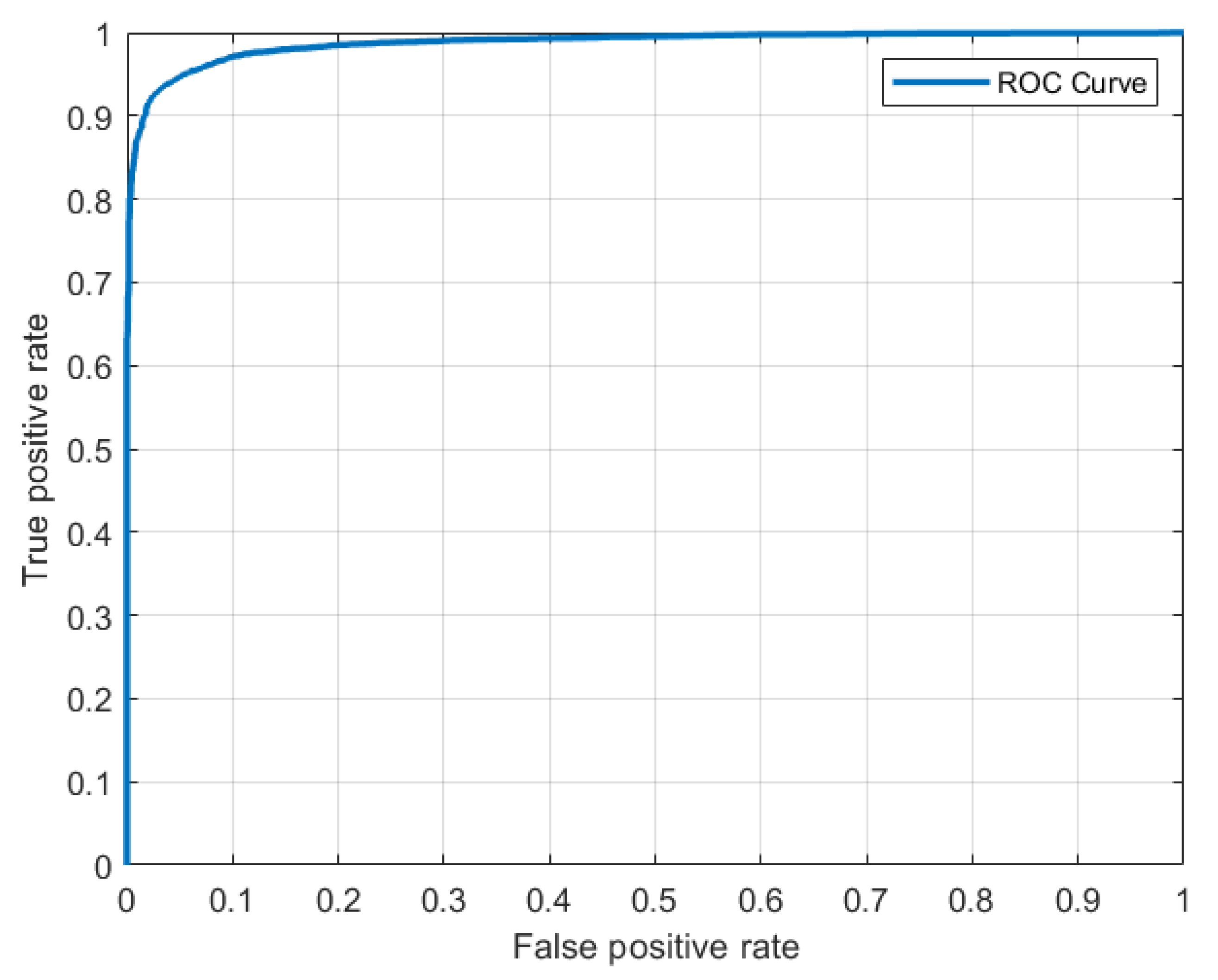
Table 2, the cascade model outperformed other popular deep neural network architectures. In comparison to the two popular deep learning models for the RxRx19 dataset, our cascade transfer learning model had the highest performance in terms of the given metrics. It must be noted that the vgg19 and GoogleNet models were pre-trained on the ImageNet dataset instead of the siRNA dataset before being retrained on the SARS-CoV-2 dataset. To further study the model’s performance, its receiver operating characteristic (ROC) curve is plotted (refer to
Figure 6). The area under the curve (AUC) is 0.98.
We then tested the cascade transfer learning model by using it to rank the efficacy of the compounds used to treat COVID-19, as shown in the last block of
Figure 5, with a score less than 0.5 indicating the promise of the drug as a lead. These test results are given in
Table 3 and
Table 4 and
Figure 7. In the figure, a low probability value, or equivalently a high efficacy score obtained by 1-probability, indicates that the corresponding compound effectively combats active viral cells. In each plot, the histograms of one high efficacy compound and one low efficacy compound are provided. The drugs identified to have the greatest impact on cell health are GS-441524, Remdesivir, CX-4945, Aloxistatin, and Calcipotriene. Of these drugs, two are anti-viral drugs, specifically GS-441524 and Remdesivir, with GS-441524 being a primary component of Remdesivir, explaining their similarly low probability (or high efficacy) scores. In terms of the histograms shown in
Figure 7, GS-441524 provides more consistent performance. The next two lowest probability scores were CX-4945 and Aloxistatin, which were both protein inhibitors. The drugs with the highest probability of classifying as Active SARS-Cov-2 were corticosteroids and regulatory drugs, which had little relation to the Cytopathic nature of COVID-19. The drugs identified in this research are similar to the drugs identified by Recursion in their study [
5] and by the study reported in [
17], as all resulted in GS-441524 and Remdesivir being isolated as clear outliers in our probability scores and also their efficacy scores. Notably, our CX-4945 probability score is much more noticeable for the same compound, although all results share the same four most effective drugs, albeit in different rankings and orders of magnitude [
28]. Of the identified compounds, CX-4945 and Aloxistatin have entered the observational clinical pretrial phase, while Remdesivir (and by association GS-441524) has been approved by the FDA as the first treatment for COVID-19.
5. Conclusions
This paper presents the preliminary results of an exploratory research on the feasibility of deep learning to predict the efficacy of compounds for drug discovery, using COVID-19 as a case study. A transfer learning approach was adopted for this task.
One novelty of the proposed approach is the way the transfer learning strategy is implemented. We first trained the DenseNet, a pre-trained deep neural network, with the siRNA dataset, larger than the SARS-CoV-2 dataset with similar characteristics. The resulting model is then used to extract features from mock cells and active viral cells provided in the SARS-CoV-2 dataset. Thus, we used transfer learning twice from the ImageNet to the siRNA image dataset and then from the siRNA dataset to the SARS-CoV-2 dataset. This cascade transfer learning approach produced superior results for the case study. Another novelty of the approach is using a SoftMax layer as the output layer for the classifier, which produces probability (equivalently efficacy) scores for classifying viral cells treated with different compounds, which allows users to analyze test results with a statistical method. Experimental results demonstrated that the model was able to identify highly promising compounds, which were consistent with those identified in the literature and the drugs that are now under clinical trials.
It must be noted that though the proposed transfer learning strategy is more explainable compared with a classification approach that outputs a hard binary decision, owing to the fact that it produces efficacy scores for candidate compounds in treating viral cells, the model on the whole is not yet transparent. For example, we have not yet been able to explain the properties of the features linked to either viral cells or mock cells. A way to improve the model’s explainability is to link the features extracted by the model to biomarkers of mock and viral cells with a functional relationship which may also be realized by a data-driven model.
Although this paper is limited in its applicability due to the requirements of obtaining a dataset to train the deep learning model, this research encourages us. The deep learning model trained only on the infected and healthy cells was then able to rank candidate compounds, among which some promising ones were under clinical trials. In theory, transfer learning serves to accelerate a drug discovery process significantly. It produces a list of compounds relatively effective on viral cells, thus providing a baseline for speeding up preclinical trials. With cases growing exponentially in a pandemic, the longer treatments and preventative measures develop, the more the situation worsens. Deep learning is a viable tool to combat future COVID-19-like outbreaks.