1. Introduction
Biogas plants in Germany receive a bonus payment for power generation, which is guaranteed by the German Renewable Energy Source Act (EEG) for a period of 20 years after installation [
1]. Many biogas plants will run out of this bonus payment during the next ten years. Thus, biogas plants have to work more efficiently for economic reasons. The biochemical methane potential (BMP) test is a common method to determine the maximum theoretical methane yield of a substrate. It has already been used for several years for dimensioning biogas plants [
2]. High accuracy of the BMP test will be necessary to precisely predict economic viability in the future.
Various protocols are available, all with the aim of achieving a high reproducibility of the BMP test. Besides the European standards (Angelidaki et al. [
3] and Holliger et al. [
4]), the VDI Standard 4630 [
5] is the most commonly used protocol. These protocols show the same basic structure: (i) performing three technical repetitions, (ii) applying prescribed inoculum/substrate ratio (ISR), (iii) measuring methane yield of pure inoculum as a blank, and (iv) measuring methane yield of standard substrates as positive control. In each of the protocols it is pointed out, how important the inoculum, the digestion system, and the working precision are to achieve a high reproducibility.
Various inter-laboratory tests were conducted to check the accuracy of the BMP by using different digestion systems, inocula, and protocols. Raposo et al. [
2] conducted an inter-laboratory test with 17 laboratories using starch, cellulose, and gelatin as substrates. The results showed a coefficient of variation (CV) of around 8%–11%. Similar results were reported by a German inter- laboratory test of KTBL/VDLUFA based on VDI Standard 4630. This test was done with different substrates and around 30 laboratories over several years. The CV excluding outliers was 8%–12% [
6]. The impact of repeating BMP tests over several years was small, it only helped to reduce the number of outliers [
7]. The Ècole Polytechnique Fèdèrale de Lausanne carried out inter- laboratory-tests with stricter specifications and a repetition after a few months [
4]. The results without eliminating the outliners revealed a CV of 15%–17% [
8]. Cresson et al. [
9] investigated differences in the CV when using different BMP measuring protocols. Free choice of the used protocol was compared to the fix protocol for each laboratory, using a mineral substrate, NaHCO3 as pH buffer and an ISR > 2. The results showed a deviation of about 20% between the free and the fixed protocol. The measurement variability was not affected by different measuring protocols. Pham et al. [
10] reported similar differences by comparing protocols of VDI 4630 [
5] and Sommer et al. [
11].
Several studies were carried out to find reasons for deviations in measurement results (
Table 1). Special focus was laid on the effect of the used inoculum. One of the main categorization criteria was the origin of the inoculum. Vrieze et al. [
12] and Regueiro et al. [
13] reported differing microbial community composition for different inocula from full scale biogas plants. Thus, inocula from different origins might affect the methane yield depending on their microbial community composition. Li et al. [
14] tested sludge from chicken manure and municipal wastewater treatment plants as inocula using similar substrates. The inoculum from wastewater treatment led to a higher biodegradability of the inoculum/substrate mixture. Pozdniakova et al. [
15] revealed converse behavior, they detected a higher methane yield for inocula from a municipal solid waste landfill plant compared to inocula from wastewater treatment and other origins, when using animal by-products as substrate. Elbeshbishy et al. [
16] investigated the methane yields of two different inocula. Higher methane yields were also achieved with inocula taken from anaerobic digesters. Dechrugsa et al. [
17] reported a significant difference in methane yields by using inocula from two different full-scale biogas plants. Another impact was reported by Chamy and Ramos [
18], where a well-adapted inoculum from digested turkey manure produced a higher methane yield when using turkey manure as a substrate. Vrieze et al. [
12] measured the methane yield of four substrates using four inocula from different origins. The effects of inocula on the results depended strongly on the substrates. Koch et al. [
19] reported similar results. A comparison of three inocula from different origins revealed repeatedly no significant effects in methane yield for three substrates. Only for cellulose—a common substrate standard—significant effects were observed, leading to the conclusion that the methane production rate was affected by the used inoculum.
Beside the source of the inoculum, there are more possibilities influencing the methane yield, such as the incubation time and the trace elements (TE). VDI 4630 [
5] prescribes the incubation of the inoculum to limit the methane production of the blanks. Other studies report no effect of incubation on their results [
12,
14]. Angelidaki et al. [
3] recommend adding TE and vitamins.
Further effects on BMP were investigated by Strömberg et al. [
20]. They examined the influence of experimental conditions and their correction to standard conditions. The biggest effect was found for samples with low methane yield, because of the high impact of the size of headspace, which determines the volume relation between produced biogas and flushing gas. The use of standard temperature showed a deviation of about 10% compared to results without correction. Also, the ambient pressure shows an impact on the biogas yield potential. It is shown that the ambient pressure can differ between 63.1 kPa (La Paz) and 103.6 kPa (sea level) [
20,
21].
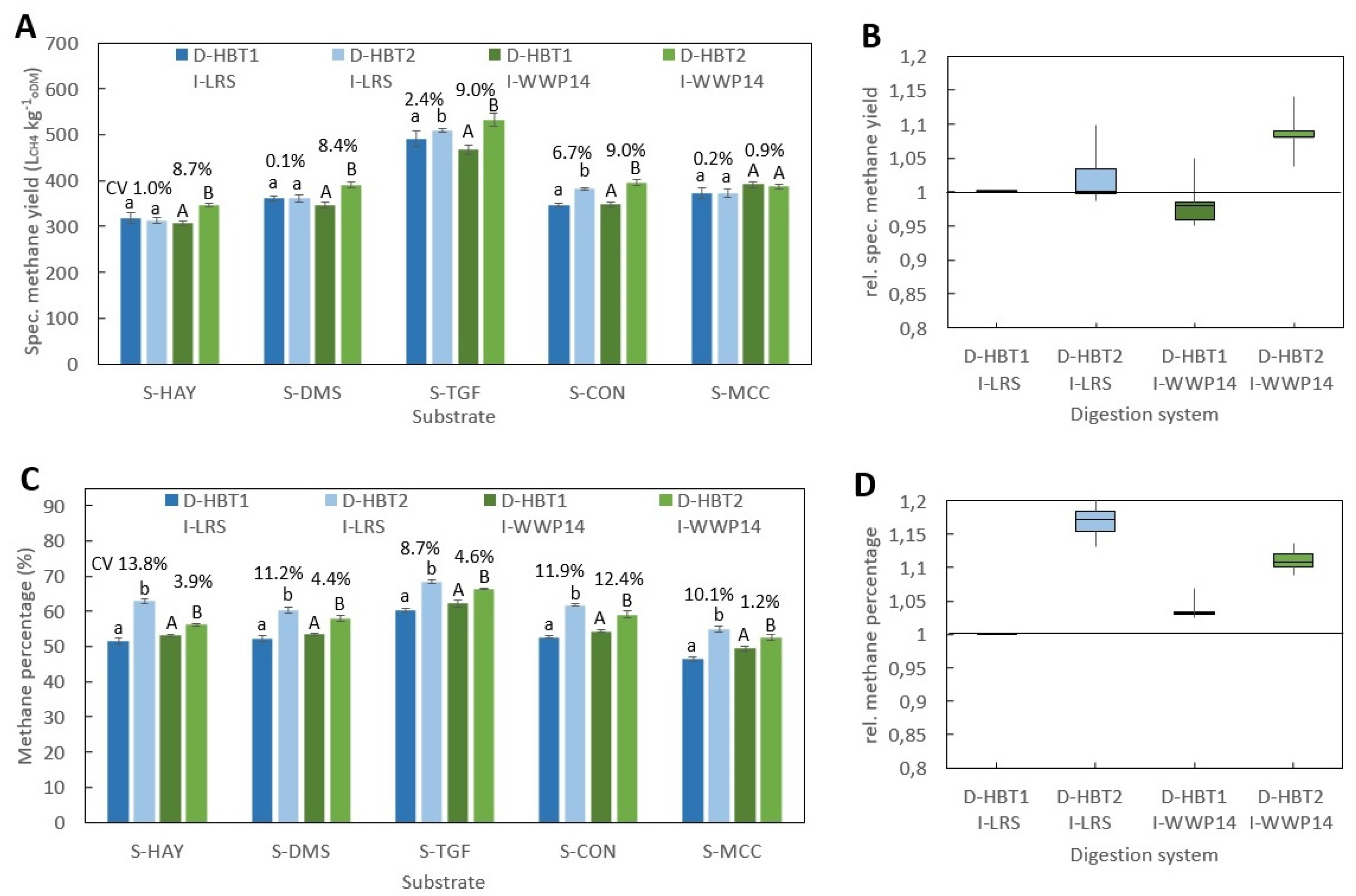
The aim of this study was to investigate the effects of different inocula and digestion systems on the specific methane yield and methane percentage of different substrates using VDI 4630 as a protocol. Unlike other publications, all investigated inocula were cultivated for several years and have been used in a national inter-laboratory test in Germany. Five long-term cultivated inocula and five substrates were tested using equipment and procedure of the standardized Hohenheim biogas yield test (D-HBT1) in comparison to four other digestion systems. To the best of our knowledge, such an extensive comparison of inocula and digestion systems has not been performed before.
2. Materials and Methods
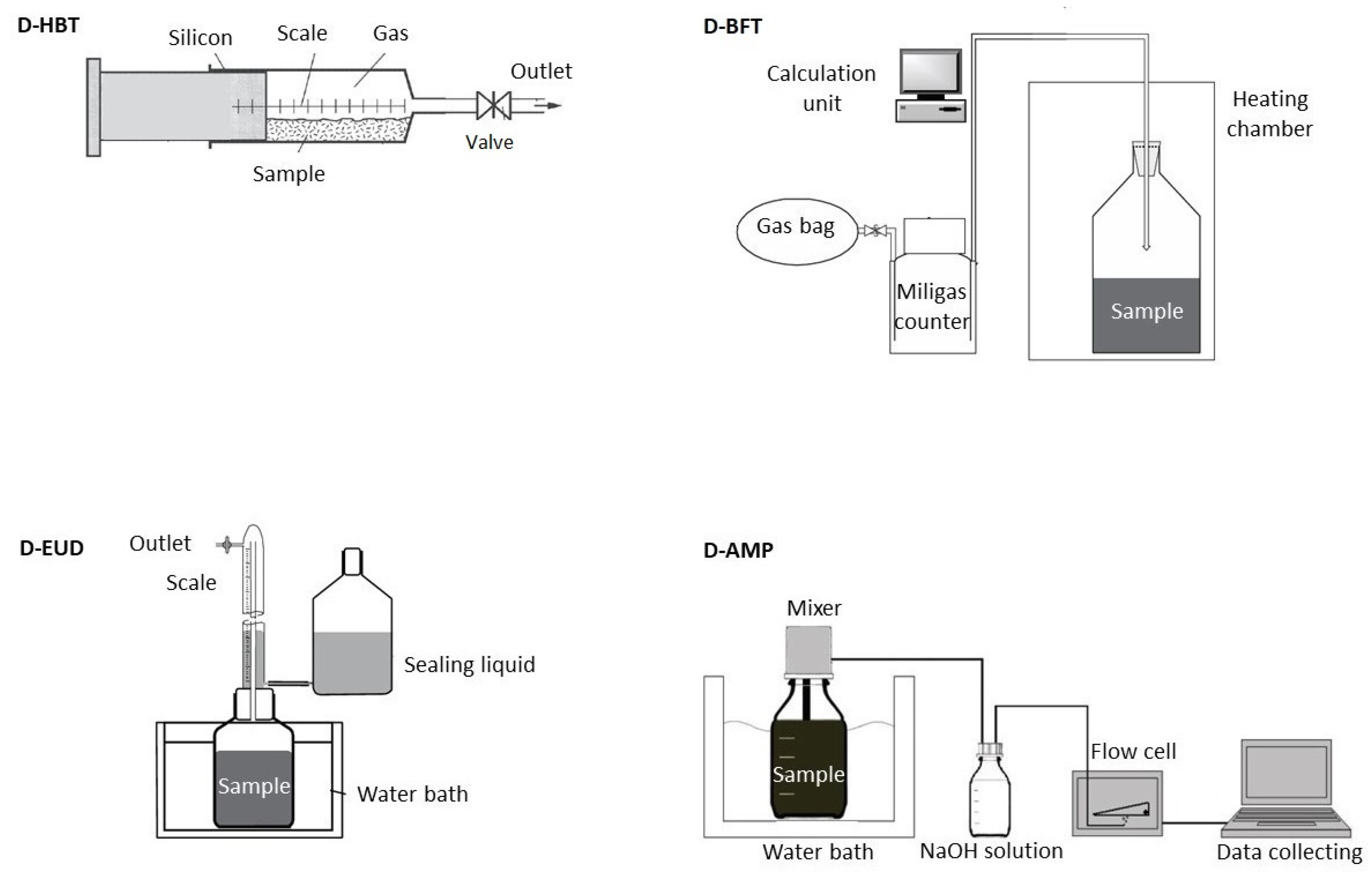
2.1. Digestion Systems
Four different digestion systems for performing the standardized BMP test according to VDI4630 [
5] were compared (
Table 2 and
Figure 1): Hohenheim biogas yield test (D-HBT), Bergedorf fermentation test (D-BFT), Eudiometer (D-EUD) and an Automatic methane potential test system (D- AMP).
2.1.1. Hohenheim Biogas Yield Test (D-HBT)
D-HBT consisted of 100 mL syringes, which are placed in a continuously rotating drum with 129 places for syringes placed in an incubator [
22]. The substrates in the syringes are agitated by the rotation of the drum at a speed of 1.2 rpm. The volume was measured manually by reading from a scale on the syringes. The methane percentage was measured by an infrared-spectrometric methane- sensor (“Advanced Gasmitter”, Pronova Analysetechnik, Berlin, Germany) [
23]. The sensor was calibrated directly before and after each measurement with a calibration gas mixture of 40% CO
2 and 60% CH
4 (G325792, Westfalen AG, Münster, Germany). The tests were performed in two variants: in dry gas (D-HBT1) and in wet gas (D-HBT2). For dry gas measurements in D-HBT1 the gas was dried with an absorbent (SICAPENT
®, Merck, Darmstadt, Germany). For the wet gas measurement, the water content of the gas was considered—by a correction of the gas volume
V0 (mL) according VDI 4360 [
5] Equation (1).
where
V (mL) and
p (hPa) are the measured volume and pressure,
pW (hPa) is the water vapor pressure at operation temperature
T (K), and
p0 (hPa) and
T0 (K) the standard pressure and temperature (1013.3 hPa, 273.15 K).
2.1.2. Bergedorf Fermentation Test (D-BFT)
The D-BFT reactor had a volume of 1.5 L and was frequently agitated manually. The gas volume was measured by a tipping cell counter (MilligasCounter, Ritter Apparatebau GmbH, Bochum, Germany). Gas was collected in a gasbag and gas quality was measured in a combined sample as soon as 1.5 L of gas was produced. The methane percentage was measured by an infrared sensor (Awite Bioenergie GmbH, Langenbach, Germany) [
24].
2.1.3. Eudiometer (D-EUD)
D-EUD (Neubert Glas GbR, Geschwenda, Germany) had a volume of 1.5 L. The headspace was flushed with nitrogen gas once a day and agitated continuously by a magnetic stirrer. The gas was collected in a tube, which was surrounded by a sealing fluid. In this tube the volume could be read by a scale bar. The gas composition was determined once at the end of the experiment with a land fill gas monitor (GA2000, Ansyco, Karlsruhe, Germany) [
25].
2.1.4. Automatic Methane Potential Test System (D-AMP)
D-AMP (AMPTS II, Bioprocess Control AB, Lund, Sweden) had a volume of 0.5 L and was agitated by a mechanical stirrer. The CO2 and H2S was stripped by using 3 M NaOH solution and then the volume of methane was measured by a flow cell. The gas lifted the flow cell, which lowered back down afterwards. The digital impulse was registered by a computer. Since the volume of biogas in this digestion system was not measured, the percentage of methane could not be determined either.
2.2. Inocula
Five different inocula were used in this study (
Table 3). All inocula were cooled down to 3–4 °C before being sent to avoid bias due to shipping. The dilutions and degassing were done after shipping.
2.2.1. Inoculum from Laboratory Reactor (I-LRS)
I-LRS was taken from a 400 L laboratory reactor, where bacteria were cultivated continuously. The inoculum was fed with maize silage, shredded wheat, soybean meal, rapeseed oil, and digestate from biogas plants in Baden-Württemberg, Germany. The organic loading rate (OLR) in terms of organic dry matter (oDM) was 0.3 kg
oDM m
−3 d
−1 and temperature was 37 °C [
23]. The hydraulic retention time (HRT) was 200 ± 25 d. Before using the inoculum, it was sieved using a mesh size of 0.5 mm.
2.2.2. Inoculum Diluted from Laboratory Reactor (I-LRD)
I-LRD was taken from a 2500 L laboratory reactor, which was fed by 80% cattle manure and 20% of a maize and grass silage mixture. OLR was 3.0 kg
oDM m
−3 d
−1, temperature was 38 ± 1 °C and HRT was 19 d [
24]. Before using, the inoculum was diluted with water in an inoculum/water ratio of 2:1 and stored at 37 °C for 7 d for degassing.
2.2.3. Inoculum from a Biogas Plant (I-BGP)
I-BGP was from a 942 m
3 biogas plant, fed with maize silage and cattle manure. HRT was 103 d, the operating temperature was 37 °C and OLR was 2.7 kg
oDM m
−3 d
−1. Prior to use, the inoculum was sieved using a mesh size of 5 mm and degassed for 5 d [
26].
2.2.4. Inoculum from Wastewater Treatment Plant (I-WWP)
I-WWP was taken from a wastewater treatment plant in northern Germany and sieved at <1 mm [
27]. I-WWP7 was stored afterwards for 7 days and I-WWP14 for 14 d, both at 37 °C for degassing.
Chemical characteristics and TE contents of the tested inocula are shown in
Table 4 and
Table 5, respectively.
2.3. Substrates
Five substrates with different nutrient compositions were chosen, where hay (S-HAY) and dried maize silage (S-DMS) represent widely used substrates and triglyceride fodder (S-TGF), concentrated fodder (S-CON) and microcrystalline cellulose (S-MCC) represent model feedstock with high content of fat, protein and fiber, respectively (
Table 6).
Hay (S-HAY) (marstall Wiesen-Cobs, marstall GmbH, Oberstaufen, Germany) is a mixture of more than 50 different grass and herb species from the first cut from Allgäu (South Germany) in the year 2012. S-HAY was dried by hot air, chopped to 16 mm, milled with a laboratory mill (Pulverisette 19, Fritsch GmbH, Markt Einersheim, Germany) and stored at −20 °C.
Maize silage (S-DMS) was taken from Unterer Lindenhof (Eningen unter Achalm, Germany). S- S-DMS was dried and milled with the laboratory mill.
Triyglyceride fodder (S-TGF) was provided as a homogenous standard material with well-known molecular structure (C
18H
32O
8N) and a known theoretical biogas yield of 609 L kg
oDM−1 [
8]. S-TGF was stored at 4 °C.
Concentrated fodder (S-CON) (Raiffeisen Kraftfutterwerke Süd GmbH, Würzburg, Germany) was milled with the laboratory mill and stored at −20 °C.
Microcrystalline cellulose (S-MCC) (CAS: 9004-34-6, Acros Organics, Pittsburgh, PA, USA) was used as standard substrate with known biogas yield of 745 L kg
−1oDM with a methane percentage of 50% [
5]. S-MCC had an average particle size of 50 µm and was stored at room temperature.
2.4. Measurement Procedure
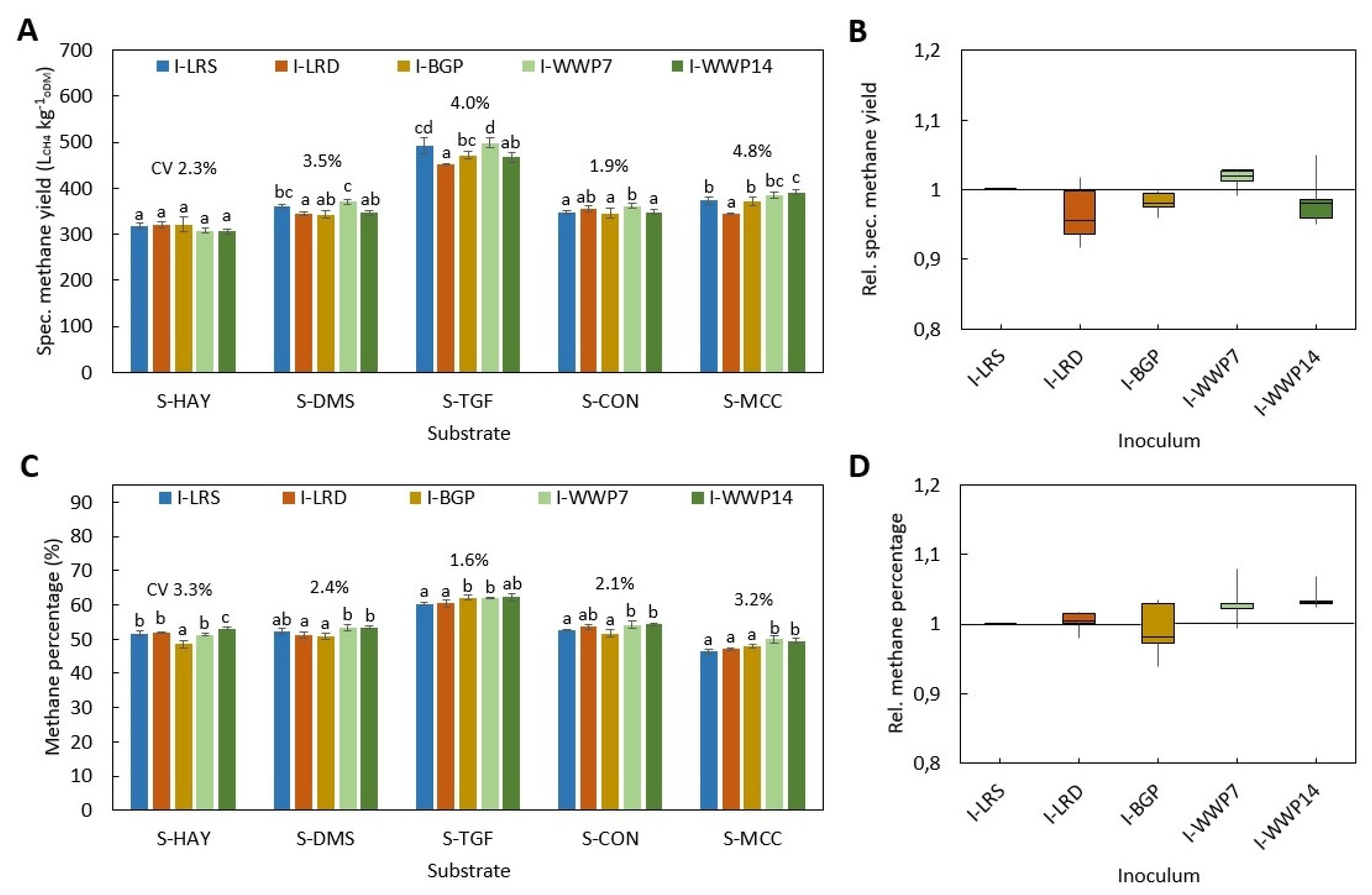
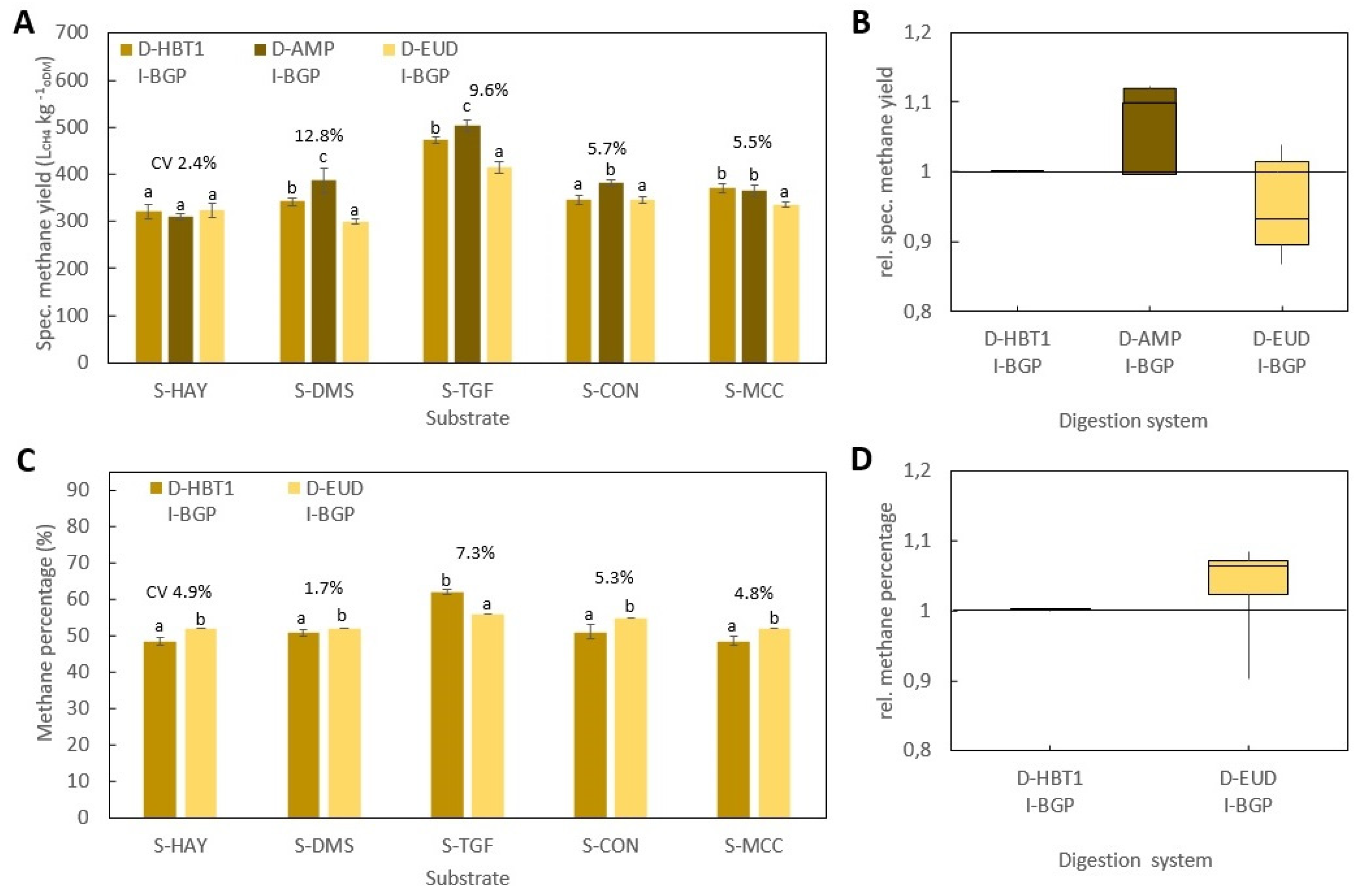
All BMP tests were performed according to VDI 4630 at ISR > 2 and a temperature of 38 ± 0.5 °C. In all digestion systems, beside D-HBT1 and D-HBT2, tests were performed in three replicates and digestion was terminated according to the 0.5% criteria, i.e., when the increase in gas production was less than 0.5% d−1 for three days. In both D-HBT, tests were performed in six replicates. Digestion time was terminated after 35 d, whereby the 0.5% criterion was met in each test run. Specific methane yield was calculated for biogas yield and methane percentage that were measured for each test run. Gas volume was corrected to standard conditions of 101.33 kPa and 0 °C.
Investigated combinations of digestion systems, substrates and inocula are listed in
Table 7. All substrates were tested in all digestion systems. However, the full range of inocula was only tested in D-HBT1, which was chosen because of the low sample volume (30 mL) required and its high repeatability. With the other digestion systems only one or two inocula could be tested due to the limited number of reactors for parallel tests (
Table 7). Additionally, trials with supplementation of TE for all inocula in D-HBT1 on substrate S-DMS were performed by adding a TE solution as described by Angelidaki et al. [
3]. In this test series, blanks for all inocula were also tested.
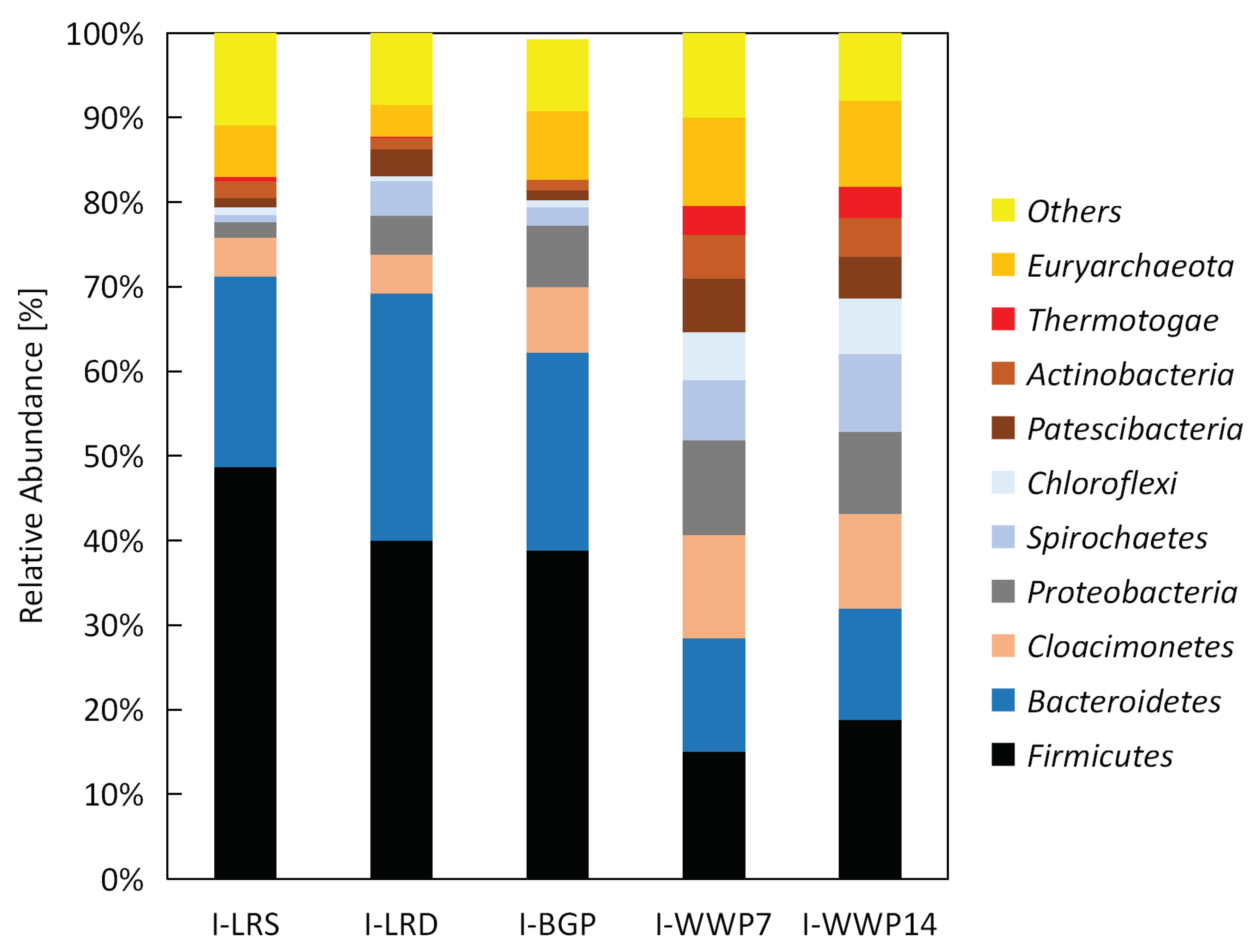
2.5. Taxonomic Profiling and Statistical Analysis
Taxonomic profiles of the microbial community within the analyzed inocula were determined by 16S RNA amplicon-sequencing. Microbial DNA was extracted with the FastDNA
® SPIN Kit for Soil (MP Biomedicals, Illkirch-Graffenstaden, France) and cleaned with Genomic DNA Clean & Concentrator TM Kit (Zymo Research, Irvine, CA, USA). The 16S RDNA gene amplicon-library was constructed by using the “16S Metagenomic Sequencing Library Preparation” protocol (Illumina, San Diego, CA, USA) and the primer pairs Pro341F and Pro805R [
29]. The sequencing was done on the Illumina MISeq platform applying the 300x bp paired-end protocol. Afterwards bioinformatics preprocessing of the sequencing data was done. Forward and reverse reads were merged with FLASH [
30], primer were removed with cutadapt [
31], while quality trimming of reads was done with sickle [
32]. Taxonomic classification was done by using the QIIME platform [
33] in combination with the SILVA 16S rDNA reference database (Release 132, 10.04.2018). Taxonomic profiles were presented as a bar chart for each inoculum. More detailed information is given in Hassa et al. [
34].
The statistical analysis of specific methane yield was performed by ANOVA post-hoc Tukey test at α = 0.05 using Microsoft Excel 2016.