Complementary Substrates-Brewery Wastewater and Piggery Effluent—Assessment and Microbial Community Profiling in a Hybrid Anaerobic Reactor
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling of Substrates
2.2. Anaerobic Digestion Experimental Set-Up
2.3. Analytical and Chromatograph Methods
2.4. Metagenomic Analysis
2.4.1. DNA Extraction and MiSeq Sequencing (Next-Generation Sequencing, NGS) of 16S rRNA Gene Amplicons
2.4.2. Data Analysis
3. Results and Discussion
3.1. Chemical Composition of Substrates
3.2. Anaerobic Digestion of the BWW and PE Mixture
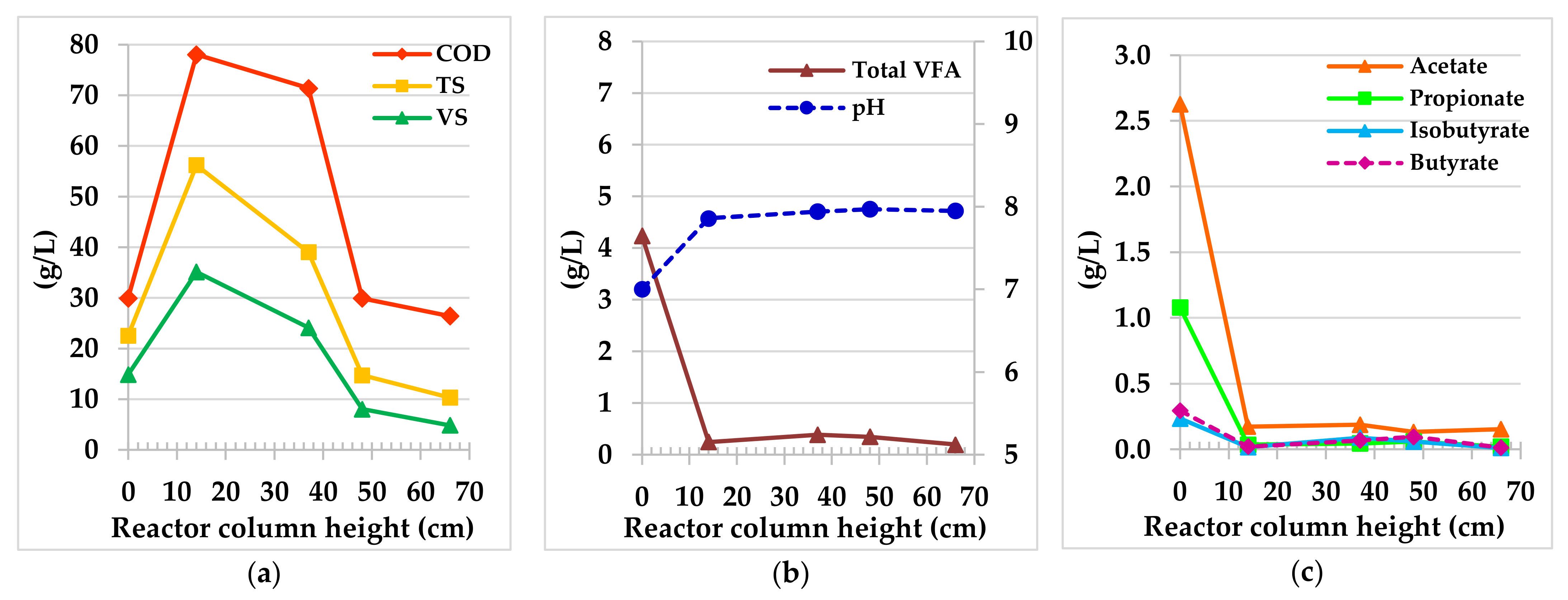
3.2.1. Hybrid Anaerobic Reactor Profile
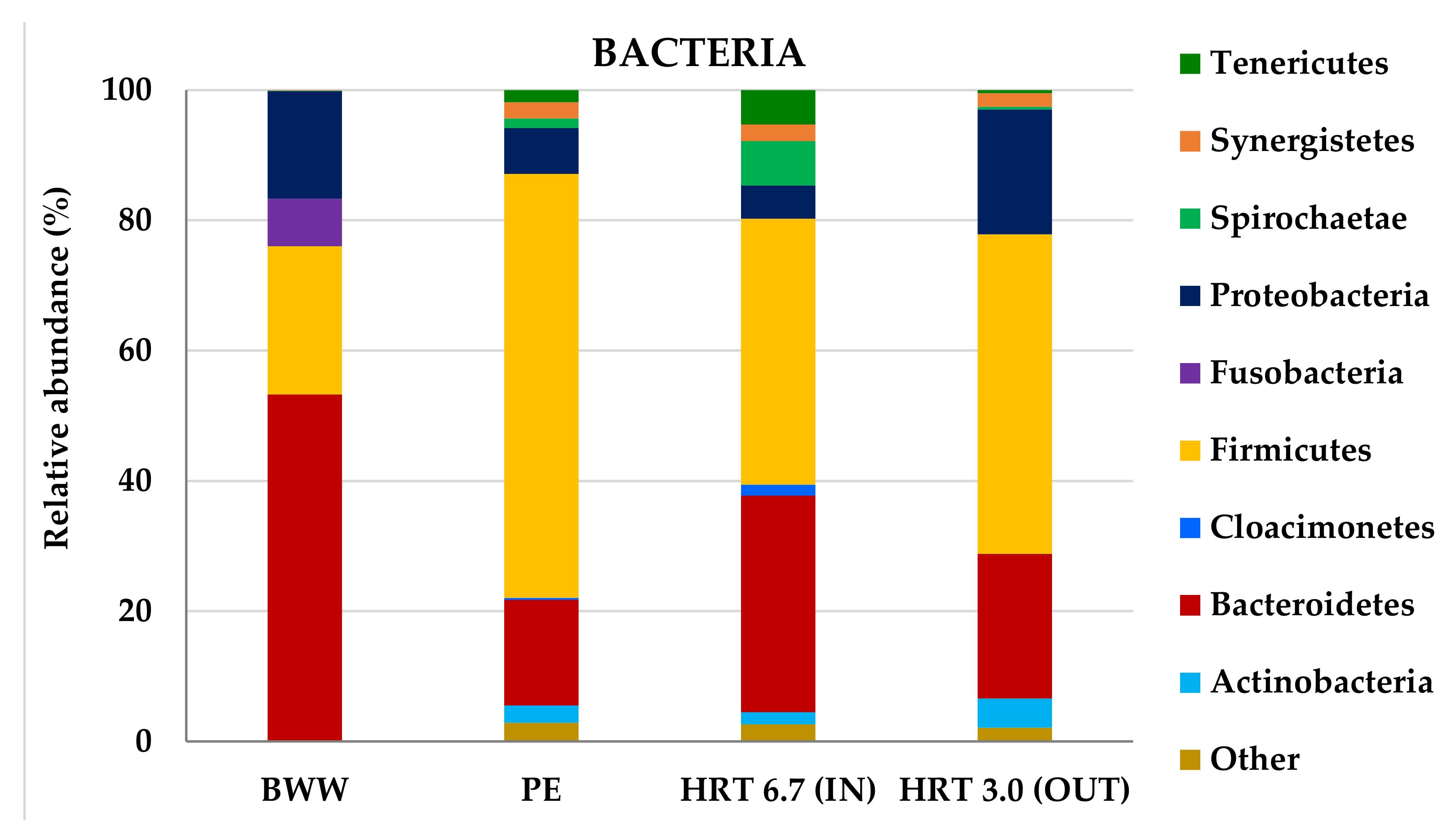
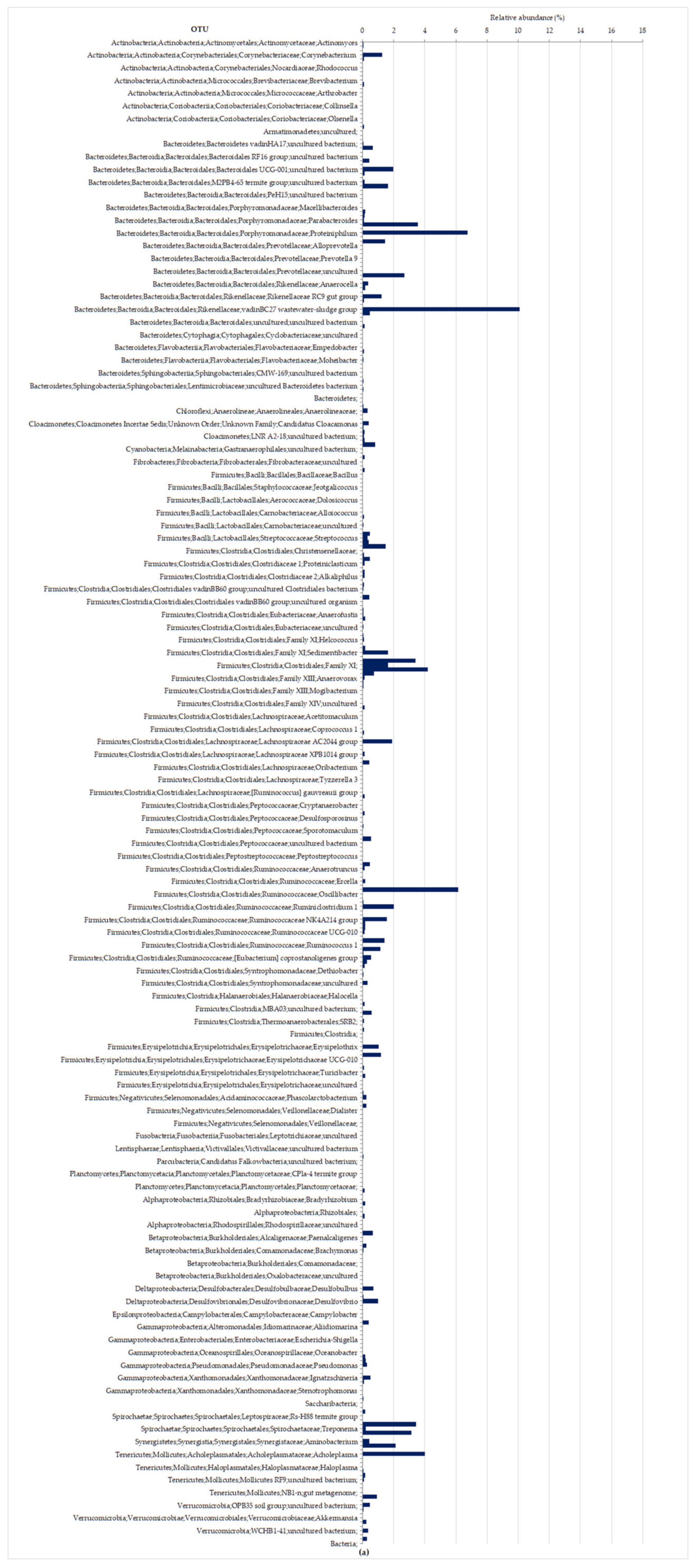
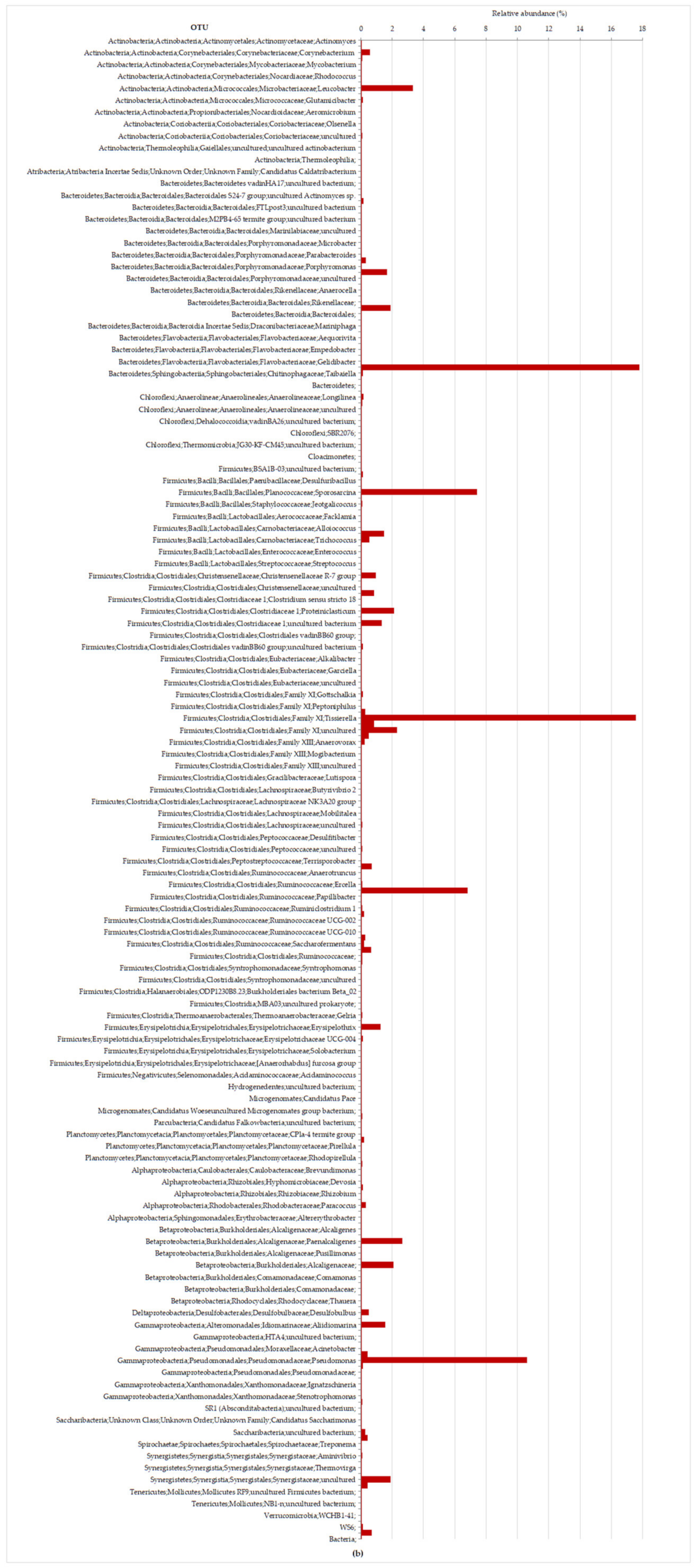
3.2.2. Molecular Characterization of Microbial Communities in the Hybrid Anaerobic Reactor
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Conway, J. Published in Statista, 12 October 2020. Available online: https://www.statista.com/statistics/270275/worldwide-beer-production/ (accessed on 27 February 2021).
- Simate, G.S.; Cluett, J.; Iyuke, S.E.; Musapatika, E.T.; Ndlovu, S.; Walubita, L.F.; Alvarez, A.E. The treatment of brewery wastewater for reuse: State of the art. Desalination 2011, 273, 235–247. [Google Scholar] [CrossRef]
- Amienyo, D.; Azapagic, A. Life cycle environmental impacts and costs of beer production and consumption in the UK. Int. J. Life Cycle Asses. 2016, 21, 492–509. [Google Scholar] [CrossRef]
- Shahbandeh, M. Published in Statista, 30 April 2020. Available online: https://www.statista.com/statistics/273232/net-pork-production-worldwide-by-country/ (accessed on 29 February 2021).
- Guia 333. 2020. Available online: https://www.3tres3.com.pt/estadisticas_suino/graficos/#5 (accessed on 29 February 2021).
- Dalibard, C. Livestock′s contribution to the protection of the environment. 1995. Available online: http://www.fao.org/ag/Aga/agap/FRG/FEEDback/War/V8180b/v8180b14.htm (accessed on 29 February 2021).
- Meegod, J.N.; Li, B.; Patel, K.; Wang, L.B. A Review of the Processes, Parameters, and Optimization of Anaerobic Digestion. Int. J. Environ. Res. Public Health 2018, 15, 2224. [Google Scholar] [CrossRef] [PubMed]
- Skerman, A.G. Methane Recovery and Use at Grantham Piggery—Addendum to Final Report; RIRDC Publication No. 13/107; RIRDC: Canbera, Australia, 2013; ISBN 978-1-74254-600-1. [Google Scholar]
- Ward, A.J.; Hobbs, P.J.; Holliman, P.J.; Jones, D.L. Optimisation of the anaerobic digestion of agricultural resources. Bioresour. Technol. 2008, 99, 7928–7940. [Google Scholar] [CrossRef] [PubMed]
- Náthia-Neves, G.; Berni, M.; Dragone, G.; Mussatto, S.I.; Forster-Carneiro, T. Anaerobic digestion process: Technological aspects and recent developments. Int. J. Environ. Sci. Technol. 2018, 15, 2033–2046. [Google Scholar] [CrossRef]
- Rajagopal, R.; Massé, D.I.; Singh, G. A critical review on inhibition of anaerobic digestion process by excess ammonia. Bioresour. Technol. 2013, 143, 632–641. [Google Scholar] [CrossRef]
- Mladenovska, Z.; Hartmann, H.; Kvist, T.; Sales-Cruz, M.; Gani, R.; Ahring, B.K. Thermal pretreatment of the solid fraction of manure: Impact on the biogas reactor performance and microbial community. Water Sci. Technol. 2006, 53, 59–67. [Google Scholar] [CrossRef]
- Neira, K.; Jeison, D. Anaerobic co-digestion of surplus yeast and wastewater to increase energy recovery in breweries. Water Sci. Technol. 2010, 61, 1129–1135. [Google Scholar] [CrossRef]
- Marques, I.P.; Texeira, A.; Rodrigues, L.; Dias, S.M.; Novais, J.M. Anaerobic co-treatment of olive mill and piggery effluents. Environ. Technol. 1997, 18, 265–274. [Google Scholar] [CrossRef]
- Marques, I.P.; Texeira, A.; Rodrigues, L.; Dias, S.M.; Novais, J.M. Anaerobic treatment of olive mill wastewater with digested piggery effluent. Water Environ. Res. 1998, 70, 1056–1061. [Google Scholar] [CrossRef]
- Marques, I.P. Anaerobic digestion treatment of olive mill wastewater for effluent re-use in irrigation. Desalination 2001, 137, 233–239. [Google Scholar] [CrossRef]
- Marques, I.P.; Sampaio, M.; Gonçalves, M. Process for the treatment of inhibiting/toxic organic waste and production of biofuels by obtaining an adapted microbial consortium. Portuguese Patent 105738, 29 October 2013. [Google Scholar]
- Sampaio, M.A.; Gonçalves, M.R.; Marques, I.P. Anaerobic digestion challenge of raw olive mill wastewater. Bioresour. Technol. 2011, 102, 10810–10818. [Google Scholar] [CrossRef] [PubMed]
- Assemany, P.; Marques, I.P.; Calijuri, M.L.; Reis, A. Complementarity of Substrates in Anaerobic Digestion of Wastewater Grown Algal Biomass. Waste Biomass Valor. 2020, 11, 5759–5770. [Google Scholar] [CrossRef]
- Eusébio, A.; Neves, A.; Marques, I.P. Structure of Microbial Communities when Complementary Effluents are Anaerobically Digested. Appl. Sci. 2021, 11, 1293. [Google Scholar] [CrossRef]
- Pérez-Pérez, T.; Pereda-Reyes, I.; Oliva-Merencio, D.; Zaiat, M. Anaerobic digestion technologies for the treatment of pig wastes. Cuba. J. Agr. Sci. 2016, 50, 343–354. [Google Scholar]
- Gonzalez-Tineo, P.A.; Durán-Hinojosa, U.; Delgadillo-Mirquez, L.R.; Meza-Escalante, E.R.; Gortáres-Moroyoqui, P.; Ulloa-Mercado, R.G.; Serrano-Palacios, D. Performance improvement of an integrated anaerobic-aerobic hybrid reactor for the treatment of swine wastewater. J. Water Process Eng. 2020, 34, 101164. [Google Scholar] [CrossRef]
- Deng, K.; Tang, L.; Lib, J.; Meng, J.; Li, J. Practicing anammox in a novel hybrid anaerobic-aerobic baffled reactor for treating high-strength ammonium piggery wastewater with low COD/TN ratio. Bioresour. Technol. 2019, 294, 122193. [Google Scholar] [CrossRef]
- Hui-ting, L.; Yong-feng, L. Performance of a hybrid anaerobic baffled reactor (HABR) treating brewery wastewater. In Proceedings of the 2010 International Conference on Mechanic Automation and Control Engineering, Wuhan, China, 26–28 June 2010. [Google Scholar]
- Gonçalves, M.R.; Freitas, P.; Marques, I.P. Bioenergy recovery from olive mill effluent in a hybrid reactor. Biomass Bioenergy 2012, 39, 253–260. [Google Scholar] [CrossRef]
- Marques, I.P. Anti-clogging packed bed for fixed bed digesters as support for microorganisms and gas-liquid separator. Portuguese Patent 106576, 28 April 2015. [Google Scholar]
- Mendonça, H.V.; Ometto, J.P.H.B.; Otenio, M.H.; Reis, A.J.D.; Marques, I.P.R. Bioenergy recovery from cattle wastewater in an UASB-AF. Water Sci. Technol. 2017, 76, 2268–2279. [Google Scholar] [CrossRef]
- Assemany, P.; Marques, I.P.; Calijuri, M.L.; da Silva, T.L.; Reis, A. Energetic valorization of algal biomass in a hybrid anaerobic reactor. J. Environ. Manag. 2018, 209, 308–315. [Google Scholar] [CrossRef]
- Standard Methods for Examination of Water and Wastewater, 22nd ed.; American Public Health Association: Washington, DC, USA; American Water Works Association: Denver, CO, USA; Water Environment Federation: Alexandria, VA, USA, 2012.
- Singleton, V.L.; Rossi, J.A., Jr. Colorimetry of Total Phenolics with Phosphomolybdic-Phosphotungstic Acid Reagents. Am. J. Eonol. Viticult. 1965, 16, 144–158. [Google Scholar]
- Zhou, J.; Bruns, M.A.; Tiedje, J.M. DNA Recovery from Soils of Diverse Composition. Appl. Environ. Microbiol. 1996, 62, 316–322. [Google Scholar] [CrossRef]
- Eusébio, A.; Tacão, M.; Chaves, S.; Tenreiro, R.; Almeida-Vara, E. Molecular assessment of microbiota structure and Dynamics along mixed olive oil and winery wastewaters biotreatment. Biodegradation 2011, 22, 773–795. [Google Scholar] [CrossRef] [PubMed]
- Illumina 16S Metagenomic Sequencing Library Preparation. 2013. Available online: https://emea.support.illumina.com/downloads/16s_metagenomic_sequencing_library_preparation.html (accessed on 1 December 2019).
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High resolution sample interference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2 Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, 1–10. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Glöckner, F.O.; Yilmaz, P.; Quast, C.; Gerken, J.; Beccati, A.; Ciuprina, A.; Bruns, G.; Yarza, P.; Peplies, J.; Westram, R.; et al. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 2017, 261, 169–176. [Google Scholar] [CrossRef]
- Parawira, W.; Kudita, I.; Nyandoroh, M.G.; Zvauya, R. A study of industrial anaerobic treatment of opaque beer brewery wastewater in a tropical climate using a full-scale UASB reactor seeded with activated sludge. Process Biochem. 2005, 40, 593–599. [Google Scholar] [CrossRef]
- Shao, X.; Peng, D.; Teng, Z.; Ju, X. Treatment of brewery wastewater using anaerobic sequencing batch reactor (ASBR). Bioresour. Technol. 2008, 99, 3182–3186. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.J.; Zheng, D.; Deng, L.W.; Wen, Q.; Liu, Y. Comparison of constructed wetland and stabilization pond for the treatment of digested effluent of swine wastewater. Environ. Technol 2014, 35, 2660–2669. [Google Scholar] [CrossRef] [PubMed]
- Boursier, H.; Béline, F.; Paul, E. Piggery wastewater characterisation for biological nitrogen removal process design. Bioresour. Technol. 2005, 96, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-S.; Han, G.-B. Pig slurry treatment by a hybrid multi-stage unit system consisting of an ATAD and an EGSB followed by a SBR reactor. Biosyst. Eng. 2012, 111, 243–250. [Google Scholar] [CrossRef]
- Yu, H.; Gu, G. Biomethanation of brewery wastewater using an anaerobic upflow blanket filter. J. Clean. Prod. 1996, 4, 219–223. [Google Scholar] [CrossRef]
- Cronin, C.; Lo, K.V. Anaerobic treatment of brewery wastewater using UASB reactors seeded with activated sludge. Bioresour. Technol. 1998, 64, 33–38. [Google Scholar] [CrossRef]
- Fernandes, T.; Fernandes, V.; Keesman, K.J.; Zeeman, G.; van Lier, J.B. Effect of ammonia on the anaerobic hydrolysis of cellulose and tributyrin. Biomass Bioenergy 2012, 47, 316–323. [Google Scholar] [CrossRef]
- Kayhanian, M. Ammonia inhibition in high-solids biogasification: An overview and practical solutions. Environ. Technol. 1999, 20, 355–365. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, X.; Zhou, J.; Yuan, Y.; Dai, Y.; Li, D.; Li, Z.; Liu, X.; Yan, Z. The dynamic changes and interactional networks of prokaryotic community between co-digestion and mono-digestions of corn stalk and pig manure. Bioresour. Technol. 2017, 225, 23–33. [Google Scholar] [CrossRef]
- Treu, L.; Tsapekos, P.; Peprah, M.; Campanaro, S.; Giacomini, A.; Corich, V.; Kougias, P.G.; Angelidaki, I. Microbial profiling during anaerobic digestion of cheese whey in reactors operated at different conditions. Bioresour. Technol. 2019, 275, 375–385. [Google Scholar] [CrossRef]
- Obaja, D.; Macee, S.; Costa, J.; Sans, C.; Mata-Alvarez, J. Nitrification, denitrification and biological phosphorus removal in piggery wastewater using a sequencing batch reactor. Bioresour. Technol. 2003, 87, 103–111. [Google Scholar] [CrossRef]
- Luo, Z.; Wang, D.; Yang, J.; Huang, H.; Su, G. Nitrogen removal from digested piggery wastewater using fermented superphosphate within the pretreatment stage and an MAP fertilizer pot test. J. Clean. Prod. 2019, 212, 372–380. [Google Scholar] [CrossRef]
- Kasinski, S. Mesophilic and Thermophilic Anaerobic Digestion of Organic Fraction Separated during Mechanical Heat Treatment of Municipal Waste. Appl. Sci. 2020, 10, 2412. [Google Scholar] [CrossRef]
- Shi, X.-S.; Dong, J.-J.; Yu, J.-H.; Yin, H.; Hu, S.-M.; Huang, S.-X.; Yuan, X.-Z. Effect of Hydraulic Retention Time on Anaerobic Digestion of Wheat Straw in the Semicontinuous Continuous Stirred-Tank Reactors. Biomed Res. Int. 2017, 2017, 2457805. [Google Scholar] [CrossRef]
- Lianhua, L.; Shuibin, H.; Yongming, S.; Xihui, K.; Junfeng, J.; Zhenhong, Y.; Dingfa, L. Anaerobic co-digestion of Pennisetum hybrid and pig manure: A comparative study of performance and microbial community at different mixture ratio and organic loading rate. Chemosphere 2020, 247, 125871. [Google Scholar] [CrossRef]
- Amaral, A.C.; Kunz, A.; Steinmetz, R.L.R.; Cantelli, F.; Scussiato, L.A.; Justi, K.C. Swine Effluent Treatment Using Anaerobic Digestion at Different Loading Rates. Eng. Agríc. 2014, 34, 567–576. [Google Scholar] [CrossRef][Green Version]
- Lo, K.V.; Liao, P.H.; Gao, Y.C. Anaerobic treatment of swine wastewater using hybrid UASB reactors. Bioresour. Technol. 1994, 47, 153–157. [Google Scholar] [CrossRef]
- Demirer, G.N.; Chen, S. Anaerobic digestion of dairy manure in a hybrid reactor with biogas recirculation. World J. Microb. Biot. 2005, 21, 1509–1514. [Google Scholar] [CrossRef]
- Nelson, M.C.; Morrison, M.; Yu, Z. A meta-analysis of the microbial diversity observed in anaerobic digesters. Bioresour. Technol. 2011, 102, 3730–3739. [Google Scholar] [CrossRef] [PubMed]
- Krause, L.; Diaz, N.N.; Edwards, R.A.; Gartemann, K.-H.; Krömeke, H.; Neuweger, H.; Pühler, A.; Runte, K.J.; Schlüter, A.; Stoye, J.; et al. Taxonomic composition and gene content of a methane-producing microbial community isolated from a biogas reactor. J. Biotechnol. 2008, 136, 91–101. [Google Scholar] [CrossRef]
- Schlüter, A.; Bekel, T.; Diaz, N.N.; Dondrup, M.; Eichenlaub, R.; Gartemann, K.-H.; Krahn, I.; Krause, L.; Krömeke, H.; Kruse, O.; et al. The metagenome of a biogas-producing microbial community of a production-scale biogas plant fermenter analysed by the 454-pyrosequencing technology. J. Biotechnol. 2008, 136, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Theuerl, S.; Klang, J.; Heiermann, M.; de Vrieze, J. Marker microbiome clusters are determined by operational parameters and specific key taxa combinations in anaerobic digestion. Bioresour. Technol. 2018, 263, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Gannoun, H.; Omri, I.; Chouari, R.; Eltaief, K.; Keskes, S.; Godon, J.J.; Hamdi, M.; Sghir, A.; Bouallagui, H. Microbial community structure associated with the high loading anaerobic codigestion of olive mill and abattoir wastewaters. Bioresour. Technol. 2016, 201, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Buhlmann, C.H.; Mickan, B.S.; Jenkins, S.N.; Tait, S.; Kahandawala, T.K.A.; Bahri, P.A. Ammonia stress on a resilient mesophilic anaerobic inoculum: Methane production, microbial community, and putative metabolic pathways. Bioresour. Technol. 2019, 275, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Hülsemann, B.; Zhou, L.; Merkle, W.; Hassa, J.; Müller, J.; Oechsner, H. Biomethane Potential Test: Influence of Inoculum and the Digestion System. Appl. Sci. 2020, 10, 2589. [Google Scholar] [CrossRef]
- Gao, R. Microbial diversity in a full-scale anaerobic reactor treating high concentration organic cassava wastewater. Afr. J. Biotechnol. 2012, 11, 6494–6500. [Google Scholar] [CrossRef]
- Guo, W.; Li, Y.; Zhao, K.; Xu, Q.; Jiang, H.; Zhou, H. Performance and Microbial Community Analysis of Anaerobic Digestion of Vinegar Residue with Adding of Acetylene Black or Hydrochar. Waste Biomass Valor. 2020, 11, 3315–3325. [Google Scholar] [CrossRef]
- Schauss, T.; Busse, H.-J.; Golke, J.; Kämpfer, P.; Glaeser, S.P. Moheibacter stercoris sp. nov., isolated from an input sample of a biogas plant. Int. J. Syst. Evol. Microbiol. 2016, 66, 2585–2591. [Google Scholar] [CrossRef]
- Zhang, R.-G.; Tan, X.; Zhao, X.-M.; Deng, J.; Lv, J. Moheibacter sediminis gen. nov., sp. nov., a member of the family Flavobacteriaceae isolated from sediment, and emended descriptions of Empedobacter brevis, Wautersiella falsenii and Weeksella virosa. Int. J. Syst. Evol. Microbiol. 2014, 64, 1481–1487. [Google Scholar] [CrossRef]
- Ariesyady, H.D.; Ito, T.; Okabe, S. Functional bacterial and archaeal community structures of major trophic groups in a full-scale anaerobic sludge digester. Water Res. 2007, 41, 1554–1568. [Google Scholar] [CrossRef]
- Goswami, R.; Chattopadhyay, P.; Shome, A.; Banerjee, S.N.; Chakraborty, A.K.; Mathew, A.K.; Chaudhury, S. An overview of physico-chemical mechanisms of biogas production by microbial communities: A step towards sustainable waste management. 3 Biotech 2016, 6, 72. [Google Scholar] [CrossRef]
- Cho, S.; Im, W.; Kim, D.; Kim, M.; Shin, H.; Oh, S. Dry anaerobic digestion of food waste under mesophilic conditions: Performance and methanogenic community analysis. Bioresour. Technol. 2013, 131, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wang, H.; Qiu, Y.; Ren, L.; Jiang, B. Microbial characteristics in anaerobic digestion process of food waste for methane production—A review. Bioresour. Technol. 2018, 248, 29–36. [Google Scholar] [CrossRef]
- Hardegen, J.; Latorre-Pérez, A.; Vilanova, C.; Günther, T.; Porcar, M.; Luschnig, O.; Simeonov, C.; Abendroth, C. Methanogenic community shifts during the transition from sewage mono-digestion to co-digestion of grass biomass. Bioresour. Technol. 2018, 265, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.J.; Lee, S.Y.; Bae, J.S.; Kang, J.G.; Kim, K.H.; Rhee, S.S.; Seo, D.C. Effect of volatile fatty acid concentration on anaerobic degradation rate from field anaerobic digestion facilities treating food waste leachate in South Korea. J. Chem. 2015, 2015, 9. [Google Scholar] [CrossRef]
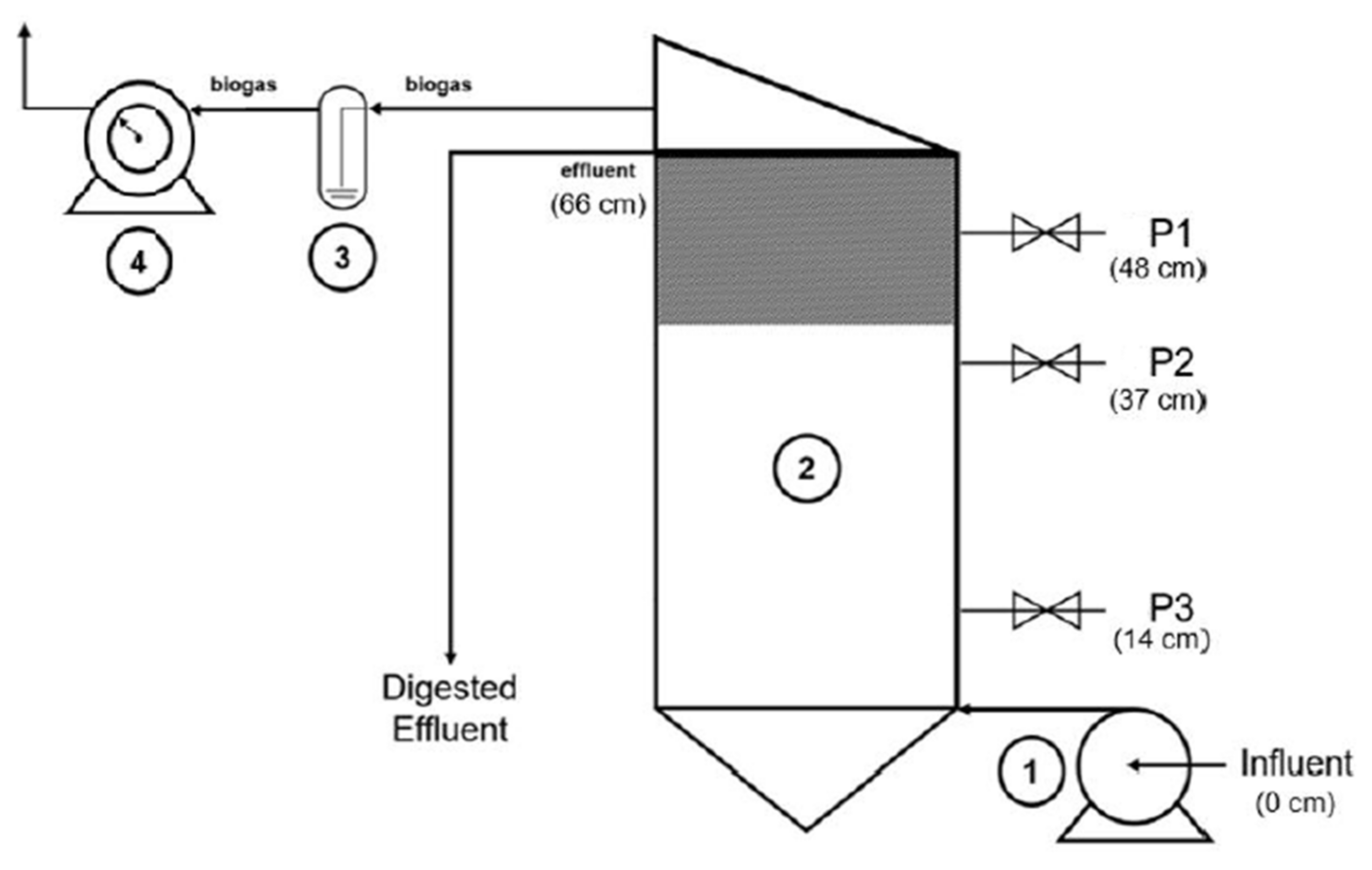
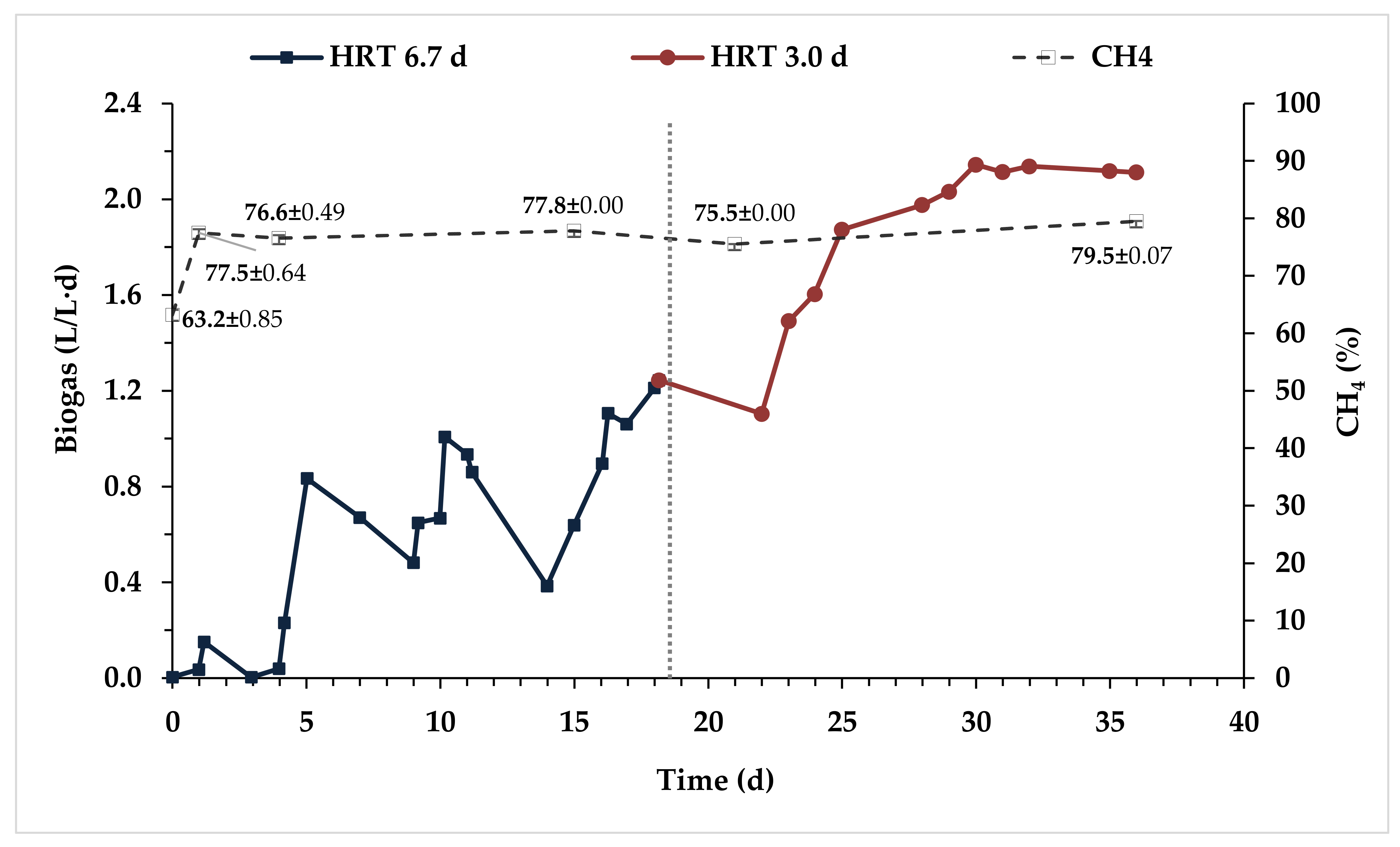


| Process Time (d) | HRT (d) | OLR (g COD/L·d) | OLR (g VS/L·d) |
|---|---|---|---|
| 0–19 | 6.7 | 4.50 | 2.22 |
| 20–37 | 3.0 | 10.0 | 4.96 |
| Parameters | Units | BWW | PE | ||||||
|---|---|---|---|---|---|---|---|---|---|
| This Work | [2] | [40] | [41] | This Work | [42] | [43] | [44] | ||
| pH | 5.1 | 3–12 | 4.5 | 3.2–3.9 | 7.3 | ||||
| COD | (g/L) | 7 ± 0 | 2–6 | 8.2–20 | 22.5–32.5 | 93 ± 5 | 0.82 ± 0.23 | 26–51 | 20–91 |
| TS | (g/L) | 4 ± 0 | 5.1–8.7 | 5.1–8.75 | 47 ± 1 | 28–54 | 23–61 | ||
| VS | (g/L) | 1 ± 0 | 32 ± 1 | 16–51 | |||||
| TKN | (mg/L) | 25 ± 0 | 25–80 | 0.02–0.03 | 320–450 | 5000 | 1500–6100 | ||
| NH4+-N | (g/L) | 0.01 ± 0 | 3 ± 0 | 0.51 ± 0.14 | 1.15–4.8 | ||||
| Total VFA | (g/L HAc) | 3.13 | 1–2.5 | 5.64 | |||||
| Acetic acid | (g/L) | 2.27 | 1.37 | ||||||
| Propionic acid | (g/L HAc) | 0.55 | 0.56 | ||||||
| Isobutyric acid | (g/L HAc) | 0.07 | 2.16 | ||||||
| Butyric acid | (g/L HAc) | 0.24 | 1.54 | ||||||
| Total phenols | (g/L caffeic acid) | 0.00 | 0.9 ± 0.0 | ||||||
| Parameters | HRT 6.7 d | HRT 3.0 d | |
|---|---|---|---|
| COD (g/L) | IN | 30 ± 1 | 30 ± 1 |
| COD (%) | R | 52 ± 2 | 12 ± 3 |
| TS (g/L) | IN | 23 ± 1 | 23 ± 1 |
| TS (%) | R | 8 ± 4 | 54 ± 2 |
| VS (g/L) | IN | 15 ± 1 | 15 ± 1 |
| VS (%) | R | 22 ± 5 | 68 ± 1 |
| TP (g/L caffeic acid) | IN | 0.4 ± 0 | 0.4 ± 0 |
| TP (%) | R | 38 ± 0 | 38 ± 1 |
| pH | IN | 7.0 | 7.0 |
| pH | OUT | 8.1 | 7.9 |
| TKN (g/L) | IN | 1.1 ± 0.0 | 1.1 ± 0.0 |
| TKN (%) | R | −4 ± 0.0 | 21 ± 0.0 |
| NH4+-N (g/L) | IN | 0.8 ± 0.0 | 0.8 ± 0.0 |
| NH4+-N (%) | R | −0.7 ± 0.0 | 6 ± 2.0 |
| Total VFA (g/L Hac) | IN | 4.24 | 4.24 |
| Total VFA (%) | R | 87 | 95 |
| Acetic acid (g/L) | IN | 2.63 | 2.63 |
| OUT | 0.24 | 0.15 | |
| Propionic acid (g/L HAc) | IN | 1.08 | 1.08 |
| OUT | 0.25 | 0.02 | |
| Isobutyric acid (g/L HAc) | IN | 0.23 | 0.23 |
| OUT | 0.03 | 0.01 | |
| Butyric acid (g/L HAc) | IN | 0.29 | 0.29 |
| OUT | 0.02 | 0.01 | |
| HRT | OLR | Biogas Yield | Methane Yield | References | ||
|---|---|---|---|---|---|---|
| (d) | (g COD/L·d) | (g VS/L·d) | (L biogas/g VSIN) | (L CH4/g VSIN) | (L CH4/g CODIN) | |
| 6.7 | 4.5 | 2.22 | 0.306 | 0.238 | 0.118 | This work |
| 3.0 | 10.0 | 4.96 | 0.245 | 0.195 | 0.096 | |
| 15 | 6.9 | 7.30 | 0.293 | 0.191 | - | [58] |
| - | 8.2 | - | - | 0.301 | [13] | |
| Samples | No. of Sequences | OTU | Shannon–Wiener Index | |
|---|---|---|---|---|
| Bacteria | Archaea | |||
| BWW | 194,323 | 334 | 3.25 | - |
| PE | 193,742 | 1623 | 3.52 | 2.95 |
| HRT 6.7 d (IN) | 168,868 | 1589 | 4.95 | 5.28 |
| HRT 3.0 d (OUT) | 220,894 | 891 | 4.03 | 1.23 |
| Sample | Relative Abundance (%) | ||
|---|---|---|---|
| Bacteria | Archaea | Unassigned | |
| BWW | 100.0 | 0.00 | 0.00 |
| PE | 99.73 | 0.27 | 0.00 |
| HRT 6.7 d (IN) | 99.26 | 0.71 | 0.03 |
| HRT 3.0 d (OUT) | 99.81 | 0.19 | 0.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eusébio, A.; Neves, A.; Marques, I.P. Complementary Substrates-Brewery Wastewater and Piggery Effluent—Assessment and Microbial Community Profiling in a Hybrid Anaerobic Reactor. Appl. Sci. 2021, 11, 4364. https://doi.org/10.3390/app11104364
Eusébio A, Neves A, Marques IP. Complementary Substrates-Brewery Wastewater and Piggery Effluent—Assessment and Microbial Community Profiling in a Hybrid Anaerobic Reactor. Applied Sciences. 2021; 11(10):4364. https://doi.org/10.3390/app11104364
Chicago/Turabian StyleEusébio, Ana, André Neves, and Isabel Paula Marques. 2021. "Complementary Substrates-Brewery Wastewater and Piggery Effluent—Assessment and Microbial Community Profiling in a Hybrid Anaerobic Reactor" Applied Sciences 11, no. 10: 4364. https://doi.org/10.3390/app11104364
APA StyleEusébio, A., Neves, A., & Marques, I. P. (2021). Complementary Substrates-Brewery Wastewater and Piggery Effluent—Assessment and Microbial Community Profiling in a Hybrid Anaerobic Reactor. Applied Sciences, 11(10), 4364. https://doi.org/10.3390/app11104364






