Abstract
Cardiac auscultation is a cost-effective and noninvasive technique for cardiovascular disease detection. Recently, various studies have been underway for cardiac auscultation using deep learning, not doctors. When training a deep learning network, it is important to secure large amount of high-quality data. However, medical data are difficult to obtain, and in most cases the number of abnormal classes is insufficient. In this study, data augmentation is used to supplement the insufficient amount of data, and data generalization to generate data suitable for convolutional neural networks (CNN) is proposed. We demonstrate performance improvements by inputting them into the CNN. Our method achieves an overall performance of 96%, 81%, and 90% for sensitivity, specificity, and F1-score, respectively. Diagnostic accuracy was improved by 18% compared to when it was not used. Particularly, it showed excellent detection success rate for abnormal heart sounds. The proposed method is expected to be applied to an automatic diagnosis system to detect heart abnormalities and help prevent heart disease through early detection.
1. Introduction
One of the most important of the organs in the body is the heart, which supplies blood to the whole body. The heart is a muscular pump, and whenever the heart contracts strongly, it sends blood throughout the body to supply oxygen and nutrients. Since the heart functions in harmony with blood vessels and muscles, electrical signals and valves, cardiac disease occurs even if one of them is abnormal. Cardiovascular disease (CVD) causes death and disabilities all around the world. According to the World Health Organization (WHO), 16% of all deaths caused by CVDs [1]. In addition, cardiovascular diseases are steadily increasing every year due to an aging population and increasing causes such as high blood pressure, and high cholesterol, stress, smoking, obesity and so on. Even if there is no past medical history or asymptomatic, CVD often occurs through asymptomatic incubation periods, which can be accompanied by organ damage. In these contexts, pre-diagnosis and treatment of heart disease have become very important.
There are many ways to check the condition of one’s heart. An electrocardiogram (ECG) is an interpretation of the heart’s electrical activity. To perform an ECG, the examiner places electrodes (a small round sensor attached to the skin) on the patient’s arms, legs, and chest. These electrodes measure the magnitude and direction of the current in the heart whenever the heart beats. A photoplethysmogram (PPG) is a method of estimating blood flow by measuring changes in light transmitted to the skin or reflected in blood vessels and bones by emitting green LED light to the skin from a light-based sensor. A wide range of methods have been studied and proposed over the years for automatic analysis of ECG [2] and PPG [3] signals. Lastly, auscultation, listening to the sound of the heart, began with Laennec [4] first listening to the sound of the heart. Auscultation is very simple and cost effective, and it is a very important medical examination method in cardiology. The heart sound is produced by the beating heart and the resulting blood flow. In particular, the pulse and sound caused by blood movement can be heard around the chest and neck. Two heart sounds are heard in healthy adults [5]. The first heart sound, S1, is heard when closing the valves systole and its frequency components lie in the range of 100–200 Hz. The second heart sound, S2 sound is heard when opening the valves diastole and has a higher frequency range of 50–250 Hz. The average interval between S1 and S2 is about 0.45 s, and the normal cardiac cycle is about 0.8 s. Other than S1, S2, there are pathologic sounds such as S3, S4, murmur, turbulent fluid, and extrasystole sounds. Figure 1 shows the representative healthy person and patient’s heart waveforms. Doctors listen to varying loudness, frequency, quality, and duration with a stethoscope, and can determine the opening and closing of heart valves as well as blood flow and turbulence through valves or defects in the heart. To do this, they need extensive knowledge and experience [6]. In fact, the diagnostic accuracy of inexperienced doctors is 20–40% and the diagnostic accuracy of professional cardiologists is about 80% [7].

Figure 1.
The representative waveforms of (a) a healthy person’s heart sound, and (b) a patient’s heart sound.
The main objective of this work is to detect cardiac abnormalities simply by the sound of the heart and prevent cardiovascular disease through early detection. In this paper, we distinguish between normal and abnormalities by detecting and classifying the sounds in heart with signal processing and entering them into a convolutional neural network (CNN) specialized in image processing. More specifically, we will be focusing on data augmentation and data generalization for better CNN performance. As a result, not only the overall accuracy, but also specificity is improved.
The paper is structured as follows. In Section 2, the proposed methods for predicting heart diseases automatically, and the background knowledge for our method, are introduced. In Section 3, the proposed methods are introduced: a method of making data suitable for learning through signal preprocessing in Section 3.2; a method of making a balanced dataset using data augmentation in Section 3.3; a method of making an input of an image processing network in Section 3.4. Section 4 presents the experiments and Section 5 shows the results. Section 6 presents conclusion of the work.
2. Related Work
In recent years, many studies have been conducted to automatically predict cardiac diseases on the basis of the heartbeat [8,9]. Recently, with the development of deep learning, many deep learning-based approaches have been studied to detect abnormal heart sound using deep neural networks (DNN) [10], recurrent neural networks (RNN) [11], and convolutional neural networks (CNN) [12,13]. As CNN extracts and learns features autonomously, it has been employed in various fields, such as image classification and speech recognition. It is also used to recognize abnormal heart sounds. In [12], CNN was used for feature extraction and classification function estimation from heart sound signals. The network of [13] was designed to classify normal and abnormal heart sounds by using CNN with the mel-frequency cepstral coefficient (MFCC) heat map as input.
As it is difficult to collect heart sound data, many previous studies have suffered from lack of data [10,14]. A large dataset is required for training deep learning networks. To solve this problem, some studies have applied data augmentation methods to increase the amount of data. Ref. [10] increased the dataset size by using two methods to solve the data shortage. The author augmented the data to prevent overfitting by performing noise injection, which adds random noise to the input and an audio transformation that slightly deforms the pitch and tempo. In [14], the authors suggested that the noise injection method was effective for covering data shortage.
When data augmentation is performed, the quality of the source data has an important influence. High-quality data means a clear signal without noise. However, noise is unavoidable during recordings, and every piece of recorded data has a different length. For effective analysis, noise reduction is necessary, and normalizing and generalizing the raw dataset is required. Ref. [15] showed an improvement of performance by conducting noise reduction in the preprocessing step. In [16], the authors mentioned the importance and effect of data generalization.
To classify data according to class, it is important to extract appropriate features for each label. There are several methods for extracting data, such as MFCC, spectrogram, and using a deep learning network. A spectrogram is a visualization of the frequency spectrum of a signal and is effective for analyzing audio signals. According to [17], the spectrogram is a suitable feature for predicting cardiovascular disease by converting a time domain signal into a spectrogram, which is a frequency domain, and using an experiment. In addition, Ref. [18] extracted spectrograms and scaled them for effective feature extraction and analysis of heart sounds.
3. Methods
The overall experimental process is shown in Figure 2. After inputting the recorded heart sound, signal preprocessing including noise reduction, normalization, and generalization is applied. After that, the signal is converted into a spectrogram and used as an input to the CNN. Finally, the features of each class are learned through the CNN and classified as either healthy or unhealthy cardiovascular behavior.
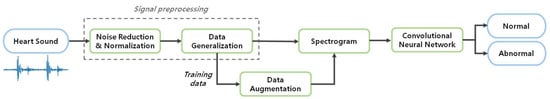
Figure 2.
Block diagram of heart sounds identification system.
3.1. Dataset
Using three kinds of heartbeat audio datasets, we created a new dataset of two classes, normal and abnormal. The PASCAL Heart Sound Challenge (HSC) dataset [19] was used for the task of heart sound segmentation and classification. The HSC consists of dataset A and dataset B. Dataset A has 176 heart sounds collected using the iStethoscope Pro iPhone app, and dataset B has 656 heart sounds recorded using a digital stethoscope. These two datasets are composed of normal, murmur, extrasystole, artifact, and extrahls classes. Apart from HSC, we collected additional audio files through the iStethoscope Pro iPhone application [20]. Divided into two classes, normal and abnormal, 10 and 50 audio files, respectively, were created. The 2016 PhysioNet computing in cardiology challenge (CinC) [21] aims to encourage the development of algorithms to classify heartbeat recordings. It includes 3240 audio files; 2575 normal and 665 abnormal.
3.2. Signal Preprocessing
3.2.1. Noise Reduction and Normalization
Noise affects recognition rate by distorting the original signal. While recording heart sounds, noises can occur such as external sounds, sweeping sounds due to body movements, breathing sounds, thermal noise and so on. Noise due to skin rubbing occurs at the beginning and end in most cases when recording actual heartbeats. To remove this unwanted sound, 15% of the audio file was first eliminated. Cardiac sounds have specific frequency ranges. S1 has a frequency of at least 100 to 200 Hz, S2 has a frequency of 50–250 Hz, and sick cardiac sound has a maximum frequency of 400 Hz. Table 1 shows the frequency range and diagnostic value according to the cardiac sound. The frequency characteristics of these components can be used as important parameters for identification. After converting to the frequency domain, the band-pass filter eliminates frequencies other than 40–400 Hz, excluding S1, S2 and the heart noise frequency ranges.

Table 1.
Types of heart sounds and corresponding frequency ranges.
Each sound in the dataset has a different amplitude range, and the quality of the sound may vary depending on the environment where the heartbeat is recorded. Therefore, it is necessary to uniform the dataset for accurate comparison. The signals must be equalized to the same amplitude range and have the same quality. To change the data to a common scale without distorting the difference in range, the signals were scaled between 0 and 1 through min-max normalization. As a result, time-domain noise in the audio files was reduced.
3.2.2. Data Generalization
The length of audio files in the dataset is diverse, ranging from 4 s to 120 s. However, CNN takes a constant size of image as an input. If the audio signals of 4 s and 120 s are made into spectrograms of the same size, the difference is large even within the same class. Additionally, the different lengths of the data make feature extraction more difficult if the data are not sufficient. Therefore, the following method was used to make constant lengths of data. Two assumptions were made for the experiment:
- Heart sounds can be judged to be about 5 s.
- Abnormal heart sounds are observed within 5 s.
To achieve a constant length of data, we assumed that it took about 5 s to judge between normal and abnormal heart sounds. However, extracystoles, where the pulse beats normally and then skips once, can occur once or twice or dozens of times a minute. Atrial fibrillation, in which the heart beats irregularly, can also be temporary or persistent. Therefore, in the experimental data, the assumption that this irregular abnormal heartbeat occurs within 5 s was added.
The number of samples in the audio file was equalized to adjust the length of the heart signal. Sampling range was determined using the cardiac sound peaks. The peaks of S1 and S2 can be found using the time interval features of S1 and S2. Figure 3 shows S1 and S2 detection through time difference. The time difference between S1 and S2 ranges from 250 to 350 ms, and between S2 and the next S1 the time ranges from 350 to 800 ms. Therefore, after finding peaks larger than a certain amplitude, the intervals between the peaks were calculated. If the previous interval was between 250 and 350 ms and the current interval is between 350 and 800 ms, then S1 and S2 can be identified [22].
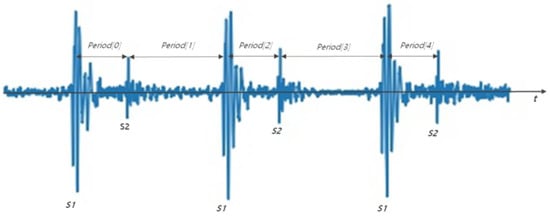
Figure 3.
S1 and S2 detection method through time difference. Period [0], Period [2], Period [4] are 250 to 350 ms, and period [1] and period [3] have values between 350 and 800 ms.
Ref. [22] was able to find S1 and S2 easily in ideal heartbeats, but there are many exceptions for diseased heartbeats. For example, as shown in Figure 4, S1 and S2 were not found when the size of a particular heartbeats were large or small, when the intervals between the extracted peaks were irregular, when the intervals of the heart signals were not constant, or if the heart signals were not clear. In this paper, we propose a method of finding peaks and make them into constant length data. Normally, S1 is louder than S2 at the apex, and softer than S2 at the base of the heart. Empirically, the louder heart sound was close to 1, and the softer heart sound had about half the amplitude of the loud sound in the normalized signal. Thus, 0.65 was set as the threshold, and then peaks larger than the threshold were selected from the normalized signals. Since the cardiac cycle is 800 ms, a peak larger than 0.65 with at least 800 ms intervals was considered the main cycle S. The coordinates of the S were obtained through the Scipy python library. Among them, the first S and sixth S coordinates were truncated to produce data. However, if the heart beats irregularly or if the murmur is large, it is impossible to find S1 using this method and to cut it at regular intervals. Therefore, when the number of peaks is shorter or longer than the average cardiac rate of a healthy person (60–100 beats per minute), that is, when the number of peaks is less than 0.5 or more than 1 per second, 5 s data from the first peak were used. For example, if the sampling rate was 2000, the first S point t to t + 2000 × 5 was used to make data. The whole process can be seen in the flow chart in Figure 5.
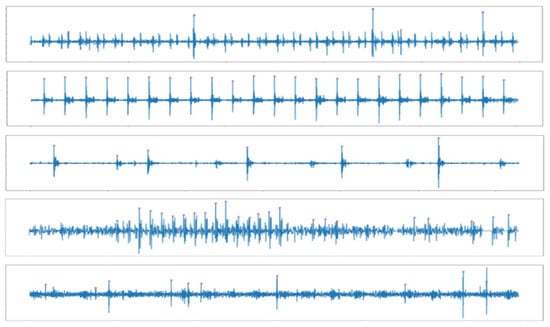
Figure 4.
Examples of not detecting S1 and S2 using the time difference method.
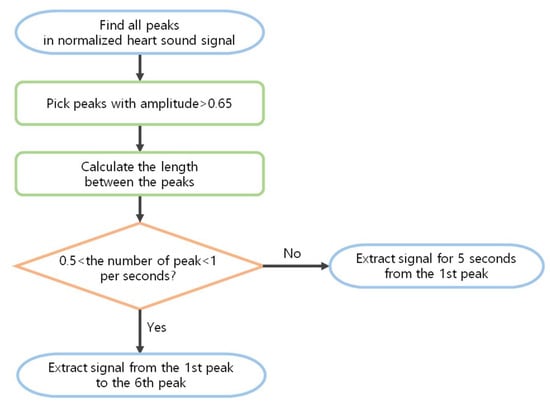
Figure 5.
Flow chart for making a constant length of data.
3.3. Data Augmentation
For medical datasets, it is often difficult to obtain the data themselves or to label them with the correct answer. In addition, since it is generally difficult to obtain patients’ data when collecting data, the amount of data for healthy people is much greater than that that of pathological data. The cardiac sound dataset is a deficient and unbalanced dataset that is lacking in terms of total amount of data, and the amount of abnormal data is insufficient in comparison to the amount of normal data. When training a deep neural network classification model, a biased classification model may appear if the number of data points of each class is not similar, and the number of data points emphasizes a specific class. Therefore, we used data augmentation to build a balanced dataset. Data augmentation is a technology that increases the amount of data while changing them through various algorithms. In this paper, data augmentation methods at the audio stage and the spectrogram stage were used. The audio stage uses a method to add random noise, move signals to the left, and flip the phase of the signal relative to the x-axis. Approaches such as pitch shifting or speed changing that could distort the audio signal were not used. In the spectrogram phase, various image augmentation methods can be used. However, augmentations such as random rotation, random brightness, and random zoom can make the spectrogram very different. Therefore, we used an effective augmentation in speech recognition. SpecAugmentation [23] has been proposed as a method for random frequency masking and time masking. Additionally, adding salt and pepper noise, similar to masking, was used to create a new spectrogram.
3.4. Feature Extraction
Data in the time domain are converted into the frequency domain through Fourier transform for signal analysis. However, the Fourier transform (FT) does not represent the transition of the signal over time. In other words, when using FT, it is impossible to know at what point the frequency changes when the frequency changes over time. A Short Time Fourier Transform (STFT) was proposed to overcome this. STFT divides time by a specific window size and performs a Fourier transform. Window length may vary depending on the application. In the spectrogram, a long window size is used to increase the frequency resolution, and a short window size is used to increase the time resolution. When applying STFT to the heart sound, the appropriate window length is 98.91% of the heart sound data length [24]. A spectrogram represents the frequency content of the audio as colors in an image. The horizontal axis is time information. The vertical axis is the frequency and the lowest frequencies at the bottom and the highest frequencies at the top. The colors are the amplitude of a specific frequency at a specific time. In this paper, time-domain audio signals of about 5 s were transformed into frequency-domain spectrogram features as Figure 6. Figure 7 shows augmented abnormal data spectrograms in train dataset.
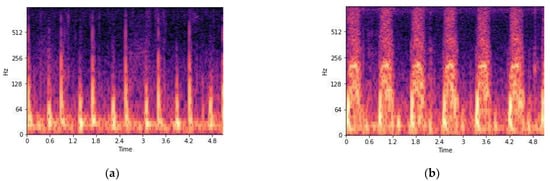
Figure 6.
(a) Normal heartbeat spectrogram and (b) abnormal heartbeat spectrogram.
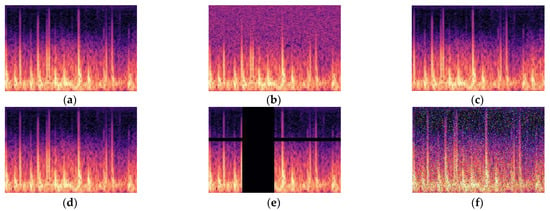
Figure 7.
Abnormal train data augmentation. (a) Normal, (b) random noise, (c) left shift, (d) x-axis symmetry, (e) masking, (f) s&p noise.
CNN is a kind of artificial neural network, and the structure of CNN can be divided into feature extraction and classification. Feature extraction uses convolution operations to maintain spatial and local information of the image. It learns the features of the image by repeating the convolutional layer, the activation function, and the pooling layer. Classification uses fully connected layers reducing the amount of computation, and the last layer outputs the score of each class. Compared with general artificial neural networks, CNNs effectively recognize image features because they have the advantage of having fewer learning parameters and use filters as shared parameters.
4. Experiments
4.1. Datasets
In this experiment, a total of 3890 cardiac sound files, 2936 normal and 909 abnormal, obtained from three datasets, were used. Training data and test data were roughly divided into 7:3, and data shorter than 5 s were excluded during the preprocessing process. The training data of the abnormal class created six spectrograms per sound through augmentation. The detailed numbers can be found in Table 2.

Table 2.
The detailed dataset used in the experiment.
4.2. Implementation Details
The network used in the experiment takes a 217 × 334 heart sound spectrogram as input. The convolution layer, pooling layer, and dropout layer were repeated four times and then two dense layers were added. This network performed a binary classification that distinguished two classes, nonsick and sick. Binary cross entropy loss and Adam optimizer were used during training. The structure of the network is shown in Figure 8.
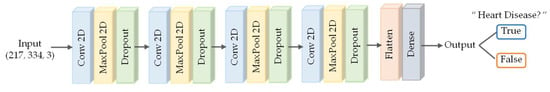
Figure 8.
Our CNN architecture.
The performance of the proposed method in the paper was evaluated using several metrics. We assessed diagnostic accuracy, as well as specificity, sensitivity, and accuracy for each class. As the classes were imbalanced, the F1-score was used as the performance metric in the paper. True Positive (TP) is a normal heart sound diagnosed as normal. False Negative (FN) is a normal heart sound, but diagnosed as an abnormal heart sound. True Negative (TN) is an abnormal heart sound diagnosed as abnormal. False Positive (FP) is an abnormal heart sound, but diagnosed as normal.
5. Results and Discussion
5.1. Ablation Study
Diagnostic performance was compared through an ablation study to find out the effect of improving the performance of data generalization (DG) and data augmentation (DA). The experiment was conducted in the same environment except for DG and DA. The amount of data used for the test was 208, which did not belong to the training or validation dataset.
Figure 9 shows the diagnostic results of each algorithm on the CNN. The test accuracy was 82% when the audio signal was directly converted into a spectrogram without DG and DA. However, the sensitivity was 43%, which means the accuracy of diseased heartbeats is low. More than half of the abnormalities were classified as normal, and it can be seen that the network could not accurately distinguish between sick and nonsick heartbeats. When augmented data were added, the test accuracy was 76%, which was lower than without the augmented data. This shows that increasing the amount of data is not always good, and the quality of the data is more important for improving performance. Although specificity was high, sensitivity was very low, and most of the cardiac abnormalities could not be detected. When data generalization was added, the accuracy was 83%. Additionally, the sensitivity improved compared to the previous cases. However, the accuracy of detecting pathologic heart beats is about 51%, which still cannot be considered as distinguishing pathologic heart sounds. When the data were created using the DG and DA proposed in the paper, the accuracy increased to 90%. Additionally, the specificity was 96% and the sensitivity was 73%, thus yielding higher diagnostic accuracy and sensitivity than other methods.
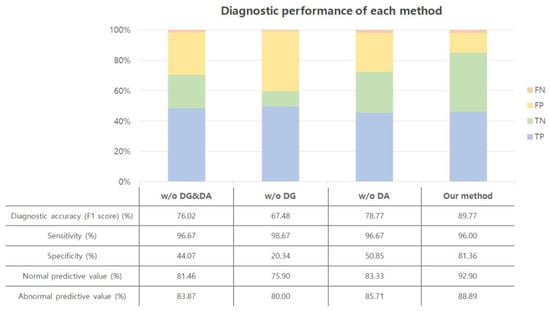
Figure 9.
Diagnostic performance. w/o: without; DG: data generalization; DA: data augmentation.
5.2. Comparison with CNN Models
We compared the performance with existing heart sound classification methods based on CNN. These methods used the PhysioNet dataset, so our model was also trained using only the PhysioNet dataset for accurate performance comparison. The performance of each method is listed in Table 3. Our method achieved better or similar performance than other methods.

Table 3.
Comparative evaluation of CNN models.
5.3. Limitations
This study had several limitations. First, the assumption that abnormal heart sounds can be observed within 5 s is different from reality. In fact, some of the abnormal data after generalization were similar to the normal data. Second, the abnormal heartbeat could be increased through data augmentation, but the types of heart disease were limited. Third, although the frequency band was used to remove noise, noise remained, hindering the ability to clearly classify heart sounds.
6. Conclusions
Detecting cardiac abnormalities through cardiac sounds is a method that has been proven after a long period of research. However, since auscultation is a method that relies on human senses, it has some drawbacks. For example, different doctors have different auscultation skills, so sometimes the diagnosis is unreliable. Recently, studies have been conducted to diagnose cardiovascular diseases based on deep learning to compensate for this. In this paper, we proposed a preprocessing method for better deep learning training. In data generalization, we used an appropriate peak processing algorithm to generate uniform forms of data. Data augmentation solved the problems of an insufficient amount of data and the imbalance of pathologic heart data, which are difficult to obtain. The proposed method improved the accuracy of auscultation and demonstrated performance improvements through accuracy comparisons in the basic CNN network. Furthermore, performance improvement can be expected when applied to other deep learning networks. The proposed method showed an improved detection success rate for unhealthy heart sounds. It is considered that this could be applied to an automatic diagnosis system. Therefore, if the heart sound can be acquired, it will help to detect patients quickly and prevent cardiovascular disease through early detection. In particular, it would be useful in areas where medical services are scarce.
In future work, we will apply better noise reduction and signal processing methods to obtain the ideal cardiac signal. In addition, studies will continue to diagnose various heart diseases that are not simply normal or abnormal.
Author Contributions
Conceptualization, Y.J.; methodology, Y.J.; software, Y.J.; validation, Y.J.; investigation, Y.J.; data curation, Y.J.; writing—original draft preparation, Y.J. and J.K.; writing—review and editing, Y.J., J.K., D.K., J.K. and K.L.; visualization, Y.J. and J.K.; supervision, Y.J.; funding acquisition, K.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by the Institute of Information and Communications Technology Planning and Evaluation (IITP) Grant funded by the Korean Government (MSIT) under Grant 2020-0-01907.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The PASCAL Heart Sound Challenge (HSC) dataset is available at http://www.peterjbentley.com/heartchallenge/ (accessed on 17 May 2021). The 2016 PhysioNet computing in cardiology challenge (CinC) dataset is available at https://physionet.org/content/challenge-2016/1.0.0/, DOI:10.22489/CinC.2016.179-154 (accessed on 17 May 2021).
Conflicts of Interest
The authors declare no conflict of interest.
References
- World Health Organization. The Top 10 Causes of Death. Available online: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death/ (accessed on 22 March 2021).
- Castells, F.; Laguna, P.; Sornmo, L.; Bollmann, A.; Roig, J.M. Principal Component Analysis in ECG Signal Processing. EURASIP J. Adv. Signal Process. 2007, 2007, 1–21. [Google Scholar] [CrossRef]
- Sepúlveda-Cano, L.M.; Gil, E.; Laguna, P.; Castellanos-Dominguez, G. Selection of nonstationary dynamic features for obstructive sleep apnoea detection in children. EURASIP J. Adv. Signal Process. 2011, 2011, 1–10. [Google Scholar] [CrossRef]
- Roguin, A. Rene Theophile Hyacinthe Laënnec (1781–1826): The man behind the stethoscope. Clin. Med. Res. 2006, 4, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Walker, H.K.; Hall, W.D.; Hurst, J.W. Clinical Methods: The History, Physical, and Laboratory Examinations, 3rd ed.; Butterworths: Boston, MA, USA, 1990. [Google Scholar]
- Roy, D.; Sargeant, J.; Gray, J.; Hoyt, B.; Allen, M.; Fleming, M. Helping family physicians improve their cardiac auscultation skills with an interactive CD-ROM. J. Contin. Educ. Health Prof. 2002, 22, 152–159. [Google Scholar] [CrossRef]
- Etchells, E.; Bell, C.; Robb, K. Does this patient have an abnormal systolic murmur? JAMA 1997, 277, 564–571. [Google Scholar] [CrossRef]
- Yaseen; Son, G.Y.; Kwon, S. Classification of Heart Sound Signal Using Multiple Features. Appl. Sci. 2018, 12, 2344. [Google Scholar] [CrossRef]
- Maglogiannis, L.; Loukis, E.; Zafiropoulos, E.; Stasis, A. Support Vectors Machine-based identification of heart valve diseases using heart sounds. Comput. Methods Programs Biomed. 2009, 95, 47–61. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Mehmood, A.; Ullah, S.; Ahmad, M.; Choi, G.S.; On, B.W. Heartbeat Sound Signal Classification Using Deep Learning. Sensors 2019, 19, 4819. [Google Scholar] [CrossRef] [PubMed]
- Messner, E.; Zohrer, M.; Pernkopf, F. Heart Sound Segmentation—An Event Detection Approach Using Deep Recurrent Neural Networks. IEEE Trans. Biomed. Eng. 2018, 65, 1964–1974. [Google Scholar] [CrossRef]
- Li, F.; Tang, H.; Shang, S.; Mathiak, K.; Cong, F. Classification of Heart Sounds Using Convolutional Neural Network. Appl. Sci. 2020, 10, 3956. [Google Scholar] [CrossRef]
- Jonathan, R.; Rui, A.; Anurag, G.; Saigopal, N.; Ion, M.; Kumar, S. Recognizing Abnormal Heart Sounds Using Deep Learning. arXiv 2017, arXiv:1707.04642. [Google Scholar]
- Poole, B.; Sohl-Dickstein, J.; Ganguli, S. Analyzing noise in autoencoders and deep networks. arXiv 2014, arXiv:1406.1831. [Google Scholar]
- Varghees, V.N.; Ramachandran, K.I. A novel heart sound activity detection framework for automated heart sound analysis. Biomed. Signal Process. Control 2014, 13, 174–188. [Google Scholar] [CrossRef]
- Stasis, A.C.; Loukis, E.N.; Pavlopoulos, S.A.; Koutsouris, D. Using decision tree algorithms as a basis for a heart sound diagnosis decision support system. In Proceedings of the 4th International IEEE EMBS Special Topic Conference on Information Technology Applications in Biomedicine, Birmingham, UK, 24–26 April 2003; pp. 354–357. [Google Scholar]
- Kang, S.H.; Joe, B.G.; Yoon, Y.Y.; Cho, G.Y.; Shin, I.S.; Suh, J.W. Cardiac Auscultation Using Smartphones: Pilot Study. JMIR mHealth uHealth 2018, 6, e49. [Google Scholar] [CrossRef]
- Zhang, W.; Han, J.; Deng, S. Heart sound classification based on scaled spectrogram and tensor decomposition. Expert Syst. Appl. 2017, 84, 220–231. [Google Scholar] [CrossRef]
- Bentley, P.; Glenn, N.; Coimbra, M.; Mannor, S. Classifying Heart Sounds Challenge. Available online: http://www.peterjbentley.com/heartchallenge (accessed on 22 March 2021).
- Bentley, P.J. iStethoscope: A demonstration of the use of mobile devices for auscultation. In Mobile Health Technologies; Humana Press: New York, NY, USA, 2015; pp. 293–303. [Google Scholar] [CrossRef]
- Liu, C.; Springer, D.; Li, Q.; Moody, B.; Juan, R.A.; Chorro, F.J.; Castells, F.; Roig, J.M.; Silva, I.; Johnson, A.E.; et al. Classification of normal/abnormal heart sound recordings: The PhysioNet/Computing in Cardiology Challenge 2016. In Proceedings of the 2016 Computing in Cardiology Conference (CinC), Vancouver, BC, Canada, 11–14 September 2016; pp. 609–612. [Google Scholar]
- Thiyagaraja, S.R.; Dantu, R.; Shrestha, P.L.; Chitnis, A.; Thompson, M.A.; Anumandla, P.T.; Sarma, T.; Dantu, S. A novel heart-mobile interface for detection and classification of heart sounds. Biomed. Signal Process. Control 2018, 45, 313–324. [Google Scholar] [CrossRef]
- Park, D.S.; Chan, W.; Zhang, Y.; Chiu, C.C.; Zoph, B.; Cubuk, E.D.; Quoc, V.L. SpecAugment: A simple data augmentation method for automatic speech recognition. arXiv 2019, arXiv:1904.08779. [Google Scholar]
- Nisar, S.; Khan, O.U.; Tariq, M. An efficient adaptive window size selection method for improving spectrogram visualization. Comput. Intell. Neurosci. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Potes, C.; Parvaneh, S.; Rahman, A.; Conroy, B. Ensemble of feature-based and deep learning-based classifiers for detection of abnormal heart sounds. In Proceedings of the 2016 Computing in Cardiology Conference (CinC), Vancouver, BC, Canada, 11–14 September 2016; pp. 621–624. [Google Scholar]
- Tschannen, M.; Kramer, T.; Marti, G.; Heinzmann, M.; Wiatowski, T. Heart sound classification using deep structured features. In Proceedings of the 2016 Computing in Cardiology Conference (CinC), Vancouver, BC, Canada, 11–14 September 2016; pp. 565–568. [Google Scholar]
- Wu, J.M.T.; Tsai, M.H.; Huang, Y.Z.; Islam, S.H.; Hassan, M.M.; Alelaiwi, A.; Fortino, G. Applying an ensemble convolutional neural network with Savitzky-Golay filter to construct a phonocardiogram prediction model. Appl. Soft Comput. 2019, 78, 29–40. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).