1. Introduction
The most common form of joint disorder in the United States is osteoarthritis (OA) [
1]. Knee OA can cause pain and is the number one disease at causing loss of ability to perform daily activities such as walking and stair climbing [
2]. Knee OA is associated with age [
3] and is characterized by the loss of articular cartilage volume [
4]. OA is viewed as a “whole-organ” disorder, manifesting damage to a range of articular structures, especially the hyaline cartilage, meniscus, periarticular bone, ligaments, and tendons [
5]. Despite its importance for public health, we have no interventions that effectively modify the OA disease process [
6]. The absence of useful biomarkers to detect OA progression is a major technological obstacle to the development of treatment and prevention of knee OA [
7].
While joint replacement is effective for treating end-stage OA, the evaluation of potential disease-modifying treatments in populations meeting current clinical criteria for OA has had limited success [
6]. In the past decade, early diagnosis and early treatment strategies in rheumatoid arthritis have reduced patient morbidity and associated costs [
8]. The early diagnosis and treatment of OA conditions may similarly improve outcomes and reduce disability and costs for OA. However, the absence of useful image biomarkers to detect OA progression has been a critical technology gap in the early diagnosis and treatment of OA [
9].
The conventional radiographs (X-rays) are commonly used for routine knee OA examinations. An X-ray of a joint with osteoarthritis typically shows a narrowing of the space between the bones of the knee joint where the cartilage has worn away. However, symptoms of knee OA may arise before the damage can be seen in standard X-rays. For example, Roemer et al. [
10] described how X-rays are unable to show certain structural phenotypes of OA and cannot detect some detrimental findings which can indicate risk of disease that would progress rapidly.
The advent of magnetic resonance imaging (MRI) offers the promise of addressing the critical technology gap by allowing quantification of structural damage in joints. For this reason, radiologists at hospitals often use the more sensitive magnetic resonance imaging (MRI) for OA early detection. Juras et al. [
11] pointed out that OA needs early detection, and MRI is a noninvasive way for detecting early biomarkers. To promote the evaluation of OA MRI biomarkers, the National Institutes of Health (NIH) launched the Osteoarthritis Initiative (OAI) cohort study, which includes four clinical centers that recruited approximately 4800 men and women (ages 45–79 years) with or at risk for knee OA. The OAI collected a wealth of data on its participants over an eight-year span. The study included annual knee MRIs for the first four years and then biannual knee MRIs for the subsequent 4 years [
12]. One of the goals of creating the OAI dataset was to discover the objective, measurable standards of disease diagnosis and progression, and to determine the predictive role of MRI changes for subsequent radiographic and clinical changes related to the development of knee OA.
The 3-dimensional (3D) MR images allow for both viewing the knee as a “whole organ” and depicting all of the tissues of the joint [
13]. While cartilage degradation and other biomarkers can be manually detected, it is time-consuming to process the volume of 3D MR images. Thus, there is a need to automate these processes with machine learning techniques.
Convolutional neural networks (CNNs) are a class of deep learning techniques that are designed to work with images and can remove the need for handcrafted feature extractors [
14]. CNNs have been used for various image classification tasks, with recent studies developing CNN models for medical image analysis. The early work of using CNNs to classify knee OA was mainly applied to radiographic (X-ray) images [
15,
16,
17,
18].
Anthony et al. employed the classical VGG-16 CNN architecture and transferred learning with X-ray images to determine the OA severity level [
15]. These images were preprocessed using an SVM and Sobel edge detector in order to locate the knee joint area. Their study used X-ray images from the OAI. A set of 4446 X-rays were used in this study, representing a total of 8892 knees. When classifying the five Kellgren and Lawrence (KL) grades, they achieved an accuracy of 59.6%. Later, in another work, the same group updated the preprocessing step to use a fully convolutional neural network (FCN) to determine the bounding box of the knee joint. The FCN method was found to be highly accurate in determining regions of interest (ROI), and when combined with a CNN for classification, the method achieved an accuracy of 61.9% [
16].
Unlike the two-stage frameworks developed in [
15,
16], a recent work [
17] proposed an end-to-end CNN architecture for knee OA severity assessment without using a neural network for preprocessing. This method used branches in its CNN that are referred to as “attention modules”, which provide an unsupervised determination of the ROI of X-ray images. Another recent work added a long short-term memory (LSTM) classifying step following the CNN layers in their network [
18]. Given the nature of LSTM for processing sequential data, additional images were generated in a preprocessing step by cropping a fixed ROI and rotating the cropped image by 5, 10, −5, and −10 degrees. The original image and augmented images were stacked, giving about 4600 images used for training and about 480 for testing. Their work also used images from the OAI and achieved an accuracy of 75.28% for the 5-category classification.
It can be seen that 3D CNNs have developed quickly and are attracting interest as a method for analyzing sequences of images or other volumetric data. In a recent study, a 3D CNN was used for classifying real-world objects [
19]. Depth information was used to create a 3D shape that was converted into a volumetric representation (voxels) to be classified by the 3D CNN. In addition, 3D CNNs have shown to be useful in medical image processing. When classifying lung nodules, working with 3D volumetric data in a 3D CNN outperformed 2D CNNs [
20]. Wang et al. applied a 3D CNN model to calculate the probability of needing a total knee replacement (TKR) within the next nine years [
21]. Their work demonstrated that the automated discovery of OA biomarkers from turbo spin echo (TSE) and double-echo steady-state (DESS) images could outperform models that use only demographic and clinical data. Another work explored this area using the popular 2D U-Net architecture for the segmentation of cartilage and meniscus in the knee, which were fed into a 3D CNN for classifying the severity of the cartilage and meniscus lesions [
22]. Given the large amount of volumetric data, another recent work for classifying knee lesions used cropping of 3 ROIs from knee MRI to reduce the dimensionality before processing by multiple 3D CNN [
23]. Aside from these applications, 3D CNNs have also been applied to segmentation problems including knee cartilage segmentation [
24] and segmentation of brain lesions [
25].
For knee OA severity classification, while previous methods used 2D CNNs to analyze X-ray images, in this work we propose a method using a 3D CNN and MR images. The details of the proposed method is introduced in the next section.
2. Materials and Methods
2.1. Method Overview
Knee MR imaging produces a 3D representation of the knee joint, utilizing a sequence of 2D images taken laterally across the knee. Given the 3D nature of MR images, 3D CNN can be advantageous in evaluating the whole sequence of images as one unit. Through the implementation of 3D kernels, information from adjacent slices could be integrated. Therefore, 3D features that may not be detectable using 2D CNN could be potentially captured.
For this study, we built a machine-learning model capable of analyzing sequences of MR images for each knee as input, with output given as one of the five KL grades. We further trained another model by relabeling the samples into OA and non-OA categories according to the clinical standard, i.e., patients with KL ≤ 1 are diagnosed as non-OA cases while patients with KL ≥ 2 are considered as OA. An overview of the proposed models is described in
Figure 1.
In addition to MRI, we also studied traditional X-ray images, with an interest of finding out which imaging modality coupled with the modern CNNs can achieve better accuracy for knee OA diagnosis. We employed several state-of-the-art 2D CNN models, including VGG 16, ResNet50, DenseNet, etc. These models were trained to classify X-ray images into five KL categories. The one with the best accuracy was selected and further applied for the binary OA/non-OA classification. The pipelines for X-ray images are similar to those illustrated in
Figure 1, except the 3D CNN is replaced by a 2D CNN and the preprocessing step for X-ray is to cut each pair of knees into individual ones. The X-ray images and MR images were obtained from the same group of patients, and the separation of training, validation, and testing sets were kept the same at the patient level for all the models trained and compared in this work.
2.2. Dataset
The dataset used in this study was from the public database Osteoarthritis Initiative (OAI). Most of the patient samples in the OAI dataset include an X-ray image; however, many do not have an accompanying MRI sequence. For this study, we used a subset of the OAI data with 1100 knees, with each knee having both MRI and X-ray available. The 3D DESS MRI data for each knee contain a sequence of 160 2D images, while there is one X-ray image containing both knees from a patient. The dataset was selected with an equal distribution among different OA severity levels (0–4) measured by the Kellgren and Lawrence (KL) grades.
A common practice in machine learning is to split the available dataset into three subsets known as the training set, validation set, and testing set [
26]. Machine learning models learn from the training set with the validation set being used during the training process to tune parameters [
27]. The testing set is not seen during the training process but rather is held back until the end of the study. The available dataset was randomized and then split into groups balanced by the KL grade with 800 training samples, 200 validation samples, and 100 testing samples. Each set contains a balanced number of samples from each of the five KL categories.
Table 1 shows the distribution of the data.
2.3. Preprocessing: Subregion Selection
Unlike natural images where useful information could appear anywhere, in medical images, features are usually located in fixed locations. As an example, the knee MR image shown in
Figure 2 contains large bone areas and many other tissues. The important indicators for knee OA are often observed near the cartilage and joint region of femur and tibia bones. Therefore, we can reduce the input dimensionality by cropping a subregion of the images.
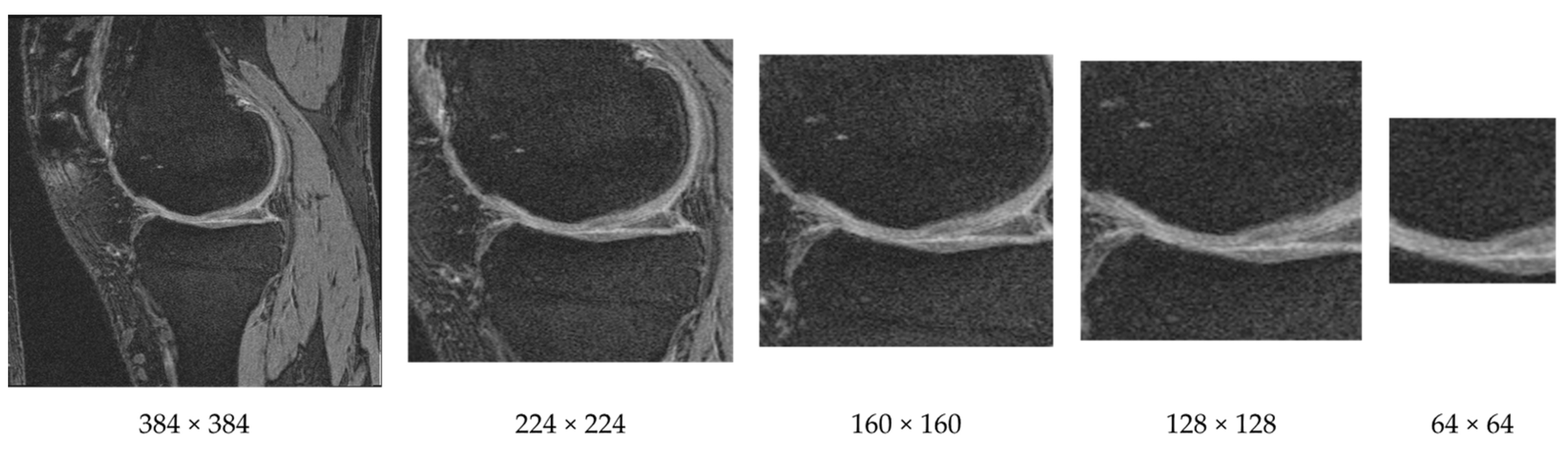
We cropped each image as a preprocessing step that helps to keep the cartilage region while removing regions that are less informative for knee OA classification. We cropped the image (original size 384 × 384) from the center using both square and rectangular regions. An example of cropping a knee MR image to various sizes is provided in
Figure 3. Through various tests we discovered that the window size of 160 × 160 achieved the highest F-measure.
2.4. Preprocessing: Slice Removal
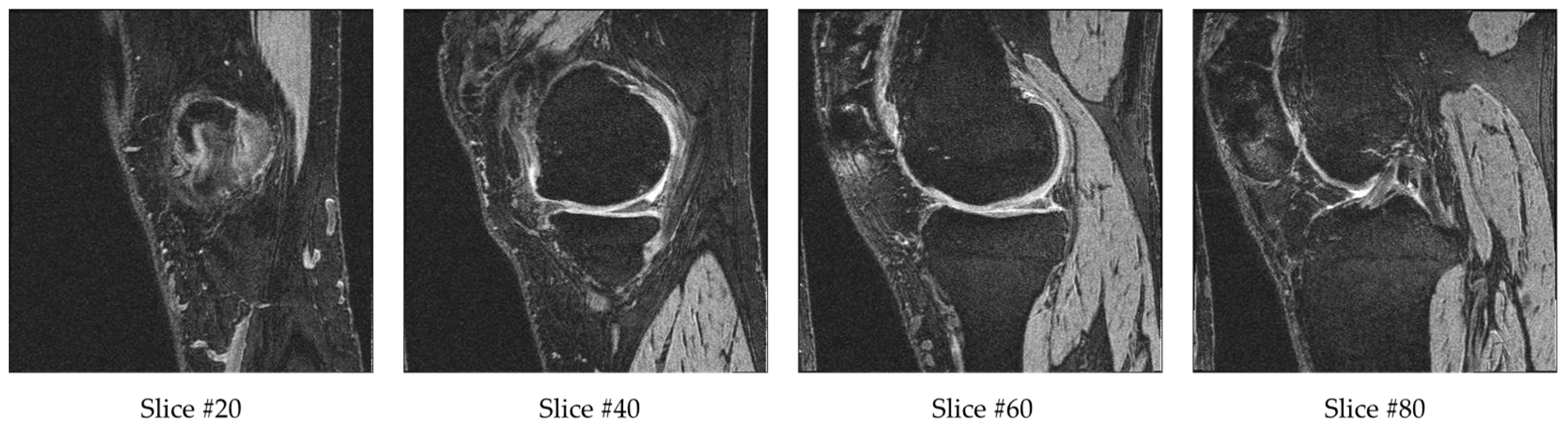
Each sample in the database contains a sequence of 160 MR images. After cropping, the input dimensions of 160 × 160 × 160 were still very high. To further reduce the input data dimensionality, we removed some of the outer and center slices. The reason for removing a few beginning and ending slices is that they do not contain bone or cartilage information. Therefore, they are not likely to contain information related to OA. The reason for removing the middle range slices is that they have ill-defined cartilage regions and blurry bone boundaries due to the transition of medial and lateral bone happening in this range. Example slices are provided in
Figure 4. Slice #20 has a small bone region starting, while slice #40 and #60 have larger bones with clearly defined bone boundaries and cartilage. Slice #80 is in the transition range, and therefore the cartilage and bone boundaries are unclear. For each sequence, we excluded the first 10 slices (1–10), middle 20 slices (71–90) and final 10 slices (151–160). The remaining 120 slices (11–70, 91–150) from the 160 slices were fed into the 3D CNN model. This is about 13% of the original 384 × 384 × 160 volume.
2.5. 3D CNN Model for MRI
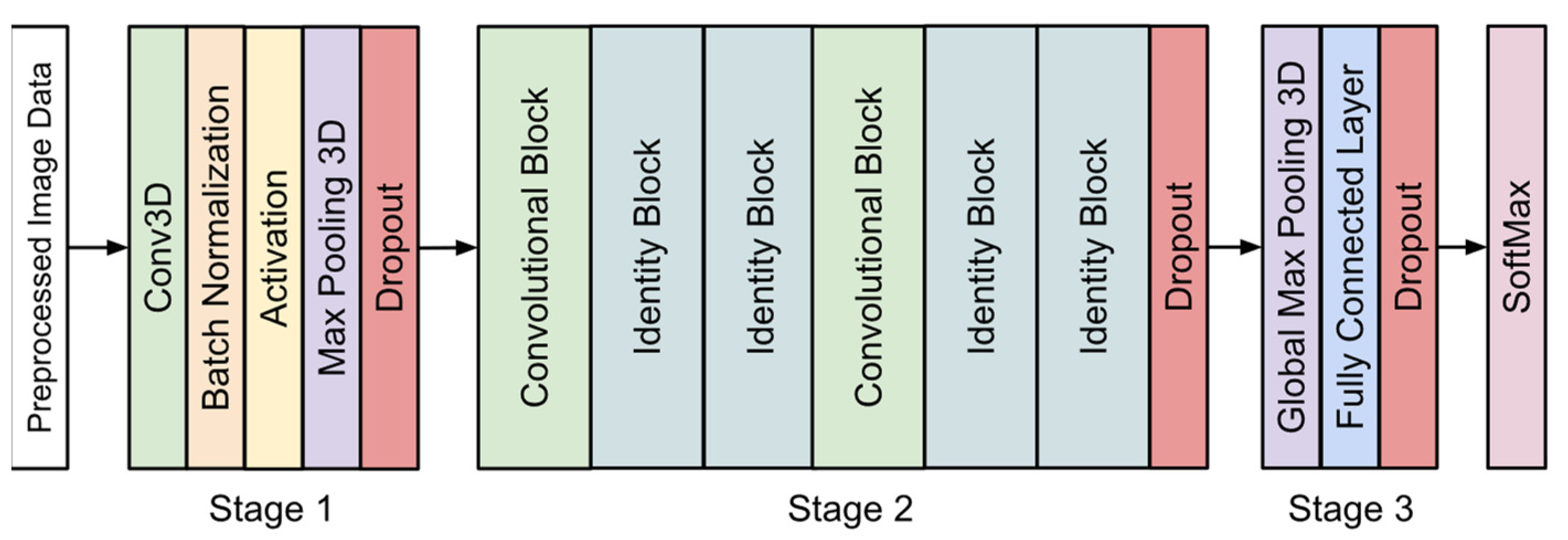
The architecture of the 3D CNN model proposed in this study is shown in
Figure 5. The structure was inspired by the work performed by Wang et al. [
21]. The most important difference between 3D CNNs and 2D CNNs is that 3D CNNs use 3D convolutional kernels to process a volumetric patch of a scan, while 2D CNNs process a single anatomical plane. The 3D convolutional kernels incorporate information from adjacent slices and are therefore able to extract 3D features, which are not detectable from 2D CNNs. As shown in
Figure 5, three stages are included in the proposed 3D CNN model before the final softmax layer. The details of the model are discussed below.
Stage 1 of the model began with a convolutional layer containing 32 kernels of size 7 × 7 × 7 with a stride of 2 × 2 × 2. This was followed by batch normalization and an activation layer using the ReLU function. A max-pooling layer was added with a window size of 2 × 2 × 2 and a stride of 2 × 2 × 2. Finally, a dropout layer was placed before the start of the residual blocks. We used dropout layers in each stage of our 3D CNN model to help reduce overfitting. Each dropout layer used a rate of 0.5, which gives each node a 50% chance of being set to 0.
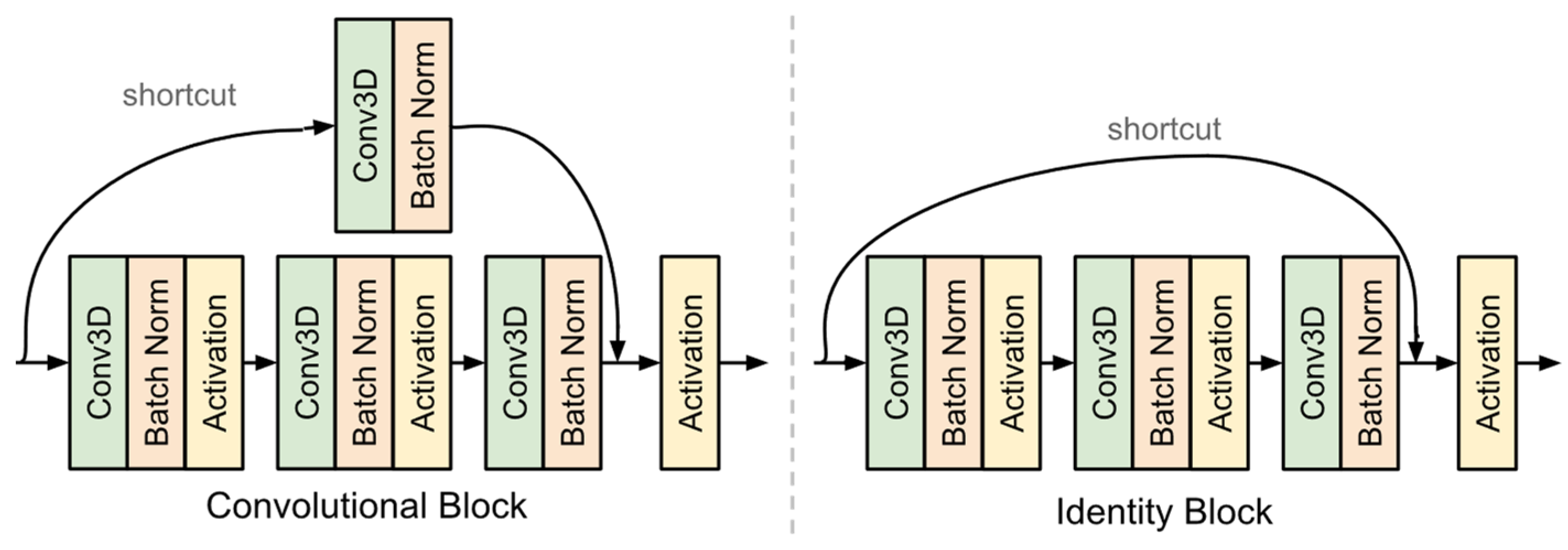
The second stage of the model contained a sequence of six residual blocks [
28]. Each residual block featured a shortcut connection from the input to the output. There were two types of blocks used in this model as shown in
Figure 6. The convolutional block features a convolutional layer in the shortcut path. This layer was used when the input dimensions were changed. The identity block did not have any layers in the shortcut path and was used when the input and output dimensions matched. This stage also ended with a dropout layer.
The final stage of the model used global max pooling, followed by a fully connected layer of size 1024 and a dropout layer. The last layer of the model uses the softmax function, which outputs the possibilities of the sample belonging to each category.
The model was implemented in Python using the Keras library with TensorFlow as a backend. The model was trained using a batch size of 15 with early stopping based on validation loss. The Adam optimization function was used with a learning rate 0.001. The training was performed on a high-performance computer with a NVIDIA Tesla V100 32 G GPU.
2.6. Classic 2D CNN Architectures for X-ray
When building our dataset, we selected patients which had an MRI volume as well as an X-ray image of the same knees. To develop the model for X-ray, we employed a variety of state-of-the-art 2D CNN architectures. VGG16 [
29] was one of the earlier deep learning models, and it showed superior performance in many applications. ResNet50 [
28] used the concept of residual blocks in which a shortcut connection is added after a series of layers. Our proposed 3D model utilizes a 3D variation of the ResNet50 convolutional and residual blocks as well. Inception-v3 [
30] is the representation of the deep learning networks with inception modules and one of the first models to make use of batch normalization. Inception-ResNet [
31] is a hybrid of Inception-v3 with residual connections. DenseNet [
32] implements dense blocks in which convolutional layers of the same size are connected to every other layer in front of them.
While an MRI volume contains just one knee, an X-ray image contains both. These X-ray images were split in half, and all the left knees were flipped so the right and left knees are aligned. The pretrained ImageNet weights were used for transfer learning. The last softmax layer was retrained using the X-ray data while the previous layers were not changed. Input images were scaled to a size of 224 × 224 or 299 × 299, depending on the architectures of different models. Since the pretrained networks were trained with RGB images, we duplicated each gray level X-ray image three times to feed it into the three input channels.
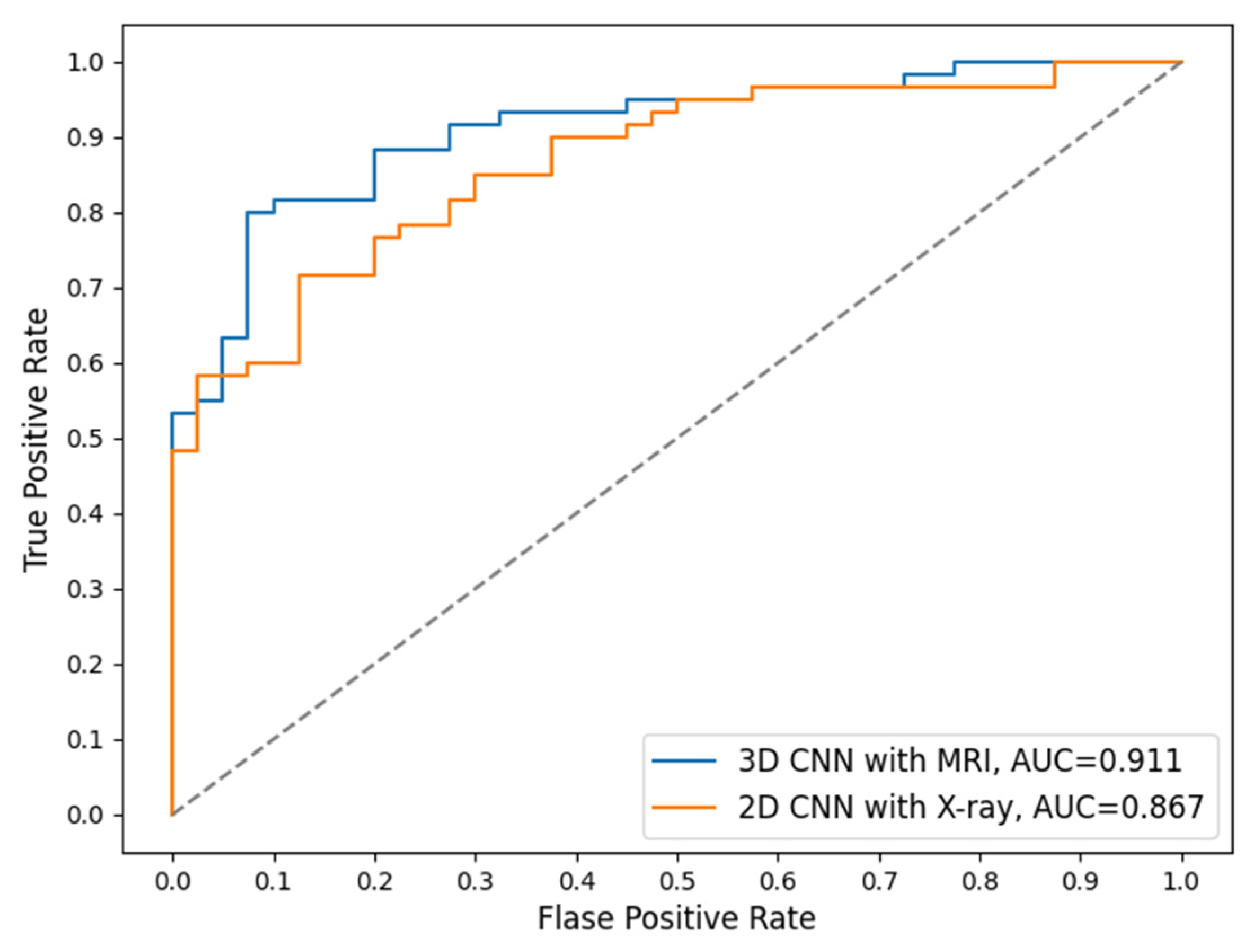
4. Discussion
Currently, X-ray is the basic routine imaging modality for examining a patient with OA potentials clinically. While X-ray is more cost-efficient than MRI, it is not as sensitive as MRI, which can show much more structure and tissue details. Therefore, MRI is considered as an alternative imaging tool, especially for detecting early osteoarthritis with slight structure change.
In the 5-category results of this study we found that MRI had higher accuracy in classifying KL = 0 and KL = 1 (
Table 6). This aligns with the previous studies that found MRI to be better at capturing detailed and small structure change and therefore more sensitive to early signs of OA development. When classifying the category KL = 4, X-rays have higher accuracy than MRI, indicating that X-rays are better at detecting OA in a more severe situation. The 2-category results in this study were consistent with those in the 5-category, in that X-ray has higher accuracy for detecting severe OA cases while MRI is more sensitive to small structure changes and early indicators of OA (
Table 7). The complementary performance of the two imaging modalities is interesting and indicates the possibility that they could be combined to develop a comprehensive and more accurate diagnosis system than using each individual imaging modality alone.
A limitation of this study is that we have a limited number of samples. This is because we have to include patients with both MRI and X-ray scanned on the same knee. Another limitation is that MRI is not widely used in clinical diagnosis due to the cost. However, MRI is a new trend of imaging to study the pathology of knee OA in many clinical trials since MRI can offer a better view of soft tissues such as cartilage, bone marrow lesions, and effusions.
Future work includes further examination of the KL categories with lower accuracy, e.g., the KL = 2 category, which was often misclassified into KL = 1 (non-OA) by the model. This may be solved with a weighting system during training. Additionally, our current preprocessing uses a fixed offset for cropping as well as a fixed range for slice removing. This could be updated as a dynamic setting for each sequence, which may retrieve more accurate information and therefore generate better classification performance. Combining the two imaging modalities for a comprehensive and more accurate model is also a promising direction.