Featured Application
Damage-associated molecular patterns (DAMP) sensors on periodontal tissue and resident cells were characterized, indicating that
nucleotide-binding oligomerization domain-like receptor 2 and toll-like receptors 1/2/4/6 could be significantly elevated in the disease state or upon stimulation. Further investigations are warranted to confirm the relevance of such DAMPs sensors in the innate defense of the cells/tissue concerned.
Abstract
Background: Toll-like receptors (TLRs) and nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) are innate, damage-associated molecular patterns (DAMP) sensors. Their expressions in human periodontal resident cells and reactions toward irritations, such as hypoxia and lipopolysaccharide (LPS), remain not well characterized. This cross-sectional study aimed to investigate and characterize TLRs, NOD1/2 and NLRP1/2 expressions at the dento-gingival junction. Methods: Immunohistochemistry screening was carried out on periodontal tissue biopsies sections, while selected DAMP sensors signal and protein expression under Escherichia coli LPS (2 µg/mL) and/or hypoxia (1% O2), 24 h, by human gingival keratinocytes (HGK) or fibroblasts (HGF) were investigated. Results: Positive TLR1/2/4/5/6, NOD1/2 and NLRP1/2 immunostaining were observed in healthy and periodontitis biopsies with apparently more pocket epithelial cells positive for TLR2, TLR4 and NOD1/2. TLR1-6, NOD1/2 and NLRP1/2 messengers were detected in gingival/periodontal biopsies as well as healthy HGK and HGF explants. LPS and/or hypoxia induced signals and protein upregulation of NOD2 in HGKs or TLR1/6 and NOD2 in HGFs. Conclusion: Transcripts and proteins of TLR1/2/4/5/6, NOD1/2 and NLRP1/2 were expressed in human periodontal tissue in health and disease. Putting all observations together, NOD2, perhaps with TLR1/2/4/6, might be considered key, damage-associated molecular pattern sensors on periodontal resident cells.
1. Introduction
Periodontitis is a common chronic oral disease caused by mixed, predominantly Gram-negative, anaerobic, pathogenic microorganisms in close proximity to periodontal tissues [1]. Human hosts have various strategies for recognizing the molecular patterns of oral microbes, including those from the periodontopathogens, which, in turn, trigger corresponding host immune reactions [2]. Periodontitis and its associated periodontium destruction is believed to be the result of damaging or inappropriate host responses induced by periodontopathogens [3].
Lipopolysaccharide (LPS) is a cell wall component of Gram-negative bacteria, which are known to cause various reactions on the human host [4]. For instance, the recognition of LPS by myeloid and/or non-myeloid cells would activate the host’s innate immune system to produce proinflammatory cytokines, including, but not limited to, tumor necrosis factors (TNF), interleukins (ILs), and cyclooxygenase-2 (COX-2) [5,6]. Systemic exposure of LPS in high quantities causes the overproduction of cytokines, which induce fever, deteriorate organ perfusion, tempt cardiovascular collapse and eventually cause fatal sepsis syndrome [7].
Toll-like receptors (TLRs) and NOD-like receptors (NLRs) are host cell receptors (surface or intracellular) responsible for various innate immune responses in mammals [8,9]. TLRs and NLRs represent two large families of proteins in charge of responses mediation via nuclear factor-κB (NF-κB)-dependent/interferon-regulatory factor-dependent signaling pathways, or NF-κB-dependent/mitogen-activated protein kinase signaling pathways [8,9]. It is speculated that the recognition of LPS is mediated by these two pattern-recognition receptors when Gram-negative pathogenic microorganisms get into close contact with host cells. Previous reports indicated that TLR2/4 are essential LPS-recognition receptors at the human periodontium [10], while in general, TLR2 appeared to be less specific than TLR4 in sensing LPS [11], the reason being that the former covers more ligands, whereas TLR4 targets LPS, specifically. TLR2 needs to form heterodimers, such as TLR1/2 or TLR2/6, to function and such heterodimers have to be formed before periodontopathogens and/or LPS can be recognized [12]. It is speculated that, nevertheless, TLR1/2 andTLR2/6 expand the receptor–ligand recognition spectrum and improve the immune reactions to Gram-negative bacteria [12]. Previous research has demonstrated the expression of TLR1-10 in 10 chronic periodontitis and healthy samples; however, the expression of those receptors in junctional epithelium was not reported [13].
Given that anaerobic, Gram-negative pathogens are key agents for periodontitis pathogenesis, the nature of the hypoxic micro-environment draws attentions of researchers interested in deciphering the complicated pathogenic process [14]. At sea level, atmospheric oxygen level at 760 mm Hg is 21% or 140–150 mm Hg, while the corresponding alveolar O2 partial pressure (pO2) is approximately 100 mm Hg in healthy human [15]. In chronic, inflamed periodontitis tissue, oxygen level could be at <50% O2 saturation or as low as 7 mm Hg [16,17], compared to 85% O2 saturation or up to 52 mm Hg at less affected gingivitis compartments [16]. Inside periodontal pockets, O2 is persistently consumed and reduced to low levels due to increased consumption via active chronic inflammatory cellular processes as well as the diminished oxygen availability through thrombosis of the involved micro-vasculature [14,18]. Therefore, hypoxia establishes and potentially sustains the survival of facultative and obligatory anaerobes, promotes periodontal inflammation and, in turn, could intensify and worsen periodontitis development. On the contrary, a hypoxic environment, however, was known to relate to cell survival, DNA repair, apoptosis, etc., driven by an array of corresponding signals transcription in [19]. For instance, the knockdown of hypoxia-inducible factor-1 (HIF-1) impaired the motility and bactericidal action of myeloid cells in [20]. At the periodontal front, hypoxia would increase the in vitro mobility of human keratinocytes on connective tissue coated with collagen I, collagen IV, or fibronectin, while 2% O2, together with periodontal microbes, enhanced IL-8 and TNF-α expressions in oral keratinocytes in [21]. Therefore, it seems that hypoxia confer an important modulatory function in the interactions between periodontopathogens and host immune responses with related mechanisms yet to be clarified.
Currently, particular TLRs expression states might appear to be related to certain hypoxic environments, despite the fact that in many cases, the exact nature of the interactions remain to be clarified [14]. For instant, TLR4 expression was reported to be up/down regulated in macrophages/endothelial cells, respectively, under low oxygen concentrations in [22]; TLR1/2 expression was observed to be enhanced in neonatal mice brains under a hypoxic environment in [23]; and TLR2/6 was found to be expressed in murine/human dendritic cells, human monocytic cells, endothelial and intestinal epithelial cells under low oxygen in [24]. Therefore, it is speculated that TLR expressions are cell type specific [25]. The influence of hypoxia on other TLR and NLR expression in human gingival resident cells, for obvious reason, remain to be investigated.
In the present study, the research group hypothesized that TLRs/NLRs play key roles in periodontal innate responses in health and disease. Hypoxia, together with LPS, may, to a certain extent, modulate the expression of damage-associated molecular pattern (DAMP) sensors in periodontal resident cells. This study, therefore, screened selected human gingival TLR/NLR expression on human periodontal biopsies, gingival keratinocyte (HGK), and human gingival fibroblast (HGF). The latter cell explants were subjected to in vitro challenges by hypoxia and/or Escherichia coli LPS.
2. Materials and Methods
This study was a convenience sample, cross-sectional investigation carried out according to the STROBES (Strengthening the Reporting of Observational Studies in Epidemiology) guidelines.
2.1. Periodontal Biopsies
The study was approved by the Institutional Review Board, the University of Hong Kong and Hospital Authority Hong Kong West Cluster (HKU/HA HKW IRB, UW 06-376 T/1401). Written consent was obtained from all study participants or, for minors, their guardians.
Inclusion criteria for participants were as follows: (1) ≤60 years old; (2) ≥2 permanent teeth per quadrant, excluding wisdom teeth; (3) non-smoker; and (4) systemically healthy. In addition, periodontitis participants had to have the following: (5) untreated periodontitis with at least one site with a probing-pocket depth (PPD) ≥6 mm, probing attachment level (PAL) ≥5 mm, and radiographic evidence of alveolar bone loss; and (6) at least one periodontally involved tooth scheduled for extraction or periodontal surgery. For periodontally healthy or control participants, additional inclusion criteria were the following: (5) full-mouth bleeding on probing ≤30%, no site with PAL >3 mm, deepest PPD ≤3 mm; (6) no radiographic evidence of alveolar bone loss nor any furcation involvement in multi-rooted tooth; and (7) crown-lengthening surgery or tooth extraction arranged for prosthodontic or orthodontic reasons. Exclusion criteria included the following: (1) female participants pregnant or lactating; and (2) participants taken systemic antibiotics within 6 months prior to recruitment. Convenient sampling of patients requiring periodontal extraction/surgery, as well as extraction due to other reasons in patients who were deemed periodontally healthy as described above, was conducted at the Faculty of Dentistry, The University of Hong Kong, Prince Philip Dental Hospital, between September 2012–April 2015, which was when recruitment, preparation and sampling was conducted.
In accordance with previous, similar reports [13,26], the current group considered that a sample size of n > 10 (one biopsy per participant) for the immunohistochemistry assay or periodontal resident cells explant study would be appropriate.
Healthy gingiva tissues were collected before periodontal surgery or tooth extraction, while periodontitis tissues were collected from participants undergoing periodontal surgery or extraction after the hygienic phase of the treatment, i.e., initial, non-surgical, mechanical, periodontal therapy, including oral hygiene education, scaling and root planing. The surgical procedure was as described in a previous report [26].
2.2. Immunohistochemistry
Healthy, human gingival and periodontitis biopsies (Table 1) were fixed overnight in a 10% neutral-buffered formalin solution before being dehydrated and embedded in paraffin. Sections of a human hepatocellular carcinoma biopsy (courtesy of Professor Nancy K. Man, Department of Surgery, LKS Faculty of Medicine, the University of Hong Kong) were used as the positive control for TLR3. Periodontal specimens were sectioned at a thickness of 4 μm and mounted onto silicon-coated slides, followed by rehydration, then were immersed in 3% hydrogen peroxide at room temperature (r.t.) for 10 min for peroxidases blocking. For antigen retrieval, the sections were immersed in a 95 °C sodium citrate buffer (0.05% tween 20–10 mM sodium citrate, pH 6.0) for 10 min, then cooled to r.t., followed by blocking with 2.5% horse serum at 37 °C for 30 min. The specimens were then incubated with anti-human TLR-1 to -8 and TLR10, NOD1/2 or NLRP1/2 primary antibodies, independently in 3% bovine serum albumin at 4 °C overnight (mouse anti-human: TLR1, TLR4 and NLRP2 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), TLR2 (Imgenex, San Diego, CA, USA), TLR3 (eBioscience, San Diego, CA, USA), TLR5 (Abnova, Taipei, Taiwan), TLR8 (Enzo Life Science, Farmingdale, NY, USA); rabbit anti-human: TLR6, NLRP1 and NOD2 (Thermo Fischer Scientific, Waltham, MA, USA), TLR7 (LifeSpan BioSciences, Inc., Seattle, WA, USA), NOD1 (R&D systems, Inc., Minneapolis, MN, USA), TLR10 (Novus Biologicals, Littleton, CO, USA)). The optimal concentration of each TLR or NLR antibody was decided by pilot studies, and was 1:100 for anti- TLR1, TLR4, TLR6, TLR7/8/10, NOD1/2 and NLRP1/2 (1:100 was set as the highest primary antibody concentration, including antibody-elicited negative staining after pilot experiments), and 1:200 for anti- TLR2, TLR3 and TLR5. The negative control sections were incubated with thesame dilution of IgGs from the corresponding animal not immunized against the target human antigen. After incubation with secondary antibodies (anti-mouse/anti-rabbit IgG Peroxidase, 1:1000, ImmPRESS® Universal Reagent, MP-7500, Vector Laboratories, Burlingame, CA, USA) at 37 °C for 1 h, the specimens were stained with diaminobenzidine (DAB) for visualization and semi-quantitative analysis. Stained sections were captured, analyzed with a digital imaging system (Leica DC 300 Ver 2.0; Leica, Wetzlar, Germany) and Leica Qwin Standard V 2.6 software (Leica, Wetzlar, Germany) [26].

Table 1.
Donors’ details concerning periodontal biopsies or cell explants used and reported in this study.
Upon preliminary experiments, no TLR9 mRNA could be detected from HGK/HGF explants nor human gingival tissue (HGT) biopsies (c.f. Section 3.4); the immunohistochemistry detection of TLR9 did not follow. All immunohistochemical investigations were repeated thrice on separate occasions with different sections from the same tissue blocks.
2.3. Cell Culture
Healthy gingiva biopsies were immersed in a 1.5 U/mL dispase solution (Gibco, Thermo Fischer Scientific, Waltham, MA, USA) in neutral Ca2+/Mg2+ free phosphate buffered saline at 4 °C for 8 h. The epithelium was then mechanically sheared from the connective tissue. Minced epithelial and subepithelial tissues were utilized for HGK or HGF primary culture, respectively. To obtain primary HGK or HGF explants, Gingival Keratinocyte Medium (1:1 mixture of Defined keratinocyte-SFM and Epilife® with calcium, Life Technologies Corporation, Grand Island, NY, USA) with 10% fetal bovine serum (FBS), or Dulbecco’s Modified Eagle Medium with 10% FBS, respectively, was used. Any HGF contamination in HGK culture was removed by brief trypsinization (0.25% trypsin (Sigma-Aldrich, St. Louis, MO, USA) without EDTA, 3 min), followed by reseeding the ‘cleaned HGK’. The same HGK purification procedure was repeated until no spindle-shape cells were detectable under microscopy. Samples of purified, early confluent explants from each donor on 35 mm dishes were carefully confirmed by carboxyfluorescein hydroxysuccinimidyl ester staining when HGK appeared as homogenous cobblestone-shaped cells, free of any contaminating spindle-shaped fibroblastic cells or HGF as homogenous spindle-shaped cells without cells of any other shapes, smaller sizes or morphology, before use. In the case of doubt, the cells concerned were not used for any experiments. HGK and HGF from individual donors were always cultured separately. The cells at third passage were harvested for in vitro experiments [26,27].
HGK and HGF cultured on individual 60 mm × 15 mm plates were treated with 2 µg/mL E. coli LPS (From O111-B4, catalog no. L3024, Sigma-Aldrich, St. Louis, MO, USA), 18% O2 (normoxia), 5% CO2 for 24 h. Both cells cultured on same conditions were also treated with 1% O2, 5% CO2 for 24 h (hypoxia). Low oxygen concentration was maintained by a hypoxia incubator (Catalog no. 3131, Forma Water-Jacketed CO2/O2 incubator, Thermo Fisher Scientific, Waltham, MA, USA). The corresponding HGK/HGF from the same donor cultured in designated media without the challenges (E. coli LPS or hypoxia) under standard conditions (18% O2, 5% CO2) were used as controls (Table 1) [27].
All experiments were repeated thrice on separate occasions with cell explants from different donors.
2.4. MTT Assay
To compare the growth rates of HGK and HGF cultured in 1% or 18% O2, HGK or HGF were seeded at a density of 3.0 × 103 per well in 96-well plates (Table 1). Wells with evenly distributed cells were used. After 24 h culture, a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed according to the manufacturer’s protocol (ATCC, Manassas, VA, USA). Briefly, HGK or HGF (treated or untreated) were incubated with 10 µL MTT for 2 h and then the MTT reagent was replaced with 100 µL dimethylsulfoxide for another 2 h. Absorbance values for each well were measured by a spectrophotometer at 570 nm (Bio-Rad Model 3550, Bio-Rad, Hercules, CA, USA). For LPS treatment, from a stock solution of 1 mg/mL E. coli LPS in Dulbecco’s phosphate-buffered saline, the correct amount of the endotoxin was added to the cell wells to achieve a final concentration of 2 µg/mL at 18% O2, 24 h [28].
All experiments were repeated three times at separate occasions, using HGK/HGF explants at the third passage.
2.5. Reverse Transcription Polymerase Chain Reaction (RT-PCR) and Quantitative Reverse Transcription PCR (RT-Qpcr) Analysis
Total RNA was purified from treated or untreated HGK or HGF and HGT biopsies (Table 1) via disruption and homogenization, using a RNeasy Mini Kit (Qiagen, Valencia, CA, USA). The human acute monocytic leukemia cell line (THP-1) was used as the positive control for TLR9. HGT were maintained in RNAlater at 4 °C overnight before proceeding with RNA extraction.
For RT-PCR, cDNA was synthesized from 1 μg of total RNA from each sample, using Superscript First-Chain Synthesis Kit (Invitrogen, Carlsbad, CA, USA). RT-PCR was performed using KAPA2G Fast HotStart Readymix (Kapabiosystems, Boston, MA, USA), and two-step RT-qPCR was carried out using cDNA from control/treated cell explants by TaqMan Real-Time PCR Master Mix (Invitrogen, Carlsbad, CA, USA) according to the protocol suggested by the manufacturer. The RT-PCR amplification process was performed at initial denaturation for 5 min at 95 °C and then 40 cycles of 95 °C for 15 s, 56–61 °C for 15 s, and 72 °C for 5 s. For RT-qPCR amplification, the initial denaturation was for 10 min at 95 °C, 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. GAPDH (RT-PCR) and β-actin (RT-qPCR) were utilized as internal controls [26,27]. The primer sequences used are shown in Table 2. Target mRNAs/transcript levels of HGK/HGF under normoxia (18% O2) at 24 h; hypoxia (1% O2) at 24 h; and µg/mL E. coli LPS challenge under normoxia at 24 h were determined with reference to the internal control (β-actin). Then, the corresponding relative target transcript levels under various test conditions were normalized against that of the same cell type under normoxia at 24 h.

Table 2.
List of primer sequences and expected product size for TLR and NLR detection 1.
Except HGT biopsies, all cells were repeatedly cultured, treated and RNA harvested three times for the RT-PCR and RT-qPCR experiments. Randomly selected positive amplicon samples, representative of all ten DAMP sensors of interest, were sequenced to confirm the identity of the related cDNA concerned. This was repeated systematically in all three independent repetitive experiments.
2.6. Western Blot
Total protein was collected from HGK and HGF with or without treatment of E. coli LPS and/or hypoxia as described under Section 2.3 (Table 1). The cells were then incubated with lysing buffer (2 μL/mL protease inhibitor (P8340, Sigma-Aldrich, St. Louis, MO, USA) in 1% Triton-X100-150 mM NaCl-20 mM Tris HCl, pH 7.4) on ice for 10 min, followed by centrifugation at 12,000× g at 4 °C for 30 min. The supernatant was collected, then the protein concentration was measured by Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). Fifty micrograms of cellular proteins extract were separated by 10% sodium dodecyl sulphate polyacrylamide gel electrophoresis followed by electro-transferal to polyvinylidene difluoride membrane (Immobilon, Sigma-Aldrich, St. Louis, MO, USA). After blocking by 5% non-fat milk at r.t. for 30 min, primary antibodies against TLR1/2/4/5/6, NOD1/2 or NLRP1/2 (mouse anti-human: TLR1, TLR4 and NLRP2 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), TLR2 (Imgenex, San Diego, CA, USA), TLR5 (Abnova, Taipei, Taiwan), rabbit anti-human: TLR6, NLRP1 and NOD2 (Thermo Fischer Scientific, Waltham, MA, USA), NOD1 (R&D systems, Inc., Minneapolis, MO, USA); at antibody concentration: TLR1/2/4, NOD1/2 and NLRP1/2: 1:500, or TLR5/6: 1:2000) in blocking buffer (5% non-fat milk in 1% Tween 20-Tris-buffered saline, pH 7.6) was added to the blot at 4 °C, overnight. The membranes were then washed by 1% Tween 20–20 mM Tris-buffered saline (pH = 7.6) r.t., 3×, 5 min, and then incubated with the corresponding secondary antibody (1:2000, Pierce goat anti-mouse/rabbit IgG, Thermo Fisher Scientific, Waltham, MA, USA) at r.t. and visualized by an enhanced chemiluminescence system (ECL) (GE healthcare, Lifescience, Marlborough, MA, USA) at r.t. via undisturbed immersing in 10 mL ECL buffer, 22 µL p-coumaric acid, 50 µL luminol and 3 µL H2O2 for 90 s [26]. β-actin (primary antibody, mouse anti-human, Thermo Fisher Scientific, Waltham, MA, USA, at 1:500, in 5% non-fat milk in 1% Tween 20-Tris-buffered saline, pH 7.6) was used as an internal control. HGK/HGF TLRs or NLRs blot densities from cells incubated under normoxia for 24 h were normalized with reference to β-actin controls, followed by semi-quantitative determination of the corresponding DAMP sensor from the same cell type after LPS or hypoxia treatment [29].
Considering the fact that the preliminary experiments showed nil detection of TLR3/7/8/10 in immunohistochemistry (c.f. Section 3.1) as well as nil TLR9 mRNA detection from HGT/HGK/HGF (c.f. Section 3.4), the presence of these proteins in HGK/HGF via Western blotting was not investigated. Independent experiments were repeated three times on separate occasions, starting with cell explants at the third passage.
2.7. Statistical Analysis
The primary outcomes of this study included the following: the location, distribution and relative quantities of the tested DAMP sensors in healthy or periodontally diseased tissue; and transcript expression of the former in healthy periodontal tissue and resident cell explants. Target DAMP sensors signal and protein expression levels from periodontal resident cells upon hypoxia and/or 2 µg/mL E. coli LPS challenge were also measured. Healthy periodontal tissue or parameters from untreated periodontal resident cells in normoxia were the control/reference.
Statistical analysis was performed using SPSS 20 (IBM Corporation, New York, NY, USA). The data were tested for normality using the Kolmogorow–Smirnoff test. Normally distributed variables were reported as mean ± SD, while medians were used to describe non-normally distributed data. Data that conformed to a normal distribution were analyzed by t-test or one-way analysis of variance (ANOVA). Data that showed considerable deviation from the normal distribution were analyzed by the Wilcoxon test. p < 0.05 was considered statistically significant.
3. Results
In total, 40 periodontal healthy patients (age 14–29 year, 25 females) and 33 chronic periodontitis patients (age 28–46, 20 females) were recruited and consented to donate periodontal tissue. All patients were systemically healthy non-smokers. The PPD of the biopsied sites was 2.3 ± 0.5 mm for healthy gingiva and 6.9 ± 0.8 mm for periodontitis pockets. PAL for biopsied periodontitis sites was 5.6 ± 0.4 mm. A number of tissue blocks (from healthy/periodontitis donors) and explants (from periodontally healthy donors) were used for TLR 3/7/8/10 immunohistochemistry or various in vitro or preliminary investigations, the data for which are not presented.
Thirteen healthy gingival and seventeen periodontitis biopsies were fixed and sectioned for immunohistopathology analysis; 27 healthy gingival biopsies were processed for HGK and HGF culture and conventional RT-PCR; and 16 healthy gingival biopsies contributed to HGT mRNA extraction (Table 1). HGK or HGF cultured on 1% or 18% O2 with or without 2 µg/mL E. coli LPS challenge showed normal cellular morphology.
3.1. Selected TLRs and NLRs Localization in Healthy Gingival and Periodontitis Biopsies
The presence and location of selected TLRs and NLRs in healthy gingival and periodontitis tissue sections were detected by immunohistochemistry (Figure 1), and semi-quantitatively, the percentage proportion of positive stain per unit section area are shown in Table 3. TLR1, TLR2, TLR4, TLR5, TLR6, NOD1, NOD2, NLRP1 and NLRP2 were detectable in both healthy gingiva and periodontitis tissues. TLR3/7/8/10 was not detectable on any section of the specimens collected (data not shown), while TLR3 was readily detectable in sections of human hepatocellular carcinoma (data not shown). TLR9 was not tested since no RNA messenger could be detected in HGK/HGF/HGT.
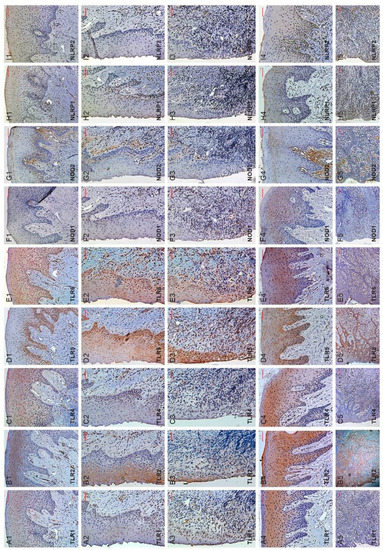
Figure 1.
TLR1/2/4/5/6, NOD1/2 or NLRP1/2 immunohistochemical detection in healthy gingival (rows 1–3) or periodontitis tissue (rows 4–5) biopsies. Specific damage-associated molecular patterns are detected as brown DAB stain. Scale bars = 100 µm. Column A-I showed micrographs of TLR1/2/4/5/6, NOD1/2 or NLRP1/2 detection, respectively. Rows 1–5 are: healthy oral epithelium (H-OE), oral sulcular epithelium (OSE), junctional epithelium (JE), periodontitis oral epithelium (P-OE) and pocket epithelium (PE), respectively. Shown are representative micrographs from healthy human gingival biopsies (n = 13) and periodontitis biopsies (n = 17) with staining from three independent experiments. TLR3/7/8/9/10 immuno-detection was attempted but had negative staining results (data not shown).

Table 3.
Percentage proportions of Toll-like and NOD-like receptor-positive staining areas in periodontal tissue biopsies 1.
TLR1, TLR2, TLR4, TLR5, TLR6, NOD1, NOD2, NLRP1 and NLRP2 appeared detectable by their corresponding antibodies both in the epithelial and connective tissue compartments of the biopsy sections. Positive DAB staining, representing TLR or NLRP detection, were mainly congregated around basal (TLR4/5/6, NOD1/2 or NLRP1/2; Figure 1(C1–C3,D1–D3,E1–E3,F1–F3,G1–G3,H1–H3,I1–I3)) and/or spinous (TLR1/2/4/6; Figure 1(A1–A3,B1–B3,C1–C3,E1–E3)) layers of healthy gingival (oral or H-OE; oral sulcular or OSE; junctional or JE) epithelium. Nevertheless, overall expressions of TLR1, NOD1/2 or NLRP1/2 appeared relatively low in healthy gingival epithelium (Figure 1(A1–A3,F1–F3,G1–G3,H1–H3,I1–I3)). Concerning sections from periodontitis biopsies, all pocket epithelium (PE) were positive with TLR1, TLR2, TLR4, TLR5, TLR6, NOD1, NOD2, NLRP1 and NLRP2 staining. However, TLR2, TLR4, TLR5, TLR6 staining appeared stronger at PE.
Regarding detection of respective DAMP sensor at sub-sulcular/pocket epithelial connective tissue, positive DAB staining indicated TLR1, TLR2, TLR4, TLR5, TLR6, NOD1, NOD2, NLRP1 and NLRP2 (Figure 1(A5–I5)) appeared similar and at times, stronger in periodontitis than healthy biopsies. The more intensely stained areas apparently were beneath the pocket epithelium with many infiltrated cells expressing the DAMP sensors of interest.
The semi-quantitative data concerning detection of DAMP sensors of interest are shown in Table 3. In brief, expression of TLR1/2/5 or NOD1 in healthy or periodontitis tissue sections showed the TLRs of concern on H-OE (former) and appeared significantly lower than that of P-OE (latter, p < 0.01). On the other hand, expressions of TLR2/4, or NOD1/2 on PE in periodontitis tissue sections seemed significantly higher than those on JE of healthy tissue sections (p < 0.05) (Table 3). The expressions of all TLRs and NLRs of interest in subepithelial connective tissue of periodontitis sections appeared higher than that of the subepithelial connective tissues from sections of healthy periodontal biopsies.
3.2. Culture and Isolation of HGK and HGF
HGK and HGF were successfully isolated from twenty-seven biopsy specimens (Table 1). In vitro experiments were conducted with specific details of the source cells meticulously recorded. At the end of the experiments no unusual observation was recorded, indicating, perhaps, minimal variations concerning the HGK/HGF explants, despite their varied host origins.
3.3. MTT Assay
MTT assay was conducted using early confluent HGK or HGF culture from randomly selected nine donors (Table 1) to test the viability of the cells. These assays indicated that under 1% O2 at 24 h, the tetrazolium dye reduction ability of HGF appeared unchanged (absorbance at 570 nm; HGK/HGF, 1% vs. 18% O2: 0.157 ± 0.060/0.862 ± 0.075 vs. 0.137 ± 0.059/0.746 ± 0.105, p = 0.08/0.08; representative data set from one of three experiments). Regarding MTT assays of HGK/HGF under LPS stimulation at normoxia, the results observed were similar to the previous report from this group (data not shown) [26,27].
3.4. Selected DAMP Sensors mRNA Detection in HGT, HGK, or HGF
Messenger RNAs encoding TLR1, TLR2, TLR3, TLR5, TLR6, NOD1, NOD2, or NLRP1 were detectable among all HGT biopsies, HGK, and HGF cell explants investigated (Table 1). TLR4 or NLRP2 mRNA were not detected in every cell sample, with TLR4 being detectable in only 12/27 HGK (44.4% healthy gingival keratinocyte explants), or NLRP2 in only 5/27 HGF (18.5% healthy gingival fibroblasts explants), respectively. However, TLR4 expression in HGF and NLRP2 expression in HGK were observable in all samples followed. TLR7, TLR8 or TLR10 mRNAs were detectable among all HGT biopsies but were not detectable in any of the HGK or HGF explants. TLR9 mRNA was undetectable in HGT, HGK, or HGF (Figure 2).
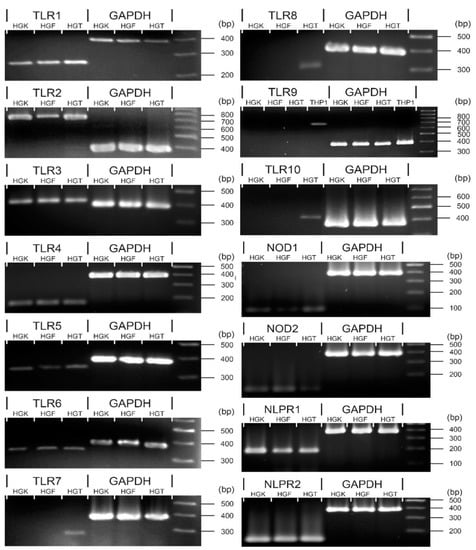
Figure 2.
Selected DAMP sensors mRNA detection from HGT, or HGK/HGF primary cultures. By reverse transcription, PCR, TLR1-6, NOD1/2 and NLRP1/2 were detectable in HGT, HGK, and HGF with TLR4 or NLRP2 detectable in some but not all samples investigated (please refer to text and Table 4 for details). TLR7/8/10 mRNAs were only detectable from HGTs. THP-1 cell line was utilized as the positive control for TLR9 mRNA detection. HGF—human gingival fibroblasts, HGK—human gingival keratinocytes, HGT—human gingival tissue.
3.5. Quantities of Selected DAMP Sensor Transcripts from HGK or HGF under Hypoxia or E. coli LPS Stimulation
The quantity of transcripts expression of DAMP sensors detectable by RT-qPCR from HGK/HGF explants, i.e., TLR1-6, NOD1, NOD2, NLRP1, or NLRP2 (Figure 2), under 1% or 18% O2, 24 h, or 2 µg/mL E. coli LPS under normoxia at 24 h, was followed (Table 4).

Table 4.
Expression level of damage-associated molecular patterns of interest upon hypoxia or 2 µg/mL E. coli lipopolysaccharide (LPS), 24 h stimulation 1.
Except TLR4 and NLRP2, the selected DAMP sensor mRNAs of interest were detectable from all HGK or HGF explants at both oxygen tensions tested at 24 h. The former, even if detectable from some cell explant samples, was persistently at low relative quantities in HGK (TLR4) or HGF (NLRP2), while the corresponding TLR4 mRNA in HGF or NLRP2 mRNA in HGK relative expression levels remained similar to other DAMP sensors investigated (Table 4). Under 2 µg/mL E. coli LPS at normoxia at 24 h, all ten DAMP sensor mRNAs of interest were detectable (Table 4).
Apparently, under 1% O2, 24 h, mRNA expression of TLR1-3, TLR6, NOD1/2 and NLRP1/2 in HGK were significantly increased at up to 4.6-fold for NOD2, while concerning HGF, TLR1-5, NOD1 and NLRP1 mRNAs seemed to be increased in hypoxia, up to 5.1-fold for TLR2 (Table 4). With the presence of 2 µg/mL E. coli LPS at normoxia at 24 h, only TLR2 and NOD2 expressions on HGK appeared significantly increased at up to 4.0 times for NOD2, compared to untreated controls. For HGF, the same LPS treatment seemed able to upregulate signals for TLR1, TLR2, TLR6 and NOD2, at up to 6.3-fold for NOD2 (Table 4).
3.6. Levels of Selected DAMP Sensor Proteins from HGK or HGF under Hypoxia and/or E. coli LPS Stimulation
DAMP sensor proteins observable from immunohistochemistry (Figure 1) were followed in HGK/HGF explants in vitro. Both cells were also subjected to 24 h 1% O2 and/or 2 µg/mL E. coli LPS treatment. In brief, TLR1-6, NOD1/2 or NLRP1/2 appeared readily detectable on both cell explants in hypoxia and/or E. coli LPS challenge (Figure 3A). Semi-quantitative analysis of DAMP sensors under various experimental conditions were also reported (Figure 3B,C). In brief, TLR1, TLR6 and NOD2 proteins appeared significantly increased after HGF treated by 1% O2 and/or 2 µg/mL E. coli LPS at 24 h (Figure 3C), while the same appeared observable for TLR6 only on HGK cells (Figure 3B). For the latter, TLR1 or NOD2 peptide appeared significantly increased upon 1% O2 or 1% O2 and 2 µg/mL E. coli LPS at 24 h treatment, respectively (Figure 3B).
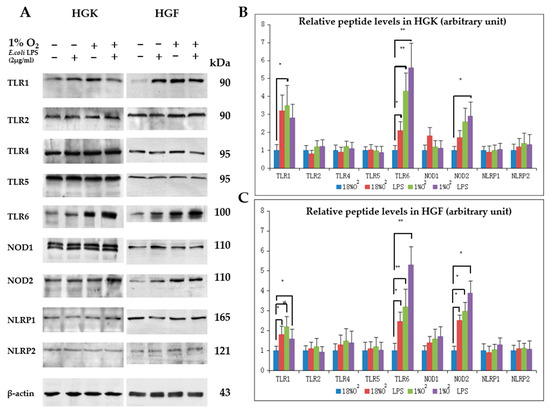
Figure 3.
Selected damage-associated molecular pattern (DAMP) sensors expression in human gingival keratinocytes (HGK) or human gingival fibroblasts (HGF) explants under 1% O2 and/or 2 µg/mL Escherichia coli LPS, 24 h. (A) Detection of TLR1, TLR2, TLR4-6, NOD1/2, NLRP1/2 from HGK/HGF cell lysates after various treatments. Shown are representative blots from three independent experiments; (B) relative HGK TLR1, TLR2, TLR4-6, NOD1/2, NLRP1/2 protein levels after various treatments; (C) relative HGF TLR1, TLR2, TLR4-6, NOD1/2, NLRP1/2 protein levels after various treatments. * p < 0.05, ** p < 0.01, one-way ANOVA with adjustments against multiple comparisons within same DAMP sensor group. Cells at passage three were used and the experiments were independently repeated three times.
4. Discussion
Periodontal resident cells utilize various defense strategies to address challenges from environmental stimuli, such as periodontopathogens [18]. Toll-like and NOD-like receptors expressing gingival resident cells could play key roles at the initial phase of innate immune responses. Many studies have investigated the expressions of these receptors in human cells and tissues [30]; however, limited studies are available reporting the expressions of such receptors in the periodontal cells and tissues in vivo [10,13].
To the best knowledge of this group, this is one of the more comprehensive reports characterizing expressions of TLRs and selected NLRs on periodontal tissue biopsies and resident cells. The current in vitro observations demonstrated that transcripts of TLR1-6, NOD1/2 and NLRP1/2 are observable in the latter. Similarly, the key proteins, i.e., TLR1/2/4/5/6, NOD1/2 and NLRP1/2, were also detected in periodontal biopsies, and HGK, and HGF with TLR2/4 and NOD1/2 appeared more readily detectable in the pocket vs. junctional epithelium in periodontal biopsies. All DAMP sensors of interest, however, were significantly enhanced in periodontal pocket connective tissue, where heavy inflammatory cells infiltrate could be readily observed.
Regarding the role of NOD-like receptors in periodontitis, the most thoroughly studied molecules are NOD1/2 [9], while not many reports are available for other NLRs. Contrary to NOD1, which is reported to be expressed almost in all cell types, NOD2 is found to be expressed in a rather restricted manner, mainly limited to macrophages, dendritic cells, epithelial cells in the intestine or oral cavity, etc. [31]. In the current study, the expressions of these four receptors were reported both in biopsies and resident cells in vitro. A moderate but enhanced stained area and the NOD1/2 intensity in the periodontitis pocket epithelium and subepithelial connective tissues, compared to that of the junctional epithelium and submucosa in healthy biopsy sections indicated their potential roles in the innate immune defense at the periodontal front. This study confirmed the existence of NOD1/2 and NLRP1/2 expressions in periodontal biopsies for either healthy or periodontitis patients.
Next, transcriptional signals for TLRs and NLRs under the influence of hypoxia or E. coli LPS were investigated. Hypoxia appeared to be associated with the upregulation of TLR2/6 transcripts and proteins in human dendritic, monocytic, endothelial and intestinal epithelial cells [24]. In this study, hypoxia associated with enhanced TLR2, NOD2 and NLRP1 transcripts but reduced TLR1/3/6 NOD1, and NLRP2 transcripts detection in HGK were observable. Upon stimulation by E. coli LPS, the upregulation of TLR2 and NOD2 only in the same cell was observed (Table 4). For HGF, hypoxia appeared to be associated more with transcript detection concerning TLR1/2/4, NOD1 and NLRP1, while that of TLR3/5 was reduced. Putting HGF under E. coli LPS, only enhanced transcript detection of TLR1/2/6 and NOD2 was observed. Such observations lined up nicely with the in vitro DAMP sensor protein expression of HGF when increased TLR1/6 and NOD2 protein were reported under hypoxia and/or E. coli LPS challenge (Figure 3C). However, this investigation also observed the unstable expression of TLR4 in HGK and NLRP2 in HGF both at normoxia and hypoxia; the authors postulated that TLR4 expression remains at a low level in healthy epithelium [26], and so it is the same with NLRP2 expression in HGF. Since it is the first report concerning NLRP2 expression in gingival cells [32], further investigation is needed.
The current study results perhaps implied the potential roles of TLR1/2/6 and NOD2 in periodontal resident cells immune responses against periodontal infection. According to the previous research studies, TLR1/2/6 is known to be activated by various ligands, such as bacteria, fungi, virus and certain endogenous substances [11], while NOD2 is reported to be a universal sensor for peptidoglycans from Gram-positive and muramyl dipeptide moiety from Gram-negative bacteria. Taking the above together, the authors postulate that TLR1/2/6 and NOD2 are potential potent receptors at the dento-gingival front responsible for periodontal innate immunity.
It is regularly believed that specific receptors might be preferentially expressed upon stimulation of corresponding target ligands, e.g., LPS for TLR4 [6]. However, in the current study, HGK or HGF TLR4 transcript and protein expression did not appear to be significantly influenced/changed under LPS stimulation. Only in 46%/38% HGK explants under normoxia/hypoxia could TLR4 transcript be detected, implying either an inherited low cellular transcription level in periodontal resident cells or perhaps that a specific yet tightly regulated mechanism is in effect [26,33]. Considering the fact that previous reports regarding the expression of TLR4 upon LPS stimulation remained ambiguous [34], the present group postulated that the diverse observation concerning TLR4 expression under LPS/hypoxia stimulation in this study was actually in line with earlier reports. There are reports describing the downregulated expression or cellular tolerance via reduced cell surface TLR4 expression under LPS stimulation [35]. However, the molecular mechanisms of endotoxin tolerance remained yet unclear. During tolerance, TLR4 is transiently suppressed or unchanged, with proximal post-receptor signaling proteins also altered, such as interleukin-1 receptor-associated kinase (IRAK), TLR4-myeloid differentiation factor 88 (MyD88) and IRAK-MyD88 association [36]. Therefore, it is hypothesized that gingival tissue under repeated exposure to Gram-negative periodontopathogens that carries LPS may lead to host cell tolerance; therefore, low expressions of TLR4 were observed in the experiment reported. This may also aid in explaining the rare distribution of TLR4 in the junctional/pocket epithelium and even the lack of expression in certain donors’ gingival primary cell culture. Previous studies have shown that TLR4 is an important receptor involved in periodontitis immune defense. However, these studies relied heavily on the use of human oral keratinocyte from oral mucosa instead of that from gingival tissue origin [37]. It is hypothesized that perhaps the expressions of TLR4 could also be location-specific or cellular type- and physiological status-dependent. Taking all information in consideration, this research team postulated that DAMP sensor regulation, such as TLR4, may not operate merely under a direct feedback mechanism, implying that various factors, such as cell types, environmental stimuli, and duration/persistence of ligand exposure, may also be involved [34,38,39]. The mechanism modulating the expression of TLR4 at resident cells at the dento-gingival junction, therefore, warrants further investigations.
On the other hand, it is indicated E. coli LPS under hypoxia-stimulated HGF, HGK TLR1/6 and NOD2 protein expression (to a certain extent), and TLR2 and NOD2 transcripts’ upregulation were readily observed under LPS challenge alone in this in vitro study. It is postulated that for HGK/HGF, TLR1/2/6 and NOD2 instead of TLR4 could be the more important/relevant DAMP sensors against E. coli LPS. TLR1/2 and TLR2/6 are not considered typical oral/periodontal LPS receptors compared to TLR4, reasons being that the former could bind to various ligands, including lipopeptides, glycolipids, fungi, virus and certain endogenous substances [40]. However, in reports concerning intestinal epithelium, TLR2 expression was reported to be relatively upregulated in intestinal epithelial cell lines upon LPS stimulation, and the same was reported in LPS-treated adipocytes [41]. One point worth noticing, however, is that LPS with relatively low endotoxic activity, such as those from Bacteroides fragilis, Chlamydia trachomatis and Pseudomonas aeruginosa, commonly transduces signals via TLR2 instead of TLR4 [4]. Highly purified Helicobacter pylori LPS also induce weak inflammatory reactions and utilize the TLR2, but not the TLR4, pathway [42].
In this research, it was noticed that TLR4/5, NOD1 and NLRP1/2 appeared constitutively expressed, especially in immunohistochemistry staining, and remained more or less unaffected by low oxygen tension and/or E. coli LPS, both at transcriptional and peptide levels. Therefore, the authors postulated that these receptors may be persistently expressed by periodontal resident cells or they are regulated by other DAMPs, such as those from Eubacterium saphenum, Eubacterium nodatum and Filifactor alocis other than low oxygen tension and/or E. coli LPS [43,44]. Interestingly, previous reports held different opinions on the regulation of Porphyromonas gingivalis on the expression of inflammasomes, such as NLRP1, and some papers described the activation of inflammasome members via P. gingivalis stimulation [32], while others published the opposite result that P. gingivalis inhibited inflammasome-involved immune responses [45]. However, both reports indicated that IL-1 plays an important role in the arena of inflammasome complex [32,45].
The current study utilized E. coli instead of P. gingivalis LPS. E. coli LPS was reported to be more readily recognized by TLR4 and TLR2 compared to P. gingivalis LPS [46], and the former could pose further influence on downstream gene expression, such as CXC chemokine ligand 5 [47]. More importantly, E. coli LPS could induce stronger cellular expressions compared to that of P. gingivalis [47]. Obviously, E. coli are less often detected in human mouths, unless associated with recurrent aphthous stomatitis [48], while P. gingivalis is an established keystone periodontopathogen [49]. Therefore, in theory, E. coli LPS could influence periodontal resident cells differently compared to P. gingivalis LPS, and the latter could facilitate experiments more accurately, mimicking the pathogenesis of periodontitis. This group hypothesized that perhaps HGF/HGK TLR1/2 or TLR2/6 heterodimeric complexes were triggered more readily instead of TLR4 in response to E. coli LPS stimulation. Further in-depth studies, including experiments utilizing LPS from periodontopathogens, are warranted to clarify the current observations.
In general, hypoxia and E. coli LPS conferred somehow similar effects on HGK/HGF expression of DAMP sensors of interests (Table 4, Figure 3). However, there are limited reports documenting whether hypoxia and LPS act on periodontal resident cells independently or in any synergistic fashion [50,51]. Further studies are, therefore, needed to decipher if hypoxia and LPS, the two elements likely to coexist in periodontal inflammation/infection, could influence each other.
Although this is a comprehensive research study investigating the expressions of TLR and selected NLR families in periodontal tissues/cells in health or disease, or in vitro, under hypoxia and/or LPS stimulation, still, it is a comparatively preliminary report of this kind. Therefore, further studies with a more elaborate design, larger sample size, etc., are warranted. For instance, all 23 members of NLRs in human periodontium in health or disease may need to be screened in further studies. Moreover, the mechanism of hypoxia upon the expressions of DAMP sensors on periodontal cells, especially the biology relating the former and expression of hypoxia inducible factor family in periodontitis, may need to be further elucidated.
Taken together, many DAMP sensors were observed to express (or not express) on healthy or diseased periodontal tissues, with HGK/HGF TLR1/2/4/6 and NOD2 appearing to be important, innate DAMP sensors, especially under hypoxia and/or LPS stimulation. This study took a small initial step toward LPS-DAMP sensor interactions under hypoxia, the anaerobic condition resembling periodontopathogenesis. Further investigations are, however, needed to explore these issues.
5. Conclusions
In summary, this study examined the expressions of TLR and selected NOD families of DAMP sensors on periodontal resident cells. In particular, a relatively comprehensive study of various DAMP sensors in healthy or diseased human periodontal tissues was conducted. The in vitro study indicated that TLR1/2/4/6 and NOD2 could somehow be stimulated by hypoxia and E. coli LPS, while TLR4/5, NOD1 and NLRP1/2 appeared constitutively expressed and remained more or less unaffected by low oxygen tension and/or E. coli LPS. Preliminary observations of this study implied that expressions of the DAMP sensors of interests appeared to be results of complex regulatory mechanisms, while certain DAMP sensor expressions were favored over others. On the whole, TLR1/2/4/6 and NOD2 appear to play important roles in HGK/HGF innate immune responses against hypoxia and LPS stimulation and may potentially be relevant in host defense against periodontal disease pathogenesis. Considering the preliminary nature of the current in vitro study, further research studies are needed to verify the current observations.
Author Contributions
Conceptualization, Y.C., S.W.T. and W.K.L.; methodology, Y.C., X.X.W. and C.H.C.N.; software, Y.C.; validation, W.K.L.; formal analysis, Y.C. and W.K.L.; investigation, Y.C.; resources, Y.C., S.W.T. and W.K.L.; data curation, Y.C.; writing—original draft preparation, Y.C.; writing—review and editing, Y.C. and W.K.L.; visualization, Y.C.; supervision, S.W.T. and W.K.L.; project administration, W.K.L.; funding acquisition, W.K.L. All authors have read and agreed to the published version of the manuscript.
Funding
The work described in this paper was substantially supported by grants from the Research Council of Hong Kong Special Administrative Region, China (HKU 17113114 and 17116819) and Small Project Funding, University Research Committee, Committee on Research and Conference Grants, The University of Hong Kong (201109176129 and 201309176135).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster (HKU/HA HKW IRB; UW 06-376 T/1401, 14 December 2006).
Informed Consent Statement
Written informed consent was obtained from all participants or their guardians involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available according to Cap. 486. Personal Data (Privacy) Ordinance, Hong Kong SAR Government stipulated by HKU/HA HKW IRB, which demands non-disclosure of any personal data of research project participants.
Acknowledgments
The authors thank Nancy K. Man, Department of Surgery, LKS Faculty of Medicine, the University of Hong Kong for providing us with sections of human hepatocellular carcinoma biopsy for TLR3 immunohistochemistry positive control. Thanks support (to Y.C.) from People’s Hospital of Longhua, Affiliated Hospital of Guangdong Medical University, Shenzhen 518109, China.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Flemmig, T.F. Periodontitis. Ann. Periodontol. 1999, 4, 32–37. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef]
- Teng, Y.-T.A. The role of acquired immunity and periodontal disease progression. Crit. Rev. Oral Biol. Med. 2003, 14, 237–252. [Google Scholar] [CrossRef]
- Erridge, C.; Pridmore, A.; Eley, A.; Stewart, J.; Poxton, I.R. Lipopolysaccharides of Bacteroides fragilis, Chlamydia trachomatis and Pseudomonas aeruginosa signal via toll-like receptor 2. J. Med. Microbiol. 2004, 53, 735–740. [Google Scholar] [CrossRef]
- Murakami, A.; Ohigashi, H. Targeting NOX, INOS and COX-2 in inflammatory cells: Chemoprevention using food phytochemicals. Int. J. Cancer 2007, 121, 2357–2363. [Google Scholar] [CrossRef]
- Rhee, S.H.; Hwang, D. Murine TOLL-like receptor 4 confers lipopolysaccharide responsiveness as determined by activation of NFκB and expression of the inducible cyclooxygenase. J. Biol. Chem. 2000, 275, 34035–34040. [Google Scholar] [CrossRef]
- Esmon, C.T. Regulation of blood coagulation. Biochim. Biophys. Acta 2000, 1477, 349–360. [Google Scholar] [CrossRef]
- Trinchieri, G.; Sher, A. Cooperation of Toll-like receptor signals in innate immune defence. Nat. Rev. Immunol. 2007, 7, 179–190. [Google Scholar] [CrossRef]
- Kanneganti, T.-D.; Lamkanfi, M.; Núñez, G. Intracellular NOD-like receptors in host defense and disease. Immunity 2007, 27, 549–559. [Google Scholar] [CrossRef]
- Sugawara, Y.; Uehara, A.; Fujimoto, Y.; Kusumoto, S.; Fukase, K.; Shibata, K.; Sugawara, S.; Sasano, T.; Takada, H. Toll-like receptors, NOD1, and NOD2 in oral epithelial cells. J. Dent. Res. 2006, 85, 524–529. [Google Scholar] [CrossRef]
- Lien, E.; Sellati, T.J.; Yoshimura, A.; Flo, T.H.; Rawadi, G.; Finberg, R.W.; Carroll, J.D.; Espevik, T.; Ingalls, R.R.; Radolf, J.D.; et al. Toll-like receptor 2 functions as a pattern recognition receptor for diverse bacterial products. J. Biol. Chem. 1999, 274, 33419–33425. [Google Scholar] [CrossRef] [PubMed]
- Farhat, K.; Riekenberg, S.; Heine, H.; Debarry, J.; Lang, R.; Mages, J.; Buwitt-Beckmann, U.; Röschmann, K.; Jung, G.; Wiesmüller, K.H.; et al. Heterodimerization of TLR2 with TLR1 or TLR6 expands the ligand spectrum but does not lead to differential signaling. J. Leuko. Biol. 2008, 83, 692–701. [Google Scholar] [CrossRef]
- Beklen, A.; Hukkanen, M.; Richardson, R.; Konttinen, Y.T. Immunohistochemical localization of Toll-like receptors 1–10 in periodontitis. Oral Microbiol. Immunol. 2008, 23, 425–431. [Google Scholar] [CrossRef]
- Zinkernagel, A.S.; Johnson, R.S.; Nizet, V. Hypoxia inducible factor (HIF) function in innate immunity and infection. J. Mol. Med. 2007, 85, 1339–1346. [Google Scholar] [CrossRef]
- Schaible, B.; Schaffer, K.; Taylor, C.T. Hypoxia, innate immunity and infection in the lung. Respir. Physiol. Neurobiol. 2010, 174, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Hanioka, T.; Takaya, K.; Shizukuishi, S. Association of oxygen tension in human periodontal pockets with gingival inflammation. J. Periodontol. 1998, 69, 1127–1130. [Google Scholar] [CrossRef] [PubMed]
- Hanioka, T.; Tanaka, M.; Takaya, K.; Matsumori, Y.; Shizukuishi, S. Pocket oxygen tension in smokers and non-smokers with periodontal disease. J. Periodontol. 2000, 71, 550–554. [Google Scholar] [CrossRef]
- Wang, X.; Chen, Y.; Leung, W.K. Role of the hypoxia-inducible factor in periodontal inflammation. In Hypoxia and Human Diseases; Zheng, J., Zhou, C., Eds.; IntechOpen Ltd.: Rijeka, Croatia, 2017; Chapter 15; pp. 285–302. [Google Scholar] [CrossRef]
- Greijer, A.; van der Groep, P.; Kemming, D.; Shvarts, A.; Semenza, G.L.; Meijer, G.A.; van de Wiel, M.A.; Belien, J.A.; van Diest, P.J.; van der Wall, E. Up-regulation of gene expression by hypoxia is mediated predominantly by hypoxia-inducible factor 1 (HIF-1). J. Pathol. 2005, 206, 291–304. [Google Scholar] [CrossRef]
- Cramer, T.; Yamanishi, Y.; Clausen, B.E.; Förster, I.; Pawlinski, R.; Mackman, N.; Haase, V.H.; Jaenisch, R.; Corr, M.; Nizet, V.; et al. HIF-1α is essential for myeloid cell-mediated inflammation. Cell 2003, 112, 645–657. [Google Scholar] [CrossRef]
- Grant, M.M.; Kolamunne, R.T.; Lock, F.E.; Matthews, J.B.; Chapple, I.L.; Griffiths, H.R. Oxygen tension modulates the cytokine response of oral epithelium to periodontal bacteria. J. Clin. Periodontol. 2010, 37, 1039–1048. [Google Scholar] [CrossRef]
- Kim, S.Y.; Choi, Y.J.; Joung, S.M.; Lee, B.H.; Jung, Y.S.; Lee, J.Y. Hypoxic stress up-regulates the expression of Toll-like receptor 4 in macrophages via hypoxia-inducible factor. Immunology 2010, 129, 516–524. [Google Scholar] [CrossRef] [PubMed]
- Stridh, L.; Smith, P.L.; Naylor, A.S.; Wang, X.; Mallard, C. Regulation of toll-like receptor 1 and-2 in neonatal mice brains after hypoxia-ischemia. J. Neuroinflamm. 2011, 8, 45. [Google Scholar] [CrossRef]
- Kuhlicke, J.; Frick, J.S.; Morote-Garcia, J.C.; Rosenberger, P.; Eltzschig, H.K. Hypoxia inducible factor (HIF)-1 coordinates induction of Toll-like receptors TLR2 and TLR6 during hypoxia. PLoS ONE 2007, 2, e1364. [Google Scholar] [CrossRef]
- Braza, F.; Brouard, S.; Chadban, S.; Goldstein, D.R. Role of TLRs and DAMPs in allograft inflammation and transplant outcomes. Nat. Rev. Nephrol. 2016, 12, 281–290. [Google Scholar] [CrossRef]
- Li, J.-P.; Chen, Y.; Ng, C.H.C.; Fung, M.L.; Xu, A.; Cheng, B.; Tsao, S.W.; Leung, W.K. Differential expression of Toll-like receptor 4 in healthy and diseased human gingiva. J. Periodontal Res. 2014, 49, 845–854. [Google Scholar] [CrossRef]
- Li, J.-P.; Li, F.Y.L.; Xu, A.; Cheng, B.; Tsao, S.W.; Fung, M.L.; Leung, W.K. Lipopolysaccharide and hypoxia-induced HIF-1 activation in human gingival fibroblasts. J. Periodontol. 2012, 83, 816–824. [Google Scholar] [CrossRef]
- Leung, W.K.; Wu, Q.; Hannam, P.M.; McBride, B.C.; Uitto, V.-J. Treponema denticola may stimulate both epithelial proliferation and apoptosis through MAP kinase signal pathways. J. Periodontal Res. 2002, 37, 445–455. [Google Scholar] [CrossRef]
- Ng, K.-T.; Li, J.-P.; Ng, K.M.; Tipoe, G.L.; Leung, W.K.; Fung, M.L. Expression of hypoxia-inducible factor-1α in human periodontal tissue. J Periodontol. 2011, 82, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Kawai, T.; Taubman, M.A. Toll-like receptor signaling in B cell-mediated RANKL-dependent periodontitis bone resorption. In Interface Oral Health Science 2011, Proceedings of the 4th International Symposium for Interface Oral Health Science; Sasaki, K., Suzuki, O., Takahashi, N., Eds.; Springer: New York, NY, USA, 2012; pp. 373–375. [Google Scholar] [CrossRef]
- Uehara, A.; Sugawara, Y.; Kurata, S.; Fujimoto, Y.; Fukase, K.; Kusumoto, S.; Satta, Y.; Sasano, T.; Sugawara, S.; Takada, H. Chemically synthesized pathogen-associated molecular patterns increase the expression of peptidoglycan recognition proteins via toll-like receptors, NOD1 and NOD2 in human oral epithelial cells. Cell Microbiol. 2005, 7, 675–686. [Google Scholar] [CrossRef]
- Aral, K.; Milward, M.R.; Kapila, Y.; Berdeli, A.; Cooper, P.R. Inflammasomes and their regulation in periodontal disease: A review. J. Periodontal Res. 2020, 55, 473–487. [Google Scholar] [CrossRef]
- McFarlin, B.K.; Flynn, M.G.; Campbell, W.W.; Stewart, L.K.; Timmerman, K.L. TLR4 is lower in resistance-trained older women and related to inflammatory cytokines. Med. Sci. Sports Exerc. 2004, 36, 1876–1883. [Google Scholar] [CrossRef] [PubMed]
- De Creus, A.; Abe, M.; Lau, A.H.; Hackstein, H.; Raimondi, G.; Thomson, A.W. Low TLR4 expression by liver dendritic cells correlates with reduced capacity to activate allogeneic T cells in response to endotoxin. J. Immunol. 2005, 174, 2037–2045. [Google Scholar] [CrossRef] [PubMed]
- Butcher, S.K.; O’Carroll, C.E.; Wells, C.A.; Carmody, R.J. Toll-like receptors drive specific patterns of tolerance and training on restimulation of macrophages. Front. Immunol. 2018, 9, 933. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Cook, J.A. Molecular mechanisms of endotoxin tolerance. J. Endotoxin Res. 2004, 10, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Fukui, A.; Ohta, K.; Nishi, H.; Shigeishi, H.; Tobiume, K.; Takechi, M.; Kamata, N. Interleukin-8 and CXCL10 expression in oral keratinocytes and fibroblasts via Toll-like receptors. Microbiol. Immunol. 2013, 57, 198–206. [Google Scholar] [CrossRef]
- Simiantonaki, N.; Kurzik-Dumke, U.; Karyofylli, G.; Jayasinghe, C.; Michel-Schmidt, R.; Kirkpatrick, C.J. Reduced expression of TLR4 is associated with the metastatic status of human colorectal cancer. Int. J. Mol. Med. 2007, 20, 21–29. [Google Scholar] [CrossRef]
- Tsukamoto, H.; Fukudome, K.; Takao, S.; Tsuneyoshi, N.; Ohta, S.; Nagai, Y.; Ihara, H.; Miyake, K.; Ikeda, Y.; Kimoto, M. Reduced surface expression of TLR4 by a V254I point mutation accounts for the low lipopolysaccharide responder phenotype of BALB/c B cells. J. Immunol. 2013, 190, 195–204. [Google Scholar] [CrossRef]
- Shah, G.; Patel, B.; Chorawala, M. Toll like receptors: An overview. Int. J. Pharmacol. Toxicol. 2014, 2, 53–61. [Google Scholar] [CrossRef][Green Version]
- Lin, Y.; Lee, H.; Berg, A.H.; Lisanti, M.P.; Shapiro, L.; Scherer, P.E. The lipopolysaccharide-activated toll-like receptor (TLR)-4 induces synthesis of the closely related receptor TLR-2 in adipocytes. J. Biol. Chem. 2000, 275, 24255–24263. [Google Scholar] [CrossRef]
- Yokota, S.-I.; Ohnishi, T.; Muroi, M.; Tanamoto, K.; Fujii, N.; Amano, K. Highly-purified Helicobacter pylori LPS preparations induce weak inflammatory reactions and utilize Toll-like receptor 2 complex but not Toll-like receptor 4 complex. FEMS Immunol. Med. Microbiol. 2007, 51, 140–148. [Google Scholar] [CrossRef]
- Beklen, A.; Sorsa, T.; Konttinen, Y. Toll-like receptors 2 and 5 in human gingival epithelial cells co-operate with T-cell cytokine interleukin-17. Oral Microbiol. Immunol. 2009, 24, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Marchesan, J.; Jiao, Y.; Schaff, R.A.; Hao, J.; Morelli, T.; Kinney, J.S.; Gerow, E.; Sheridan, R.; Rodrigues, V.; Paster, B.J.; et al. TLR4, NOD1 and NOD2 mediate immune recognition of putative newly identified periodontal pathogens. Mol. Oral Microbiol. 2016, 31, 243–258. [Google Scholar] [CrossRef]
- Shibata, K. Historical aspects of studies on roles of the inflammasome in the pathogenesis of periodontal diseases. Mol. Oral Microbiol. 2018, 33, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Andrukhov, O.; Ertlschweiger, S.; Moritz, A.; Bantleon, H.P.; Rausch-Fan, X. Different effects of P. gingivalis LPS and E. coli LPS on the expression of interleukin-6 in human gingival fibroblasts. Acta Odontol. Scand. 2014, 72, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Barksby, H.; Nile, C.J.; Jaedicke, K.M.; Taylor, J.J.; Preshaw, P.M. Differential expression of immunoregulatory genes in monocytes in response to Porphyromonas gingivalis and Escherichia coli lipopolysaccharide. Clin. Exp. Immunol. 2009, 156, 479–487. [Google Scholar] [CrossRef]
- Yang, Z.; Cui, Q.; An, R.; Wang, J.; Song, X.; Shen, Y.; Wang, M.; Xu, H. Comparison of microbiomes in ulcerative and normal mucosa of recurrent aphthous stomatitis (RAS)-affected patients. BMC Oral Health 2020, 20, 128. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G.; Darveau, R.P.; Curtis, M.A. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 2012, 10, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Gölz, L.; Memmert, S.; Rath-Deschner, B.; Jäger, A.; Appel, T.; Baumgarten, G.; Götz, W.; Frede, S. LPS from P. gingivalis and hypoxia increases oxidative stress in periodontal ligament fibroblasts and contributes to periodontitis. Mediat. Inflamm. 2014, 2014, 986264. [Google Scholar] [CrossRef]
- Gölz, L.; Memmert, S.; Rath-Deschner, B.; Jäger, A.; Appel, T.; Baumgarten, G.; Götz, W.; Frede, S. Hypoxia and P. gingivalis synergistically induce HIF-1 and NF-κB activation in PDL cells and periodontal diseases. Mediat. Inflamm. 2015, 2015, 438085. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).