Abstract
Hydrocarbons can have very harmful effects on organisms and the environment, and conventional techniques for their removal are expensive and require the use of chemicals and long-term actions. Trichoderma is an ascomycete genus known to be active on different recalcitrant substrates, since it can produce a set of nonspecific extracellular enzymes generally involved in the degradation of lignin. However, the literature concerning the use of Trichoderma to degrade hydrocarbons is still limited. In this work we aimed to investigate the ability of Trichoderma to exploit used engine oil as its sole carbon source for prospective bioremediation of contaminated substrates. Four different strains belonging to Trichoderma asperellum and Trichoderma harzianum species were tested. The fungi were inoculated in direct contact with used engine oil, and after 45 days the samples were analyzed by gas chromatography/mass spectrometry (GC/MS). The results showed that all strains (except Trichoderma asperellum F1020) significantly changed the oil composition, decreasing the aromatic fraction in favor of the aliphatic one. T. harzianum F26, especially, showed a significant reduction of the BTEX (benzene, toluene, ethylbenzene, and the three xylene isomers) and alkylbenzenes fraction and an increase in short-chain aliphatics C1–C20. Enzymatic tests for laccase and peroxidase were also carried out, demonstrating that every strain seems to express a different mode of action.
1. Introduction
Fuel oils are commonly used worldwide. Despite significant improvements in handling, transportation, and containment, they still enter water and soil environments. The most severe damage to natural ecosystems has been reported after accidental releases [1]. Engine oil is a complex mixture of hydrocarbons, engine additives, and metals like aluminum, chromium, copper, iron, lead, manganese, nickel, and silicon. Engine oil contains hundreds of aliphatic, linear, or branched and aromatic hydrocarbons [2], most of which are toxic to living organisms [3,4]. Hydrocarbons can accumulate in animal organs directly or indirectly through the food chain [5]. In humans, prolonged exposure to or contact with high concentrations of hydrocarbons can cause liver or kidney disease, bone marrow damage, reproductive disorders, and increased risk of cancer [6,7]. Oil products can also have important repercussions on the environment and plants. In fact, they can create a thick layer on plant organs, reducing their metabolic activities and physical capacities [8].
Conventional treatments to remove hydrocarbons from the environment are efficient but very expensive and often require the use of chemicals in operational steps that alter the structure and composition of soil and subsoil, as well as changing the autochthonous microbial population [9,10]. An unconventional and environmentally friendly method of treating hydrocarbon-contaminated soils is bioremediation, which is the exploitation of natural microbial metabolism and co-metabolism to degrade toxic contaminants [4]. For these reasons, bioremediation is a “green” strategy, simple to design, and cost-effective.
Filamentous fungi are potential agents of degradation because they possess branched hyphae with which they can reach the deepest portions of contaminated substrates, digesting it through the secretion of extracellular enzymes [11]. Most fungi produce enzymes that include manganese peroxidase (MnP), laccases (Lac), cytochrome P450 monooxygenases, and epoxide hydrolases [12]. These extracellular enzymes, usually involved in the oxidation of lignin, are not substrate-specific and can therefore oxidize a wide range of xenobiotics, including pesticides, plastics, and hydrocarbons. Moreover, fungi can grow under different and extreme environmental conditions [13,14].
Among the different species of filamentous fungi, Trichoderma represents a genus with high potential. Trichoderma species belong to the Hypocreales order of the phylum Ascomycota, usually present in soils and plant roots [15]. This genus is one of the most widely distributed fungal groups in terrestrial and aquatic ecosystems [16]. It is resistant to most agrochemicals [17], heavy metals, pesticides, and polycyclic aromatic hydrocarbons (PAHs) [18]. The genus Trichoderma is often used as a biocontrol agent of plant pathogens since its species are mycoparasites and antibiotic producers. They can regulate the growth and incidence of microorganisms that cause diseases in several horticultural plants [19,20]. During the last few decades, many research teams started to focus their attention on the possibility of exploiting the properties of Trichoderma in the bioremediation of petroleum hydrocarbons. Chaîneau et al. [21] showed that some Trichoderma species contributed to the degradation of a saturated hydrocarbons fraction in a petroleum sample. T. harzianum, T. pseudokoningii, and T. viride can utilize pyrene as a carbon source [22,23]. Hadibarata and Tachibana [24] demonstrated that the strain Trichoderma S019 can degrade 73% of n-eicosane when glucose is applied as a carbon source. Other researchers reported the ability of T. hamatum, T. harzianum, T. koningii, T. viride, T. virens, and T. asperellum in the degradation of low-molecular-weight PAHs such as naphthalene and phenanthrene, or more complex PAHs such as anthracenebenzo[a]anthracene, benzo[a]fluoranthene, benzo[a]pyrene, and chrysene [12,25,26]. Argumedo-Delira et al. [27] reported the tolerance of 11 Trichoderma isolates to crude oil, naphthalene, phenanthrene, and benzo[a]pyrene using in vitro systems. Probable mechanisms for PAH degradation have been hypothesized for Trichoderma, including the production of laccases [28], peroxidases (Per) [29], and dioxygenases [30].
In light of these results, in this work we aimed to investigate the ability and the properties of different strains of Trichoderma (belonging to T. asperellum and T. harzianum) to be used as bioremediation agents, exploiting used engine oil as the sole carbon source. The strains were isolated from different environments and substrates. The degree of transformation of the motor oil used was evaluated by gas chromatography coupled to mass spectrometry (GC/MS), a technique commonly used for this purpose [31,32,33] and applied in this study to identify the classes of hydrocarbons mainly attacked by the different Trichoderma species.
2. Materials and Methods
2.1. Used Engine Oil
Used engine oil was chosen for the growth and degradation tests; it was a complex mixture of hydrocarbons, engine additives, and metals, provided by an Italian private company. Its composition was defined by gas chromatography coupled to mass spectrometry (GC/MS).
2.2. Biological Material
Trichoderma Strains and Their Phylogenetic Relationships
Four strains of Trichoderma (F12, F26, F1020, and F58) belonging to the Mycology Laboratory of the University of Pavia were utilized in this study. The fungal strains were isolated from four different environments: F12 and F26 from hydrocarbon-contaminated soils in Ecuador and Pakistan, respectively; F1020 from the rhizosphere of a plant growing in a hydroponic setting (toxin free) in Italy; and F58 from soil polluted with pesticides. The Mycology Laboratory previously used F12 and F1020 for tolerance tests with different types of hydrocarbon substances. They were chosen for this work to study their potential in the treatment of used engine oil. The strains were grown on potato dextrose agar (PDA) Petri dishes and incubated at 27 °C for one week. After this time, all the fungal strains were identified by morpho-dimensional examination under a light microscope, based on the nature of the mycelium and the different reproductive structures. Molecular characterization was performed to confirm the morphological identification. The fungal genomic DNA was extracted using the NucleoSpin Plant II by Macherey-Nagel (Bethlehem, PA, USA), with one hour of incubation at 65 °C after the addition of RNase, and then subjected to PCR amplification of the internal transcribed spacer (ITS) region of the ITS1-5.8S-ITS2 rDNA gene. The PCR reaction was performed on a Thermocycler Bio-Rad T100 in 20 µl reaction mixture containing 1× DREAM Taq Green PCR MasterMix reaction buffer (Thermo Scientifics, Pittsburg, PA, USA), 2 µl (5 µM) of each primer, 2 µl of DNA sample, and 4 µL of Nuclease Free water. The PCR program was as follows: denaturation by heating for 5 min at 95 °C, then 35 cycles of 30 s at 95 °C, 45 s at 50 °C, and 1 min at 72 °C, and a final elongation step for 10 min at 72 °C. The primers used were ITS1 (5′-TCCGTAGG TGAACCTGCGG-3′) and ITS4 (5′ TCCTCCGCTTATTGATATGC-3′) [34]. PCR products were purified with ExoSAP-IT (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocol. The amplified and purified DNA was sent to BMR Genomics (Padova, Italy), and the sequences were compared with target sequences using BLAST online (https://blast.ncbi.nlm.nih.gov/) and MEGA X 10.1.7.
The DNA sequences generated in this work, together with representative ITS sequences of other Trichoderma strains, were aligned using MEGA X [35]. The ITS sequences of 28 strains belonging to seven species were downloaded from Genebank. To show the phylogenetic positions of our strains, strains belonging to the Trichoderma clade Pachybasium A and the Pachybasium B clade Lixii/catoptron were included in this analysis. The maximum likelihood tree was obtained with 100 bootstrap replications, using the Jukes–Cantor model. The rates among sites were gamma-distributed with invariant sites (G+I).
2.3. Engine Oil Tolerance Test on the Fungal Strains
The ability of the Trichoderma strains to tolerate and grow on used engine oil was tested on Bushnell Haas mineral agar medium (BHA) Petri dishes complemented with 1% (v/v) of used engine oil. All the fungi were inoculated in the Petri dishes, and each one was replicated three times; BHA dishes with the inoculum but without the used engine oil were considered as control. After one week at 27 °C, the dishes were visually examined to compare the growth on the controls and the growth on the BHA and used engine oil. For evaluation of the tolerance, the dish coverage percentage by fungal mycelia growing on used engine oil compared to the coverage in dishes without used engine oil was assessed. In fact, Trichoderma is a genus that shows nonregular and non-diametric growth. In the early stages of growth, its development is submerged in the medium; it only secondarily emerges and on the surface develops scattered conidiophores aggregated into fascicles or pustules [36]. For these reasons, it is difficult to determine the growth of the strains based on their radius of growth; alternatively, to evaluate the coverage percentage of the dish can be helpful.
2.4. Analysis of Used Engine Oil Degradation
The Trichoderma strains were tested for their ability to degrade used engine oil by analyzing its composition before and after the fungal action. A 5 mm disk of each Trichoderma strain was collected from the outer part of the fungal colony grown for seven days on PDA Petri dishes, and each was aseptically inoculated into a 20 mL sterile glass bottle with a crimp seal filled with 5 mL of Bushnell Haas mineral medium (BH) and 1% (v/v) of used engine oil. The degradation study bottles were incubated at room temperature and 80 revolutions per minute (rpm) for 45 days. The amounts of hydrocarbon were determined by extracting the residual oil at the inoculum moment (T0) and after 45 days (T45), with four replicates for fungal strain each time. T0 was considered as control. At the moment of analysis, diethyl ether was added to the degradation bottles in a ratio of 1:1. The samples were shaken vigorously for 1 min, and the organic layer was allowed to separate for 5 min. At this point, 1 mL of the supernatant was collected from the degradation bottles, placed in a 1.5 mL Eppendorf tube, and centrifuged for 3 min at 3000 rpm. The supernatant was then transferred to a 1 mL GC/MS glass vial. The extracted oil was evaluated by GC/MS carried out on a Thermo Scientific DSQII single-quadrupole GC/MS system. The injection in the GC/MS system was performed at 250 °C in split mode. The oven temperature program was 70 °C for 1 min, 70–120 °C at 5 °C/min, 120–260 °C at 8 °C/min, and hold at 260 °C for 5 min. A Restek Rxi-5Sil MS 30 m × 0.25 mm × 0.25 µm film thickness capillary column was used with helium as the carrier gas at a constant flow rate of 1.0 mL/min. The transfer line temperature was 270 °C and the ion source temperature was 250 °C. Electron ionization mode was used with 70 eV, and the ions were registered in full scan mode in a mass range of m/z 35–800 amu. The chromatogram acquisition, detection of mass spectral peaks, and waveform processing were performed using Xcalibur MS Software Version 2.1 (Thermo Scientific Inc., Waltham, MA, USA). The assignment of chemical structures to chromatographic peaks was done based on comparison with the databases for the GC/MS National Institute of Standard and Technology (NIST) Mass Spectral Library (NIST 08) and Wiley Registry of Mass Spectral Data (8th Edition). The percentage content of each component was directly computed from the peak areas in the GC/MS chromatogram.
2.5. Colorimetric Screening for the Detection of Ligninolytic Enzymatic Activities
The presence of ligninolytic enzymes useful for the degradation of used engine oil was tested by three different approaches using Petri dishes supplemented with different substrates (Remazol Brilliant Blue R (RBBR), gallic acid, and guaiacol). Fungal cultures in malt extract agar (MEA) without the addition of the different substrates were used as controls and incubated under the same conditions. Three replicates of each culture were analyzed in each experiment. Each test was evaluated visually, and the difference between the tested dishes and the control ones was estimated qualitatively, giving a mark from 0 (no colorimetric difference with the control) to +++ (colorimetric difference considerably higher than the control).
Petri dishes were used with MEA and 5 g/L of gallic acid as a culture medium to determine the ligninolytic enzyme production [37,38]. The production of a brown shaded color produced in the agar has previously been strongly correlated with the ability of fungi to oxidize gallic acid by ligninolytic enzymes [39]. The strains were inoculated in the medium and incubated for seven days at 26 °C. Three replicates of each culture were analyzed. The Petri dishes were visually examined daily to monitor the production of a brown halo due to the oxidation of gallic acid [40].
Also, other Petri dishes with MEA and 0.2% (v/v) of guaiacol were used as a screening method to determine the Lac and Per production in the fungi. Guaiacol is a phenolic natural product, and the oxidation of guaiacol to its reddish-brown-colored form in agar is indicative of Lac and Per activity in fungi [41]. A plug of 7 mm diameter of each fungus was inoculated in the above medium, and cultures were incubated for seven days at 26 °C. Petri dishes were visually examined daily to monitor the production of a reddish-brown color due to the oxidation of guaiacol.
An RBBR dye-decolorization test was used to determine whether Trichoderma strains were capable of degrading PAHs and can also suggest the presence of Lac [37]. This is possible because RBBR is an anthracene derivative structurally similar to certain PAHs, and it is strongly decolorized by lignin-degrading fungi [42,43]. MEA Petri dishes supplemented with 50 mg/L of RBBR were used as culture medium. Each strain was inoculated onto the medium and incubated for seven days at 26 °C. Petri dishes were visually examined daily [44].
2.6. Statistical Analysis
GC/MS data were processed statistically using Past3 software [45], and one-way multivariate analysis of variance (MANOVA) or one-way nonparametric MANOVA (PERMANOVA) tests were applied to the data, according to the normality and homoscedasticity of the data, tested via Mardia’s multivariate skewness and kurtosis, Doornik–Hansen test, and Box’s M test (with significance at p < 0.05). The differences between each class at Time 0 and Time 45 were assessed using the two-tailed (Wilcoxon) Mann–Whitney U test. GC/MS data were also evaluated by principal component analysis (PCA).
3. Results
3.1. Identification of Trichoderma Strains and Analysis of Their Phylogenetic Relationships
The morphological analysis of the selected strains based on observations under a light microscope of the conidia and conidiophore morphology led to the identification of the four strains as F1020 T. asperellum and F12, F26, and F58 T. harzianum (Table 1). Moreover, the sequencing of the ITS regions, compared with target sequences with BLAST, confirmed this morphological identification. The tree obtained in the phylogenetic analysis (Figure 1) contains 32 taxa. Our strains of T. harzianum (F26, F58, and F12) formed a clade together with the other strains of T. harzianum obtained from Genebank. In particular, T. harzianum F26 and T. harzianum F58 seem to be closely related from a phylogenetic point of view. On the other hand, T. asperellum 1020 is grouped with other T. asperellum strains and other strains from the Trichoderma Pachybasium A complex (T. hamatum and T. pubescens).

Table 1.
Strains of Trichoderma identified by morphological and molecular identification.
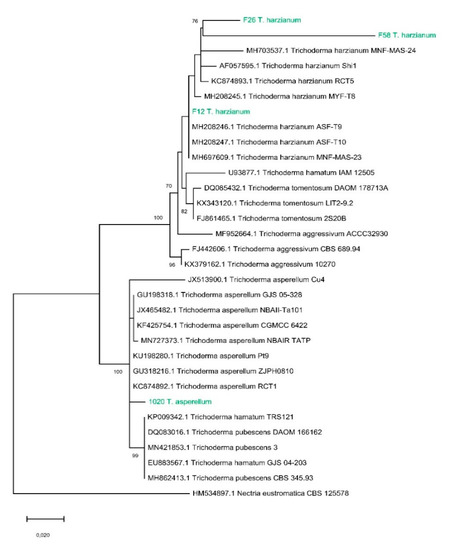
Figure 1.
Phylogenetic tree based on maximum likelihood analysis of the internal transcribed spacer (ITS) sequences. Nectria eustromatica was used as the outgroup. Maximum likelihood bootstrap values greater than 70% are given at the nodes. The four strains used in this work are highlighted in green.
3.2. Used Engine Oil Tolerance Test on Trichoderma Strains
After one week of incubation at 27 °C, the dishes were visually examined. All the fungi inoculated on BHA and used engine oil showed more abundant growth than did those on BHA alone (Table 2).

Table 2.
Results of the tolerance test on Trichoderma strains. The table reports the fungal growth on BHA + 1% (v/v) used engine oil compared to the BHA control and calculated as the percentage of dish coverage by fungal mycelia.
3.3. Used Engine Oil Composition
The hydrocarbon composition of the used engine oil was determined by GC/MS analysis (Figure 2). The compounds were grouped into the following classes according to Hostettler et al. [46]: C1–C20 aliphatics, C20–C50 aliphatics, BTEX (benzene, toluene, ethylbenzene, and the three xylene isomers), Alkyl Benzenes, Alkyl Indenes, Alkyl Tetralines, Alkyl Biphenyls, Polycyclic Aromatics, and Other Compounds (Table S1). The most abundant families in used motor oil are BTEX, C20–C50 aliphatics, and PAHs, which together make up 67% of the present compounds.
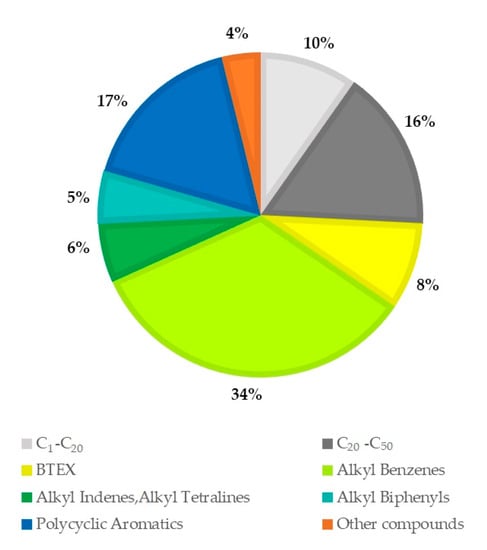
Figure 2.
Composition of the used engine oil determined by GC/MS analysis.
3.4. Analysis of Used Engine Oil Fungal Degradation
The percentage composition of the engine oil used by fungi was compared between T0 and T45 for each fungal strain by GC/MS analysis. The peaks detected on the chromatogram, indicative of the various compounds present in the used engine oil, were initially grouped into the macroclasses “Aliphatic”, “Aromatics”, and “Other Hydrocarbons” in order to identify the most degraded families of compounds. The three strains of T. harzianum (F12, F26, and F58) all significantly modified the composition of the used engine oil macroclasses (MANOVA or PERMANOVA, p < 0.05) (Figure 3). T. asperellum F1020 was the only strain that did not significantly modify the oil composition (Figure 3). The compounds were then grouped into the following microclasses: C1–C20 aliphatics, C20–C50 aliphatics, BTEX, Alkyl Benzenes, Alkyl Indenes, Alkyl Tetralines, Alkyl Biphenyls, Polycyclic Aromatics, and Other Compounds. The only fungal strain that produced significant changes in these groups after 45 days of action was T. harzianum F26 (PERMANOVA, p < 0.05, Figure 4), which reduced the presence of BTEX from 4.33% ± 0.38% to 2.98% ± 0.50% and that of Alkyl Benzenes from 14.45% ± 0.87% to 10.99% ± 1.57% significantly, while C1–C20 compounds increased significantly from 3.44% ± 0.17% to 4.49% ± 0.14% (two-tailed Mann–Whitney U test, p < 0.05) (Figure 5). The chromatograms are shown in Figures S1 and S2.
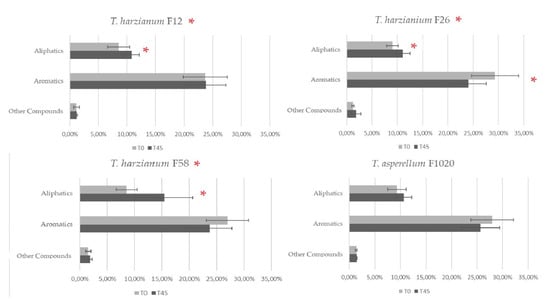
Figure 3.
Degradation of used engine oil by T. harzianum F12, F26, and F58 and T. asperellum F1020. The compounds were grouped in the macroclasses Aliphatics, Aromatics, and Other Compounds. A red asterisk next to the strain name indicates statistically significant data tested by multivariate analysis of variance (MANOVA) or nonparametric MANOVA (PERMANOVA). A red asterisk next to the bar indicates statistically significant data via two-tailed (Wilcoxon) Mann–Whitney U test between T0 and T45 for the macroclass.
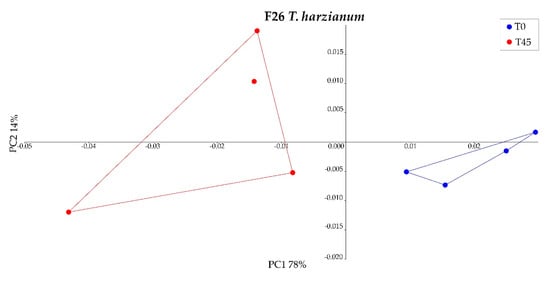
Figure 4.
Principal component analysis (PCA) of used engine oil microclass degradation by T. harzianum F26 at Time 0 (T0) and after 45 days (T45).
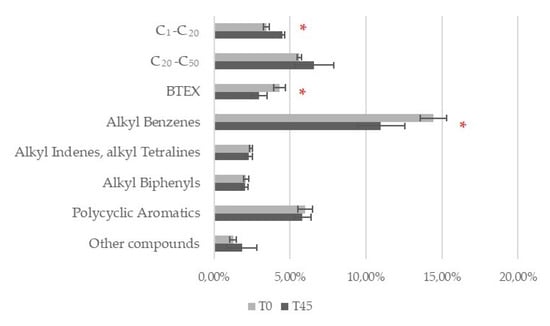
Figure 5.
Degradation of used engine oil by T. harzianum F26 at Time 0 (T0) and after 45 days (T45). The compounds were grouped into microclasses. A red asterisk indicates statistically significant data (two-tailed (Wilcoxon) Mann–Whitney U test).
3.5. Results of the Colorimetric Screening for the Detection of Fungal Enzymatic Activities
The gallic acid test was performed to detect ligninolytic secretion enzymes. Among the Trichoderma strains tested, only T. asperellum F1020 (Figure 6A) produced an intense dark shade in the agar around the growing colony, while T. harzianum F12 produced only a slight shade. T. harzianum F26 (Figure 6B) and T. harzianum F58 showed no activity. A different condition was reported for the guaiacol test. The four strains responded positively, indicating their ability to produce Lac and Per. Once again, T. asperellum F1020 (Figure 6C) and T. harzianum F12 were the most active, confirming the gallic acid test. T. harzianum F26 and T. harzianum F58 (Figure 6D) also showed a slight coloration of the agar. For the RBBR test, all the Trichoderma strains were able to decolorize the RBBR medium; T. harzianum F26 was the most active (Figure 6E). T. asperellum F1020 showed no activity (Figure 6F); this seems to confirm the results of GC/MS. The results of the colorimentric test are shown in Table 3.
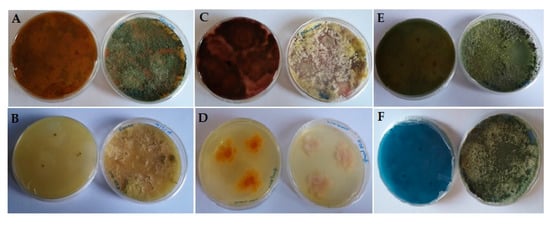
Figure 6.
Colorimetric screening. (A) Gallic acid test for T. asperellum F1020 (very high activity); (B) gallic acid test for T. harzianum F26 (no activity); (C) guaiacol test for T. asperellum F1020 (very high activity); (D) guaiacol test for T. harzianum F58 (no activity); (E) Remazol Brilliant Blue R (RBBR) test for T. harzianum F26 (very high activity); (F) RBBR test for T. asperellum F1020 (no activity). It is possible to appreciate the dark halos around the colonies for the gallic acid and guaiacol tests and the decolorization of the medium for the RBBR test.

Table 3.
Results of the colorimetric testing of Trichoderma strains.
4. Discussion
Over the years, many studies have focused on the selection of fungal strains capable of degrading hydrocarbons and the potential of Trichoderma in this field [26,27,47,48]. The aim of our work, however, was to compare the bioremediation actions of Trichoderma strains from different areas of the world and isolated from contaminated and noncontaminated substrates. Moreover, while most of the works already published showed the ability of Trichoderma to degrade crude oil or PAH mixtures [49,50,51], our investigation focused on the biodegradation of used engine oil. This pollutant is certainly very easy to find in the environment, as it is used daily all over the world, so finding appropriate bioremediation remedies for used engine oil could have a great beneficial impact.
The tolerance test showed that all strains could grow on used engine oil, meaning that the pollutant does not totally inhibit their growth and that they could use it as a nutrient source. However, there were differences in the growth of the strains. T. harzianum F26 was the best performing, while T. asperellum F1020 was the worst. Moreover, in the GC/MS analysis of hydrocarbon degradation after 45 days, T. asperellum F1020 was the only one that did not produce statistically significant changes in the used engine oil macroclasses, while T. harzianum F26, F12, and F58 changed them significantly. This result can be explained by the isolation substrate origin: T. asperellum F1020 was isolated from toxin-free hydroponic plant cultivation; on the contrary, the three strains of T. harzianum were isolated from contaminated substrates with pesticides or hydrocarbons and could be naturally adapted to the use of recalcitrant substances for their growth.
The literature reports several cases of T. asperellum being able to degrade hydrocarbons efficiently. For example, Zafra et al. [26] reported the case of T. asperellum removing large amounts of PAHs in soils contaminated with 1000 mg kg−1 of a mixture of phenanthrene, pyrene, and benzo[a]pyrene, degrading 74%, 63%, and 81%, respectively, in 14 days. Similarly, Husaini et al. [52] showed how T. asperellum TUB F-1067 (SA4), T. asperellum Tr48 (SA5), and T. asperellum TUB F-756 (SA6) strains can grow on a minimal substrate containing 1% (v/v) of used engine oil. However, these strains had been isolated from soil heavily contaminated with crude oil or used motor oil. Probably, highly tolerant organisms, having the capacity to use hydrocarbons as the only source of carbon, are more likely to adapt to a more polluted environment [53].
We also analyzed the composition of the used engine oil by grouping the compounds into microclasses. In this case, the only strain that showed statistically significant results was T. harzianum F26, which showed better degrading action on the BTEX and alkylbenzene family, increasing the content of C1–C20 alkanes. An explanation for this may be that T. harzianum F26 is more selective and prefers the degradation of BTEX and alkylbenzenes, while the other strains have more widespread and less marked degradation in all microclasses, making it more challenging to appreciate the results from a statistical point of view. Many authors have indicated T. harzianum as one of the Trichoderma species able to degrade unsaturated hydrocarbons, BTEX, and resins, as well as PAHs [21,54]. This species, in fact, can degrade up to about 10% of anthracene at a concentration of 400 mg kg−1 and 24.7% of pyrene at a concentration of 10 mg kg−1 [55]. The results obtained by the action of T. harzianum F26 partially agree with the literature as it acts more on BTEX and alkylbenzenes. Other works, however, have also demonstrated its ability to grow even on a mixture of complex hydrocarbons, such as crude oil, up to a concentration of 3% [56].
Finally, the colorimetric tests were aimed at identifying the ligninolytic enzymatic activities often associated with hydrocarbon degradation. Regarding the gallic acid test, only T. asperellum F1020 and T. harzianum F12 showed indications of ligninolytic enzyme production, and the guaiacol test showed that T. asperellum F1020 and T. harzianum F12 were able to produce Lac and Per. However, the guaiacol test also revealed that T. harzianum F26 and T. harzianum F58 showed slight activity. This result suggests that the guaiacol test may be more sensitive to the production of Lac and Per than the gallic acid test, even detecting milder activity. The RBBR test was used to determine the strains’ ability to degrade PAHs and indicate the presence of Lac [41]. T. harzianum F58 and T. harzianum F12 showed positive results, but T. harzianum F26 was the most active. F1020 showed no activity, consistent with the results obtained by GC/MS. The production of Lac and Per enzymes by these Trichoderma species has already been demonstrated by other authors [43,55]. Zafra et al. [26] reported that fungal catechol 1,2 dioxygenase, laccase, and peroxidase are involved in the degradation of PAHs by T. asperellum.
Considering the results of the colorimetric screening, T. asperellum F1020 was an excellent producer of Lac and Per. However, this result was not demonstrated in the GC/MS test, suggesting the action of other enzymes in the degradation of used engine oil. Many authors have shown the metabolic pathways activated by fungi to degrade hydrocarbons, and what emerges is that microfungi, in particular, mainly use cytochrome P450 (CYP) monoxygenases (EC:1.14.13.12) to degrade recalcitrant substances [11,56,57,58,59,60]. Moreover, as already mentioned, adaptation to the pollutant is fundamental for its efficient degradation. On the contrary, the strains that generated significant results under GC/MS did not provide equally evident signals in colorimetric tests. The reasons for this may be that T. harzianum F12, T. harzianum F26, and T. harzianum F58 could take greater advantage of the CYP complex mentioned above or could increase their production of Lac and Per in the presence of used engine oil. In fact, the colorimetric tests were performed on MEA without the pollutant present, as required by the protocol, but this may not have stimulated the production of the enzymes of interest. It is known that the concertation of lignin products and organic pollutants can induce the regulation of Lac and Per, especially in white-rot fungi [61,62], but a similar effect has been noted by Jonathan et al. [57] in T. harzianum. They found that the concentrations of Lac and Per produced by a strain of T. harzianum increased when the fungus grew in a polluted medium of crude oil compared with in oil-free medium.
Further research should focus on exploring the enzymatic mechanisms of hydrocarbon degradation in depth, especially for the strain T. harzianum F26, which was the most successful.
The experiments carried out in this work showed that the Trichoderma strains tested tolerate the presence of used engine oil and that they can use it for their growth. The degradation efficiency, however, varies greatly depending on the single strains and species, as demonstrated in GC/MS tests. The causes may be different environmental growing conditions and substrate types, but especially adaptation to the presence of toxic agents such as pesticides, PAHs, and chlorinated agents [62,63,64,65].
T. harzianum F26 was the strain that best responded to all the tests, and it was hypothesized that this strain might have developed adaptive mechanisms to be able to degrade this type of hydrocarbon pollutant.
Further studies are certainly needed to obtain more information on the chemical processes involved in bioremediation by Trichoderma. For this reason, an approach that integrates genetic, molecular, biochemical, and ecological studies may be a useful tool in understanding the behavior of this fungus and its metabolic processes. Besides this, genetic engineering could improve the degradative pathways and substrate specificity in Trichoderma and allow the prospective development of modified strains capable of degrading specific families of pollutants.
In conclusion, we can summarize the results of this work in three main messages:
- The efficiency of degradative activities seems to vary greatly depending on different species and single strains.
- The use of strains isolated from contaminated environments provides an advantage in bioremediation activities. This may depend on the adaptation of fungi to stress conditions that limit their growth and on improved enzymatic response to the degradation of recalcitrant substances.
- T. harzianum F26 should be subject to further studies, including field studies, to deepen and enhance its potential as a bioremediation agent.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-3417/10/9/3152/s1, Table S1: Used engine oil fingerprint based on characteristic ions (m/z). Division based on Hostelletler at al. [46], Figure S1. Chromatograms of the used engine oil at T0 and after 45 gg of Trichoderma treatment. The compounds were grouped in the macroclasses of aliphatics (black line), aromatics (red line) and other compounds (blue line). Treatment with T. harzianum F12 at T0 (A), and at T45 (B); treatment with T. harzianum F26 at T0 (C) and at T45 (D); treatment with T. harzianum F58 at T0 (E) and at T45 (F); treatment with T. asperellum F1020 at T0 (G) and at T45 (H), Figure S2. Chromatograms of the used engine oil treated with T. harzianum F26. The compounds were grouped in the microclasses of aliphatics C1-C50 (black line), BTEX (red line), Alkyl Benzenes (green line), Alkyl Indenes and Alkyl Tetralines (blue line), Alkyl Biphenyls (yellow line), Polycyclic Aromatics (purple blue) and other compounds (light blue line).
Author Contributions
Conceptualization: C.D., M.E.E.T., L.N., S.T., B.M.; methodology: C.D., M.E.E.T., L.N., B.M., F.C.; software: L.N., M.E.E.T., C.D.; data curation: M.E.E.T., L.N., C.D.; writing—original draft preparation: C.D., L.N.; writing—review and editing: L.N., C.D., S.T., G.C.; visualization: M.E.E.T., C.D., L.N.; supervision: S.T.; project administration: S.T.; funding acquisition: S.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research is part of a PhD project funded by Eni S.p.A.
Acknowledgments
We acknowledge Scuola di Alta Formazione Dottorale (SAFD) of the University of Pavia, Italy and ENI S.p.A.., and Ernesto Delgado Fernández of the Universidad Politecnica Salesiana, Cuenca, Ecuador, for kindly having made available the Trichoderma strain from Ecuador.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chaîneau, C.H.; Rougeux, G.; Yéprémian, C.; Oudot, J. Effects of nutrient concentration on the biodegradation of crude oil and associated microbial populations in the soil. Soil Biol. Biochem. 2005, 37, 1490–1497. [Google Scholar] [CrossRef]
- Wang, Z.; Fingas, M.; Blenkinsopp, S.; Sergy, G.; Landriault, M. Comparison of oil composition changes due to biodegradation and physical weathering in different oils. J. Chromatogr. A 1998, 809, 89–107. [Google Scholar] [CrossRef]
- Edewor, T.I.; Adelowo, O.O.; Afolabi, T.J. Preliminary studies into the biological activities of a broad spectrum disinfectant formulated from used engine oil. Pollut. Res. 2004, 234, 581–586. [Google Scholar]
- Wu, Y.; Lou, Y.; Zou, D.; Ni, J.; Liu, W.; Teng, Y.; Li, Z. Bioremediation of polycyclic aromatic hydrocarbons contaminated soil with Molilinia sp.: Degradation and microbial community analysis. Biodegradation 2008, 19, 247–257. [Google Scholar] [CrossRef]
- Mackay, D.; Fraser, A. Bioaccumulation of persistent organic chemicals: Mechanisms and models. Environ. Pollut. 2000, 111, 375–391. [Google Scholar] [CrossRef]
- Lloyd, A.C.; Cackette, T.A. Diesel engines: Environmental impact and control. J. Air Waste Manag. Assoc. 2001, 51, 809–847. [Google Scholar] [CrossRef]
- Mishra, S.; Jyot, J.; Kuhad, R.C.; Lal, B. Evaluation of inoculum addition to stimulate in situ bioremediation of oily-sludge-contaminated soil. Appl. Environ. Microbiol. 2001, 67, 1675–1681. [Google Scholar] [CrossRef]
- Amadi, A.; Abbey, S.D.; Nma, A. Chronic effects of oil spill on soil properties and microflora of a rainforest ecosystem in Nigeria. Water. Air Soil Pollut. 1996, 86, 1–11. [Google Scholar] [CrossRef]
- Asemoloye, M.D.; Ahmad, R.; Jonathan, S.G. Synergistic action of rhizospheric fungi with Megathyrsus maximus root speeds up hydrocarbon degradation kinetics in oil polluted soil. Chemosphere 2017, 187, 1–10. [Google Scholar] [CrossRef]
- Spini, G.; Spina, F.; Poli, A.; Blieux, A.L.; Regnier, T.; Gramellini, C.; Varese, G.C.; Puglisi, E. Molecular and microbiological insights on the enrichment procedures for the isolation of petroleum degrading bacteria and fungi. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Daccò, C.; Girometta, C.; Asemoloye, M.D.; Carpani, G.; Picco, A.M.; Tosi, S. Key fungal degradation patterns, enzymes and their applications for the removal of aliphatic hydrocarbons in polluted soils: A review. Int. Biodeterior. Biodegrad. 2020, 147. [Google Scholar] [CrossRef]
- Cerniglia, C.E.; Sutherland, G.R. Degradation of polycyclic aromatic hydrocarbons by fungi. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin, Germany, 2010; pp. 2079–2110. [Google Scholar]
- Magan, N. Fungi in extreme environment. In The Mycota Environmental and Microbial Relationships; Esser, K., Lemke, P.A., Eds.; Springer-Verlag: Berlin/Heidelberg, Germany, 2007; Volume IV, pp. 99–114. [Google Scholar]
- Deshmukh, R.; Khardenavis, A.A.; Purohit, H.J. Diverse metabolic capacities of fungi for bioremediation. Indian J. Microbiol. 2016, 56, 247–264. [Google Scholar] [CrossRef] [PubMed]
- Lieckfeldt, E.; Samuels, G.J.; Nirenberg, H.; Petrini, O. A morphological and molecular perspective of Trichoderma viride: Is it one or two species? Appl. Environ. Microbiol. 1999, 65, 2418–2428. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Druzhinina, I.; Kubick, C.P.; Xu, T. Trichoderma biodiversity in China: Evidence for a north to southern distribution of species in East Asia. FEMS Microbiol. Lett. 2005, 251, 251–257. [Google Scholar] [CrossRef]
- Ezzi, M.I.; Lynch, J.M. Biodegradation of cyanide by Trichoderma spp. and Fusarium spp. Enzyme Microb. Technol. 2005, 36, 849–854. [Google Scholar] [CrossRef]
- Tripathi, P.; Singh, P.C.; Mishra, A.; Chauhan, P.S.; Dwivedi, S.; Bais, R.T.; Tripathi, R.D. Trichoderma: A potential bioremediator for environmental cleanup. Clean Technol. Environ. Policy 2013, 15, 541–550. [Google Scholar] [CrossRef]
- Ávila-Miranda, M.E.; Herrera-Estrella, A.; Peña-Cabriales, J.J. Colonization of the rhizosphere, rhizoplane and endorhiza of garlic (Allium sativum L.) by strains of Trichoderma harzianum and their capacity to control allium white-rot under field conditions. Soil Biol. Biochem. 2006, 38, 1823–1830. [Google Scholar] [CrossRef]
- Rojo, F.G.; Reynoso, M.M.; Ferez, M.; Chulze, S.N.; Torres, A.M. Biological control by Trichoderma species of Fusarium solani causing peanut brown root rot under field conditions. Crop Prot. 2007, 26, 549–555. [Google Scholar] [CrossRef]
- Chaîneau, C.H.; Morel, J.; Dupont, J.; Bury, E.; Oudot, J. Comparison of the fuel oil biodegradation potential of hydrocarbon-assimilating microorganisms isolated from a temperate agricultural soil. Sci. Total Environ. 1999, 227, 237–247. [Google Scholar] [CrossRef]
- Ravelet, C.; Krivobok, S.; Sage, L.; Steiman, R. Biodegradation of pyrene by sediment fungi. Chemosphere 2000, 40, 557–563. [Google Scholar] [CrossRef]
- Saraswathy, A.; Hallberg, R. Degradation of pyrene by indigenous fungi from a former gasworks site. FEMS Microbiol. Lett. 2002, 210, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Hadibarata, T.; Tachibana, S. Microbial degradation of n-eicosane by filamentous fungi. In Interdisciplinary Studies on Environmental Chemistry e Environmental Research in Asia; Obayashi, Y., Isobe, T., Subramanian, A., Suzuki, S., Tanabe, S., Eds.; Terrapub: Tokyo, Japan, 2009; pp. 323–329. [Google Scholar]
- Atagana, H.I. Biodegradation of PAHs by fungi in contaminated soil containing cadmium and nickel ions. Afr. J. Biotechnol. 2009, 21, 5780–5789. [Google Scholar] [CrossRef]
- Zafra, G.; Moreno-Montano, A.; Absalon, A.E.; Cortes-Espinosa, D.V. Degradation of polycyclic aromatic hydrocarbons in soil by a tolerant strain of Trichoderma asperellum. Environ. Sci. Pollut. Res. 2015, 22, 1034–1042. [Google Scholar] [CrossRef] [PubMed]
- Argumedo-Delira, R.; Alarcon, A.; Ferrera-Cerrato, R.; Almaraz, J.J.; Pena-Cabriales, J.J. Tolerance and growth of 11 Trichoderma strains to crude oil, naphthalene, phenanthrene and benzo[a]pyrene. J. Environ. Manag. 2012, 95, S291–S299. [Google Scholar] [CrossRef] [PubMed]
- Cazares-Garcia, S.V.; Vazquez-Garciduenas, M.S.; Vazquez-Marrufo, G. Structural and phylogenetic analysis of laccases from Trichoderma: A bioinformatic approach. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Cristica, M.; Manoliu, A.; Barbaneagra, T.; Ciornea, E. Compared analysis of catalase and peroxidase activity in cellulolytic fungus Trichoderma reesei grown on medium with different concentrations of grinded wheat and barley straws. Sci. Ann. Alexandru Ioan Cuza Univ. Iaşi N. Ser. Sect. IIA Genet. Mol. Biol. 2011, 12, 89. [Google Scholar]
- Hadibarata, T.; Tachibana, S.; Itoh, K. Biodegradation of phenanthrene by fungi screened from nature. Pak. J. Biol. Sci. 2007, 10, 2535–2543. [Google Scholar] [CrossRef]
- Marchand, C.; St-Arnaud, M.; Hogland, W.; Bell, T.H.; Hijri, M. Petroleum biodegradation capacity of bacteria and fungi isolated from petroleum-contaminated soil. Int. Biodeterior. Biodegrad. 2017, 116, 48–57. [Google Scholar] [CrossRef]
- Ameen, F.; Moslem, M.; Hadi, S.; Al-Sabri, A.E. Biodegradation of diesel fuel hydrocarbons by mangrove fungi from Red Sea Coast of Saudi Arabia. Saudi J. Biol. Sci. 2016, 23, 211–218. [Google Scholar] [CrossRef]
- Yao, L.; Teng, Y.; Luo, Y.; Christie, P.; Ma, W.; Liu, F.; Wu, Y.; Luo, Y.; Li, Z. Biodegradation of polycyclic aromatic hydrocarbons (PAHs) by Trichoderma reesei FS10-C and effect of bioaugmentation on an aged PAH-contaminated soil. Bioremediat. J. 2015, 19, 9–17. [Google Scholar] [CrossRef]
- Martin, K.J.; Rygiewicz, P.T. Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol. 2005, 5. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Gams, W.; Bissett, J. Morphology and identification of Trichoderma. In Trichoderma and Gliocladium. Basic Biology, Taxonomy and Genetics; Kubicek, C.P., Harman, G.E., Eds.; Taylor & Francis: London, UK, 1998; Volume 1, pp. 3–34. [Google Scholar]
- Scharf, M. Termites as targets and models for biotechnology. Ann. Rev. Entomol. 2015, 60, 77–102. [Google Scholar] [CrossRef] [PubMed]
- Yanto, D.H.Y.; Tachibana, S. Biodegradation of petroleum hydrocarbons by a newly isolated Pestalotiopsis sp. NG007. Int. Biodeterior. Biodegrad. 2013, 85, 438–450. [Google Scholar] [CrossRef]
- Shleev, S.V.; Morozova, O.V.; Nikitina, O.V.; Gorshina, E.S.; Rusinova, T.V.; Serezhenkov, V.A.; Burbaev, D.S.; Gazaryan, I.G.; Yaropolov, A.I. Comparison of physico-chemical characteristics of four laccases from different basidiomycetes. Biochimie 2004, 86, 693–703. [Google Scholar] [CrossRef]
- Lee, H.; Jang, Y.; Choi, Y.S.; Kim, M.J.; Lee, J.; Lee, H.; Hong, J.H.; Min Lee, Y.; Kim, G.H.; Kim, J.J. Biotechnological procedures to select white rot fungi for the degradation of PAHs. J. Microbiol. Methods 2014, 97, 56–62. [Google Scholar] [CrossRef]
- Kumar, V.V.; Rapheal, V.S. Induction and purification by three-phase partitioning of aryl alcohol oxidase (AAO) from Pleurotus ostreatus. Appl. Biochem. Biotechnol. 2011, 163, 423–432. [Google Scholar] [CrossRef]
- Hammel, K.E. Oxidation of aromatic pollutants by lignin-degrading fungi and their extracellular peroxidases. In Metal Ions in Biological Systems; Sigel, H., Sigel, A., Eds.; Marcel Dekker, Inc.: New York, NY, USA, 1992; pp. 41–60. [Google Scholar]
- Batista-García, R.A.; Kumar, V.V.; Ariste, A.; Tovar-Herrera, O.E.; Savary, O.; Peidro-Guzmán, H.; González-Abradelo, D.; Jackson, S.A.; Dobson, A.D.W.; Sánchez-Carbente, M.d.R.; et al. Simple screening protocol for identification of potential mycoremediation tools for the elimination of polycyclic aromatic hydrocarbons and phenols from hyperalkalophile industrial effluents. J. Environ. Manag. 2017, 198, 1–11. [Google Scholar] [CrossRef]
- Lee, H.; Choi, Y.S.; Kim, M.J.; Huh, N.Y.; Kim, G.H.; Lim, Y.W.; Kang, S.M.; Cho, S.T.; Kim, J.J. Degrading ability of oligocyclic aromates by Phanerchaete sordida selected via screening of white rot fungi. Folia Microbiol. 2010, 55, 447–453. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontologia Electronica 2001, 4, 9. [Google Scholar]
- Hostettler, F.D.; Lorenson, T.D.; Bekins, B.A. Petroleum fingerprinting with organic markers. Environ. Forensics 2013, 14, 262–277. [Google Scholar] [CrossRef]
- Zafra, G.; Cortés-Espinosa, D.V. Biodegradation of polycyclic aromatic hydrocarbons by Trichoderma species: A mini review. Environ. Sci. Pollut. Res. 2015, 22, 19426–19433. [Google Scholar] [CrossRef] [PubMed]
- Silva, I.S.; Grossman, M.; Durrant, L.R. Degradation of polycyclic aromatic hydrocarbons (2–7 rings) under microaerobic and very-low-oxygen conditions by soil fungi. Int. Biodeterior. Biodegrad. 2009, 63, 224–229. [Google Scholar] [CrossRef]
- Hamzah, A.; Zarin, M.A.; Hamid, A.A.; Omar, O.; Senafi, S. Optimal physical and nutrient parameters for growth of Trichoderma virens UKMP-1M for heavy crude oil degradation. Sains Malaysiana 2012, 41, 71–79. [Google Scholar]
- Cobas, M.; Ferreira, L.; Tavares, T.; Sanroman, M.A.; Pazos, M. Development of permeable reactive biobarrier for the removal of PAHs by Trichoderma longibrachiatum. Chemosphere 2013, 91, 711–716. [Google Scholar] [CrossRef]
- Husaini, A. Bioremediation of crude oil by different fungal genera. Asian J. Plant Biol. 2014, 2, 11–18. [Google Scholar]
- Husaini, A.; Roslan, H.A.; Hii, K.S.Y.; Ang, C.H. Biodegradation of aliphatic hydrocarbon by indigenous fungi isolated from used motor oil contaminated sites. World J. Microbiol. Biotechnol. 2008, 24, 2789–2797. [Google Scholar] [CrossRef]
- Matsubara, M.; Lynch, J.M.; De Leij, F.A.A.M. A simple screening procedure for selecting fungi with potential for use in the bioremediation of contaminated land. Enzyme Microb. Technol. 2006, 39, 1365–1372. [Google Scholar] [CrossRef]
- Romero, M.C.; Salvioli, M.L.; Cazau, M.C.; Arambarri, A.M. Pyrene degradation by yeasts and filamentous fungi. Environ. Pollut. 2002, 117, 159–163. [Google Scholar] [CrossRef]
- Kadri, T.; Rouissi, T.; Kaur Brar, S.; Cledon, M.; Sarma, S.; Verma, M. Biodegradation of polycyclic aromatic hydrocarbons (PAHs) by fungal enzymes: A review. J. Environ. Sci. 2017, 51, 52–74. [Google Scholar] [CrossRef]
- Prenafeta-Boldú, F.X.; de Hoog, G.S.; Summerbell, R.C. Fungal communities in hydrocarbon degradation. In Microbial Communities Utilizing Hydrocarbons and Lipids: Members, Metagenomics and Ecophysiology. Handbook of Hydrocarbon and Lipid Microbiology; McGenity, T., Ed.; Springer: Cham, Switzerland, 2019; pp. 1–36. [Google Scholar]
- Jonathan, S.G.; Asemoloye, M.; Ahmad, R.; Olawuyi, O.J.; Adejoye, D. Response of a newly identified Trichoderma harzianum ky488466 to crude oil pollution and its expression of peroxidase genes. SSRN 2017, 1–10. [Google Scholar] [CrossRef]
- Durairaj, P.; Malla, S.; Nadarajan, S.P.; Lee, P.G.; Jung, E.; Park, H.H.; Kim, B.G.; Yun, H. Fungal cytochrome P450 monooxygenases of Fusarium oxysporum for the synthesis of ω-hydroxy fatty acids in engineered Saccharomyces cerevisiae. Microb. Cell Fact. 2015, 14, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Asemoloye, M.D.; Jonathan, S.G.; Ahmad, R. Synergistic plant-microbes interactions in the rhizosphere: A potential headway for the remediation of hydrocarbon polluted soils. Int. J. Phytoremediat. 2019, 2, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Janusz, G.; Kucharzyk, K.H.; Pawlik, A.; Staszczak, M.; Paszczynski, A.J. Fungal laccase, manganese peroxidase and lignin peroxidase: Gene expression and regulation. Enzyme Microb. Technol. 2013, 52, 1–12. [Google Scholar] [CrossRef]
- Yang, J.; Li, W.; Bun Ng, T.; Deng, X.; Lin, J.; Ye, X. Laccases: Production, expression regulation, and applications in pharmaceutical biodegradation. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Casieri, L.; Anastasi, A.; Prigione, V.; Varese, G.C. Survey of ectomycorrhizal, litter-degrading, and wood-degrading basidiomycetes for dye decolorization and ligninolytic enzyme activity. Int. J. Gen. Mol. Microbiol. 2010, 98, 483–504. [Google Scholar] [CrossRef]
- Jeon, J.R.; Baldrian, P.; Murugesan, K.; Chang, Y.S. Laccase-catalysed oxidations of naturally occurring phenols: From in vivo biosynthetic pathways to green synthetic applications. Microb. Biotechnol. 2012, 5, 318–332. [Google Scholar] [CrossRef]
- Liu, J.; Liu, W.; Cai, Y.; Liao, X. Laccase production by Trametes hirsuta, characterization, and its capability of decoloring chlorophyll. Pol. J. Microbiol. 2014, 63, 323–333. [Google Scholar] [CrossRef]
- Si, J.; Peng, F.; Cui, B. Purification, biochemical characterization and dye decolorization capacity of an alkali-resistant and metal-tolerant laccase from Trametes pubescens. Bioresour. Technol. 2013, 128, 49–57. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).