Multilevel Gene Regulation Using Switchable Transcription Terminator and Toehold Switch in Escherichia coli
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Construction and E. coli Strains Used
2.2. Media, Chemicals, and Other Reagents
2.3. Flow Cytometry Analysis
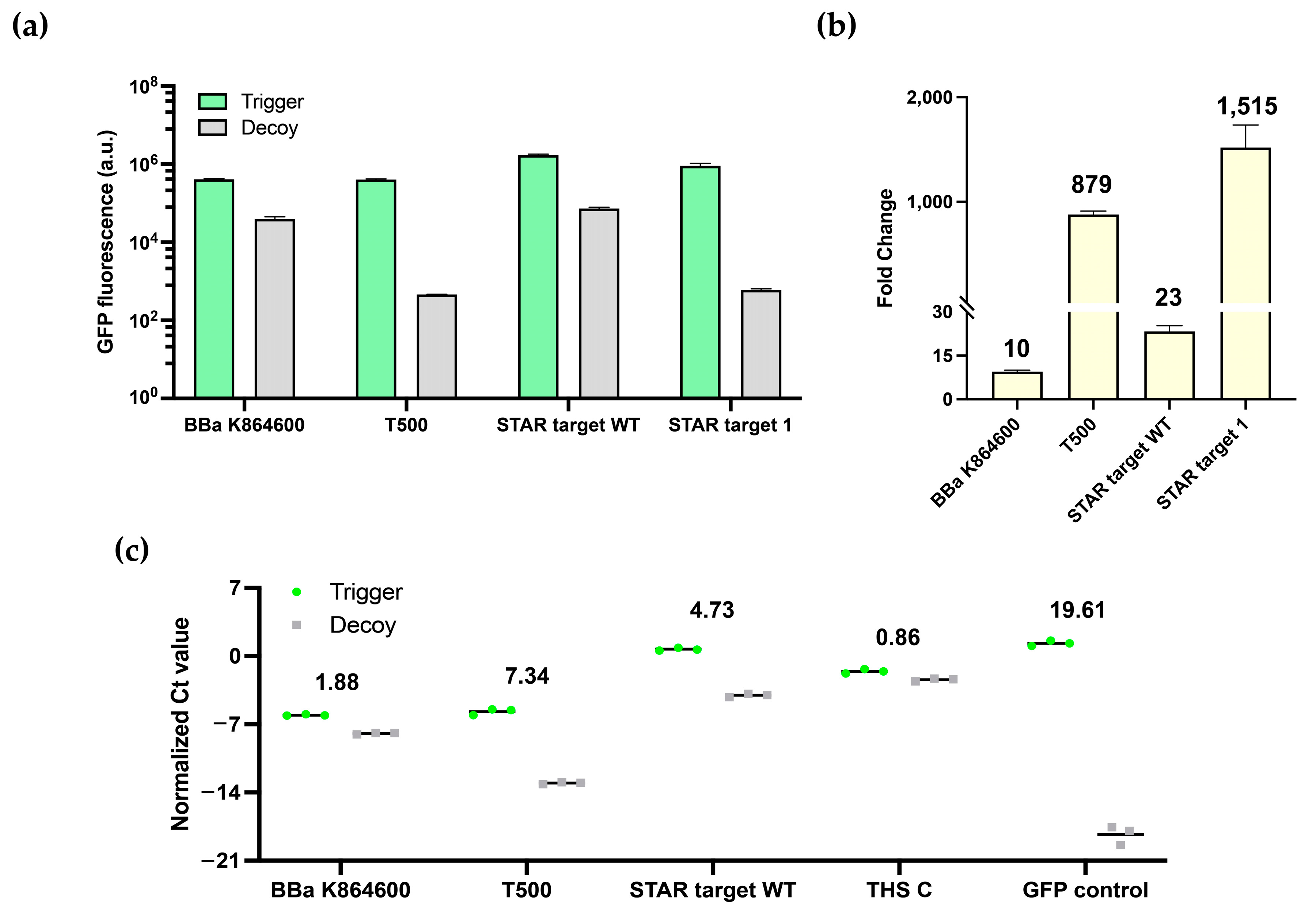
2.4. Quantitative Reverse-Transcription PCR for Transcription Termination Verification
2.5. Plate-Reader Analysis for Multilayered Circuits
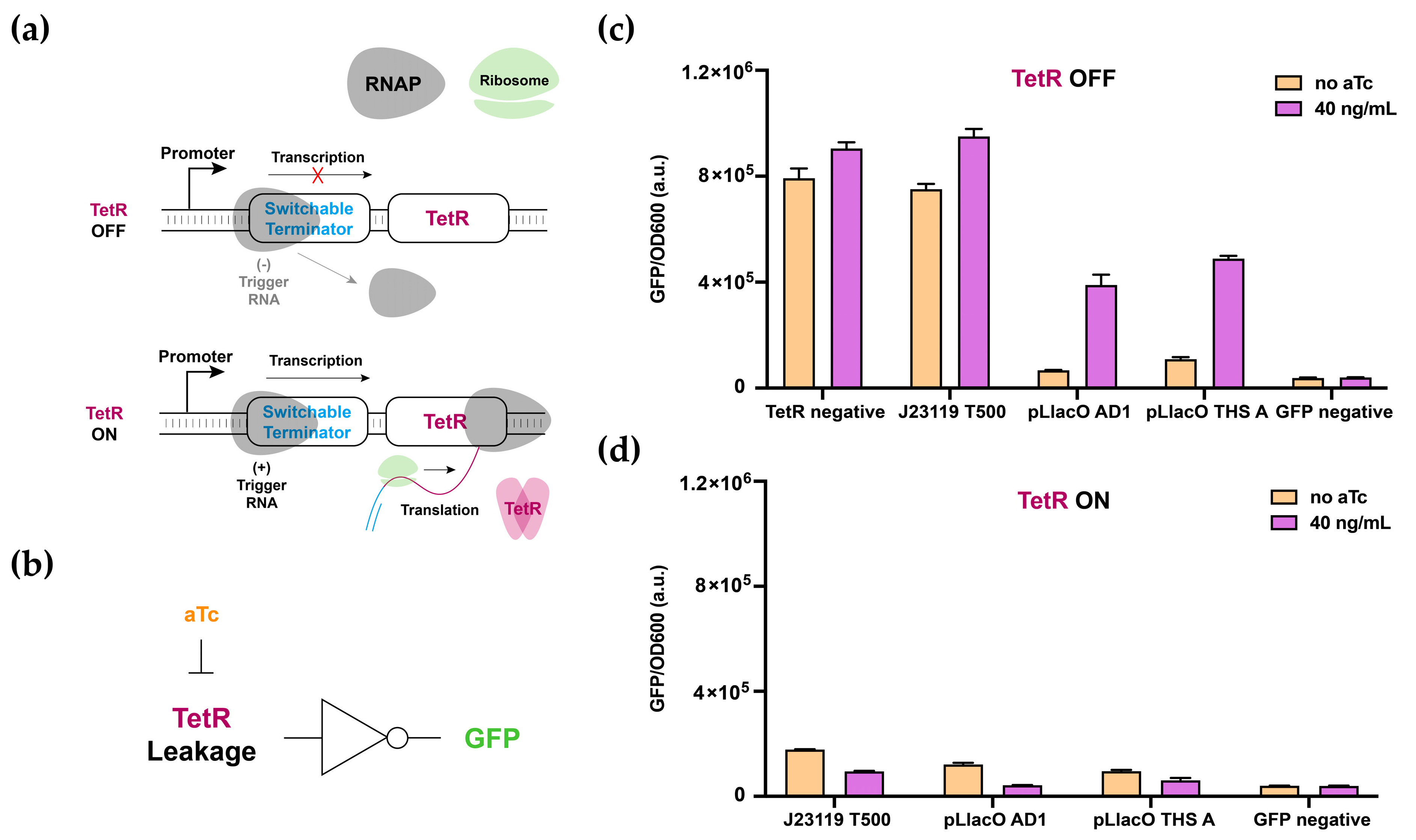
3. Results
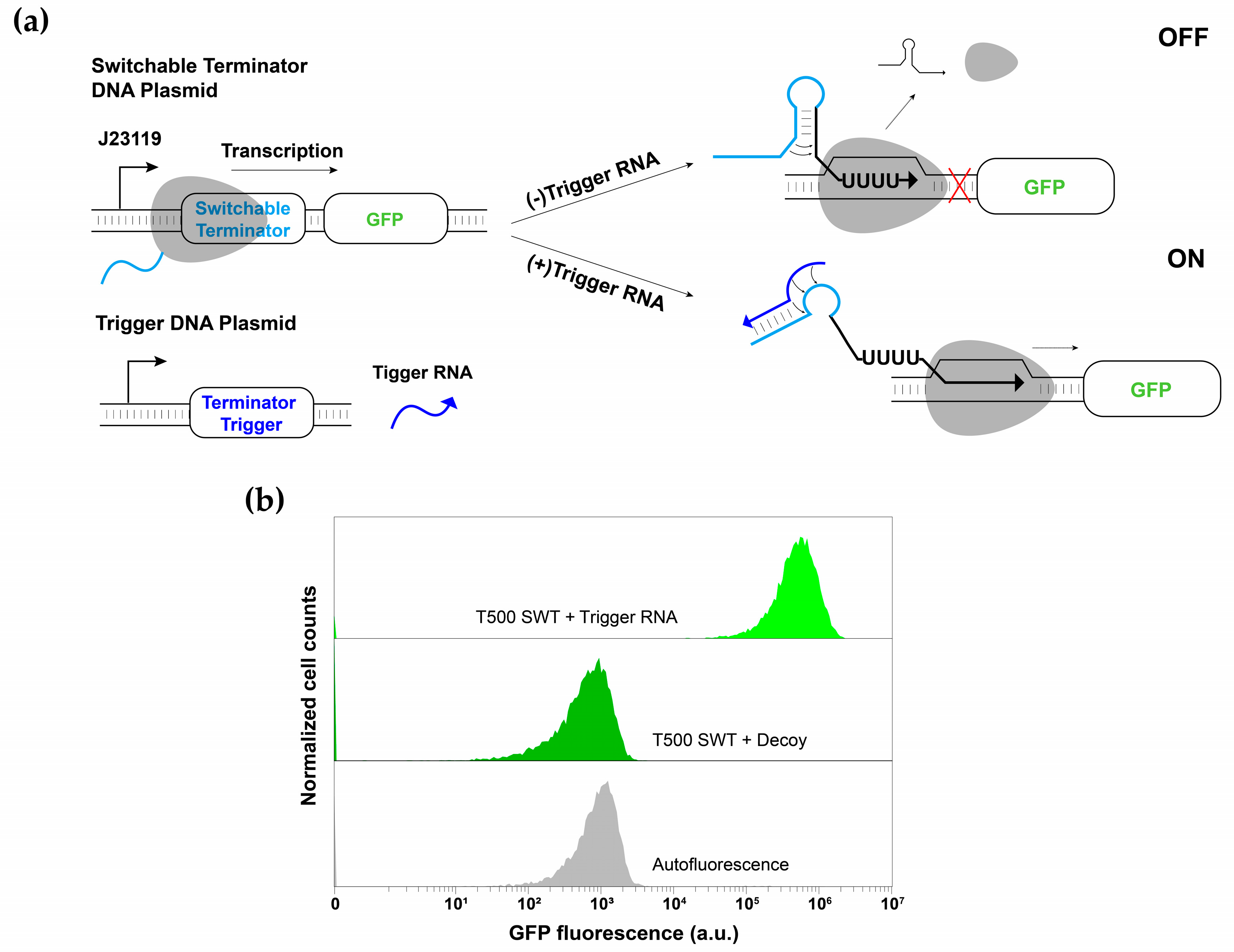
3.1. Switchable Terminator Design
3.2. Characterization of Switchable Terminators
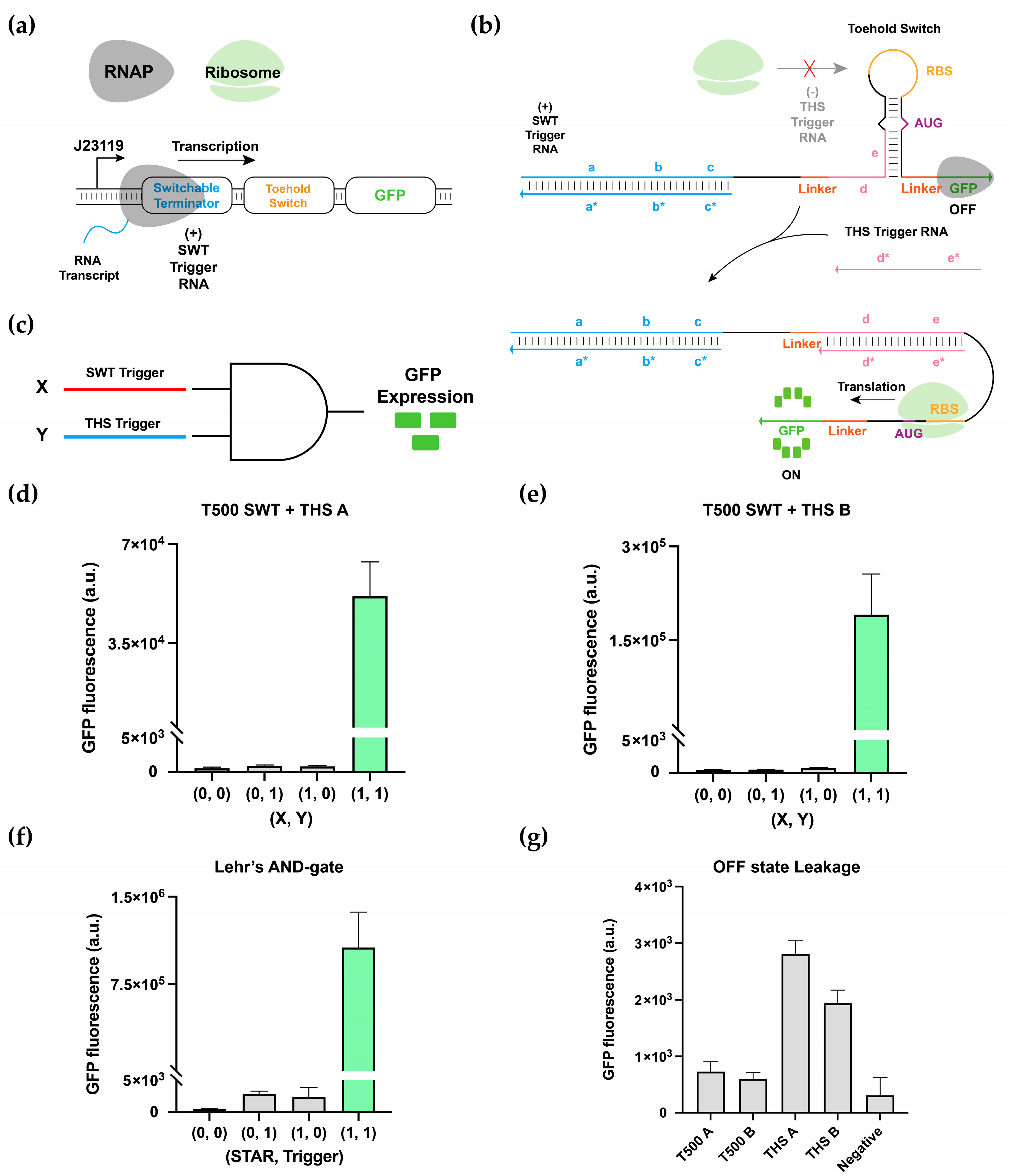
3.3. Multilevel Regulatory System and 2-Input AND-Gate Implementation
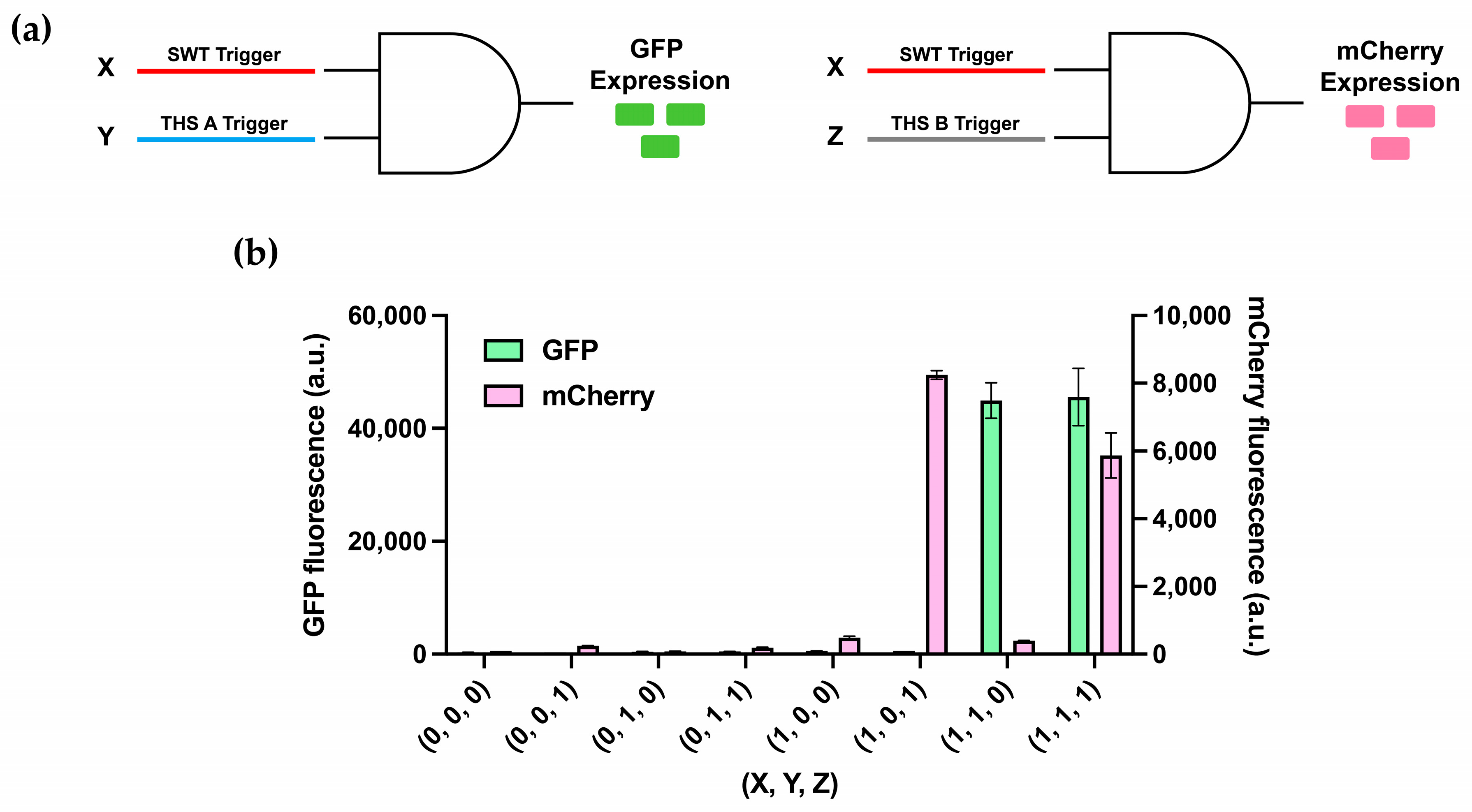
3.4. Multiplexing of 2-Input AND-Gates
3.5. Multilayered Circuit Construction Using SWT
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cameron, D.E.; Bashor, C.J.; Collins, J.J. A brief history of synthetic biology. Nat. Rev. Microbiol. 2014, 12, 381–390. [Google Scholar] [CrossRef]
- Cheng, A.A.; Lu, T.K. Synthetic biology: An emerging engineering discipline. Annu. Rev. Biomed. Eng. 2012, 14, 155–178. [Google Scholar] [CrossRef]
- Elowitz, M.B.; Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 2000, 403, 335–338. [Google Scholar] [CrossRef] [PubMed]
- Win, M.N.; Smolke, C.D. Higher-order cellular information processing with synthetic RNA devices. Science 2008, 322, 456–460. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; White, K.S.; Winfree, E. Construction of an in vitro bistable circuit from synthetic transcriptional switches. Mol. Syst. Biol. 2006, 2, 68. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Li, H.; Miao, Y.; Liu, Y.; Wang, E. Implementation of half adder and half subtractor with a simple and universal DNA-based platform. NPG Asia Mater. 2013, 5, e76. [Google Scholar] [CrossRef]
- Sun, Z.Z.; Yeung, E.; Hayes, C.A.; Noireaux, V.; Murray, R.M. Linear DNA for rapid prototyping of synthetic biological circuits in an Escherichia coli based TX-TL cell-free system. ACS Synth. Biol. 2014, 3, 387–397. [Google Scholar] [CrossRef]
- Grünberg, R.; Serrano, L. Strategies for protein synthetic biology. Nucleic Acids Res. 2010, 38, 2663–2675. [Google Scholar] [CrossRef]
- Davidson, E.A.; Ellington, A.D. Synthetic RNA circuits. Nat. Chem. Biol. 2007, 3, 23–28. [Google Scholar] [CrossRef]
- Nielsen, A.A.; Segall-Shapiro, T.H.; Voigt, C.A. Advances in genetic circuit design: Novel biochemistries, deep part mining, and precision gene expression. Curr. Opin. Chem. Biol. 2013, 17, 878–892. [Google Scholar] [CrossRef]
- Jagadevan, S.; Banerjee, A.; Banerjee, C.; Guria, C.; Tiwari, R.; Baweja, M.; Shukla, P. Recent developments in synthetic biology and metabolic engineering in microalgae towards biofuel production. Biotechnol. Biofuels 2018, 11, 185. [Google Scholar] [CrossRef] [PubMed]
- Bereza-Malcolm, L.T.; Mann, G.l.; Franks, A.E. Environmental sensing of heavy metals through whole cell microbial biosensors: A synthetic biology approach. ACS Synth. Biol. 2015, 4, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Trosset, J.-Y.; Carbonell, P. Synthetic biology for pharmaceutical drug discovery. Drug Des. Dev. Ther. 2015, 9, 6285–6302. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.Y.; Turberfield, A.J.; Yurke, B.; Winfree, E. Engineering entropy-driven reactions and networks catalyzed by DNA. Science 2007, 318, 1121–1125. [Google Scholar] [CrossRef]
- Lee, J.; Kladwang, W.; Lee, M.; Cantu, D.; Azizyan, M.; Kim, H.; Limpaecher, A.; Gaikwad, S.; Yoon, S.; Treuille, A. RNA design rules from a massive open laboratory. Proc. Natl. Acad. Sci. USA 2014, 111, 2122–2127. [Google Scholar] [CrossRef]
- Isaacs, F.J.; Dwyer, D.J.; Ding, C.; Pervouchine, D.D.; Cantor, C.R.; Collins, J.J. Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 2004, 22, 841–847. [Google Scholar] [CrossRef]
- Rodrigo, G.; Landrain, T.E.; Jaramillo, A. De novo automated design of small RNA circuits for engineering synthetic riboregulation in living cells. Proc. Natl. Acad. Sci. USA 2012, 109, 15271–15276. [Google Scholar] [CrossRef]
- Rosenfeld, N.; Alon, U. Response delays and the structure of transcription networks. J. Mol. Biol. 2003, 329, 645–654. [Google Scholar] [CrossRef]
- Laalami, S.; Zig, L.; Putzer, H. Initiation of mRNA decay in bacteria. Cell. Mol. Life Sci. 2014, 71, 1799–1828. [Google Scholar] [CrossRef]
- Na, D.; Yoo, S.M.; Chung, H.; Park, H.; Park, J.H.; Lee, S.Y. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat. Biotechnol. 2013, 31, 170–174. [Google Scholar] [CrossRef]
- Qi, L.S.; Arkin, A.P. A versatile framework for microbial engineering using synthetic non-coding RNAs. Nat. Rev. Microbiol. 2014, 12, 341–354. [Google Scholar] [CrossRef]
- Zadeh, J.N.; Steenberg, C.D.; Bois, J.S.; Wolfe, B.R.; Pierce, M.B.; Khan, A.R.; Dirks, R.M.; Pierce, N.A. NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 2011, 32, 170–173. [Google Scholar] [CrossRef]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef]
- Chappell, J.; Watters, K.E.; Takahashi, M.K.; Lucks, J.B. A renaissance in RNA synthetic biology: New mechanisms, applications and tools for the future. Curr. Opin. Chem. Biol. 2015, 28, 47–56. [Google Scholar] [CrossRef]
- Liang, J.C.; Bloom, R.J.; Smolke, C.D. Engineering biological systems with synthetic RNA molecules. Mol. Cell 2011, 43, 915–926. [Google Scholar] [CrossRef]
- Neupert, J.; Karcher, D.; Bock, R. Design of simple synthetic RNA thermometers for temperature-controlled gene expression in Escherichia coli. Nucleic Acids Res. 2008, 36, e124. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Hu, Y.; Ding, T.; Xie, C.; Wang, Z.; Chen, Z.; Yang, J.; Zhang, C. Aptamer-based regulation of transcription circuits. Chem. Commun. 2019, 55, 7378–7381. [Google Scholar] [CrossRef] [PubMed]
- Etzel, M.; Mörl, M. Synthetic riboswitches: From plug and pray toward plug and play. Biochemistry 2017, 56, 1181–1198. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, A.; Maeda, M. An artificial aptazyme-based riboswitch and its cascading system in E. coli. ChemBioChem 2008, 9, 206–209. [Google Scholar] [CrossRef] [PubMed]
- Chappell, J.; Takahashi, M.K.; Lucks, J.B. Creating small transcription activating RNAs. Nat. Chem. Biol. 2015, 11, 214–220. [Google Scholar] [CrossRef]
- Green, A.A.; Silver, P.A.; Collins, J.J.; Yin, P. Toehold switches: De-novo-designed regulators of gene expression. Cell 2014, 159, 925–939. [Google Scholar] [CrossRef] [PubMed]
- Stanton, B.C.; Nielsen, A.A.; Tamsir, A.; Clancy, K.; Peterson, T.; Voigt, C.A. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nat. Chem. Biol. 2014, 10, 99–105. [Google Scholar] [CrossRef]
- Chappell, J.; Westbrook, A.; Verosloff, M.; Lucks, J.B. Computational design of small transcription activating RNAs for versatile and dynamic gene regulation. Nat. Commun. 2017, 8, 1051. [Google Scholar] [CrossRef]
- Verosloff, M.; Chappell, J.; Perry, K.L.; Thompson, J.R.; Lucks, J.B. PLANT-Dx: A molecular diagnostic for point-of-use detection of plant pathogens. ACS Synth. Biol. 2019, 8, 902–905. [Google Scholar] [CrossRef]
- Glasscock, C.J.; Biggs, B.W.; Lazar, J.T.; Arnold, J.H.; Burdette, L.A.; Valdes, A.; Kang, M.-K.; Tullman-Ercek, D.; Tyo, K.E.; Lucks, J.B. Dynamic Control of Gene Expression with Riboregulated Switchable Feedback Promoters. ACS Synth. Biol. 2021. [Google Scholar] [CrossRef]
- Green, A.A.; Kim, J.; Ma, D.; Silver, P.A.; Collins, J.J.; Yin, P. Complex cellular logic computation using ribocomputing devices. Nature 2017, 548, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Callura, J.M.; Dwyer, D.J.; Isaacs, F.J.; Cantor, C.R.; Collins, J.J. Tracking, tuning, and terminating microbial physiology using synthetic riboregulators. Proc. Natl. Acad. Sci. USA 2010, 107, 15898–15903. [Google Scholar] [CrossRef] [PubMed]
- Lehr, F.-X.; Hanst, M.; Vogel, M.; Kremer, J.; Göringer, H.U.; Suess, B.; Koeppl, H. Cell-free prototyping of AND-logic gates based on heterogeneous RNA activators. ACS Synth. Biol. 2019, 8, 2163–2173. [Google Scholar] [CrossRef]
- Bartoli, V.; Meaker, G.A.; Di Bernardo, M.; Gorochowski, T.E. Tunable genetic devices through simultaneous control of transcription and translation. Nat. Commun. 2020, 11, 2095. [Google Scholar] [CrossRef] [PubMed]
- Greco, F.V.; Pandi, A.; Erb, T.J.; Grierson, C.S.; Gorochowski, T.E. Harnessing the central dogma for stringent multi-level control of gene expression. Nat. Commun. 2021, 12, 1738. [Google Scholar] [CrossRef]
- Lee, Y.J.; Kim, S.-J.; Moon, T.S. Multilevel regulation of bacterial gene expression with the combined STAR and antisense RNA system. ACS Synth. Biol. 2018, 7, 853–865. [Google Scholar] [CrossRef] [PubMed]
- Westbrook, A.M.; Lucks, J.B. Achieving large dynamic range control of gene expression with a compact RNA transcription-translation regulator. Nucleic Acids Res. 2017, 45, 5614–5624. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-J.; Liu, P.; Nielsen, A.A.; Brophy, J.A.; Clancy, K.; Peterson, T.; Voigt, C.A. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat. Methods 2013, 10, 659–664. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Moon, T.S. Design rules of synthetic non-coding RNAs in bacteria. Methods 2018, 143, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Larson, M.H.; Greenleaf, W.J.; Landick, R.; Block, S.M. Applied force reveals mechanistic and energetic details of transcription termination. Cell 2008, 132, 971–982. [Google Scholar] [CrossRef] [PubMed]
- Cambray, G.; Guimaraes, J.C.; Mutalik, V.K.; Lam, C.; Mai, Q.-A.; Thimmaiah, T.; Carothers, J.M.; Arkin, A.P.; Endy, D. Measurement and modeling of intrinsic transcription terminators. Nucleic Acids Res. 2013, 41, 5139–5148. [Google Scholar] [CrossRef] [PubMed]
- McDowell, J.C.; Roberts, J.W.; Jin, D.J.; Gross, C. Determination of intrinsic transcription termination efficiency by RNA polymerase elongation rate. Science 1994, 266, 822–825. [Google Scholar] [CrossRef]
- iGEM Parts. Terminators/Catalog. Available online: http://parts.igem.org/Terminators/Catalog (accessed on 29 April 2021).
- Wandera, K.G.; Collins, S.P.; Wimmer, F.; Marshall, R.; Noireaux, V.; Beisel, C.L. An enhanced assay to characterize anti-CRISPR proteins using a cell-free transcription-translation system. Methods 2020, 172, 42–50. [Google Scholar] [CrossRef]
- Yeung, E.; Ng, A.; Kim, J.; Sun, Z.Z.; Murray, R.M. Modeling the effects of compositional context on promoter activity in an E. coli extract based transcription-translation system. In Proceedings of the 53rd IEEE Conference on Decision and Control, Los Angeles, CA, USA, 15–17 December 2014; pp. 5405–5412. [Google Scholar]
- Shin, J.; Noireaux, V. Efficient cell-free expression with the endogenous E. Coli RNA polymerase and sigma factor 70. J. Biol. Eng. 2010, 4, 8. [Google Scholar] [CrossRef]
- Kamionka, A.; Bogdanska-Urbaniak, J.; Scholz, O.; Hillen, W. Two mutations in the tetracycline repressor change the inducer anhydrotetracycline to a corepressor. Nucleic Acids Res. 2004, 32, 842–847. [Google Scholar] [CrossRef] [PubMed]
- Dragosits, M.; Nicklas, D.; Tagkopoulos, I. A synthetic biology approach to self-regulatory recombinant protein production in Escherichia coli. J. Biol. Eng. 2012, 6, 2. [Google Scholar] [CrossRef]
- Lim, H.G.; Kwak, D.H.; Park, S.; Woo, S.; Yang, J.-S.; Kang, C.W.; Kim, B.; Noh, M.H.; Seo, S.W.; Jung, G.Y. Vibrio sp. dhg as a platform for the biorefinery of brown macroalgae. Nat. Commun. 2019, 10, 2486. [Google Scholar] [CrossRef] [PubMed]
- Jeong, D.; Klocke, M.; Agarwal, S.; Kim, J.; Choi, S.; Franco, E.; Kim, J. Cell-free synthetic biology platform for engineering synthetic biological circuits and systems. Methods Protoc. 2019, 2, 39. [Google Scholar] [CrossRef] [PubMed]
- Weaver, K.E.; Chen, Y.; Miiller, E.M.; Johnson, J.N.; Dangler, A.A.; Manias, D.A.; Clem, A.M.; Schjodt, D.J.; Dunny, G.M. Examination of Enterococcus faecalis toxin-antitoxin system toxin Fst function utilizing a pheromone-inducible expression vector with tight repression and broad dynamic range. J. Bacteriol. 2017, 199, e00065-17. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Chan, C.T.; Slomovic, S.; Collins, J.J. Next-generation biocontainment systems for engineered organisms. Nat. Chem. Biol. 2018, 14, 530–537. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.T.; Lee, J.W.; Cameron, D.E.; Bashor, C.J.; Collins, J.J. ‘Deadman’ and ‘Passcode’ microbial kill switches for bacterial containment. Nat. Chem. Biol. 2016, 12, 82–86. [Google Scholar] [CrossRef]
- Kushwaha, M.; Rostain, W.; Prakash, S.; Duncan, J.N.; Jaramillo, A. Using RNA as molecular code for programming cellular function. ACS Synth. Biol. 2016, 5, 795–809. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, S.; Kim, J.; Kim, J. Multilevel Gene Regulation Using Switchable Transcription Terminator and Toehold Switch in Escherichia coli. Appl. Sci. 2021, 11, 4532. https://doi.org/10.3390/app11104532
Hong S, Kim J, Kim J. Multilevel Gene Regulation Using Switchable Transcription Terminator and Toehold Switch in Escherichia coli. Applied Sciences. 2021; 11(10):4532. https://doi.org/10.3390/app11104532
Chicago/Turabian StyleHong, Seongho, Jeongwon Kim, and Jongmin Kim. 2021. "Multilevel Gene Regulation Using Switchable Transcription Terminator and Toehold Switch in Escherichia coli" Applied Sciences 11, no. 10: 4532. https://doi.org/10.3390/app11104532
APA StyleHong, S., Kim, J., & Kim, J. (2021). Multilevel Gene Regulation Using Switchable Transcription Terminator and Toehold Switch in Escherichia coli. Applied Sciences, 11(10), 4532. https://doi.org/10.3390/app11104532




