Spatial Modelling of Bacterial Diversity over the Selected Regions in Bangladesh by Next-Generation Sequencing: Role of Water Temperature
Abstract
1. Introduction
2. Methodology
2.1. Collection of Water Samples
2.2. DNA Extraction and Next-Generation Sequencing
- a tube (2 mL) containing glass beads (ø0.1 and ø0.5 mm) was selected to store the water samples (membranes in 70% ethanol);
- suspension was centrifuged (at 8000 g) for 10 minutes;
- after centrifugation, the supernatant was redundant and the pellet was re-proscribed in pure water (200 μL);
- the glass bead tube containing the membrane received resuspension;
- phenol-chloroform protocol was followed to perform the DNA extraction process as previously described by [13];
- isopropanol containing 1 μL glycogen precipitated the template DNA at −20 °C for 30 minutes;
- the pellet was washed with 70% ethanol after centrifugation at 15,000 g for 15 minutes;
- after drying, the template DNA was finally dissolved in 30 μL TE buffer, and the concentration and purity of the template DNA were measured by a spectrophotometer (NanoDrop 2000, Thermo Fisher Scientific, Waltham, MA, USA) then stored at −20 °C for further analysis.
2.3. Spatial Modelling and Geographical Weighted Regression for Bacterial Distribution
2.4. Model Validation
3. Results
3.1. Identification of Bacteria
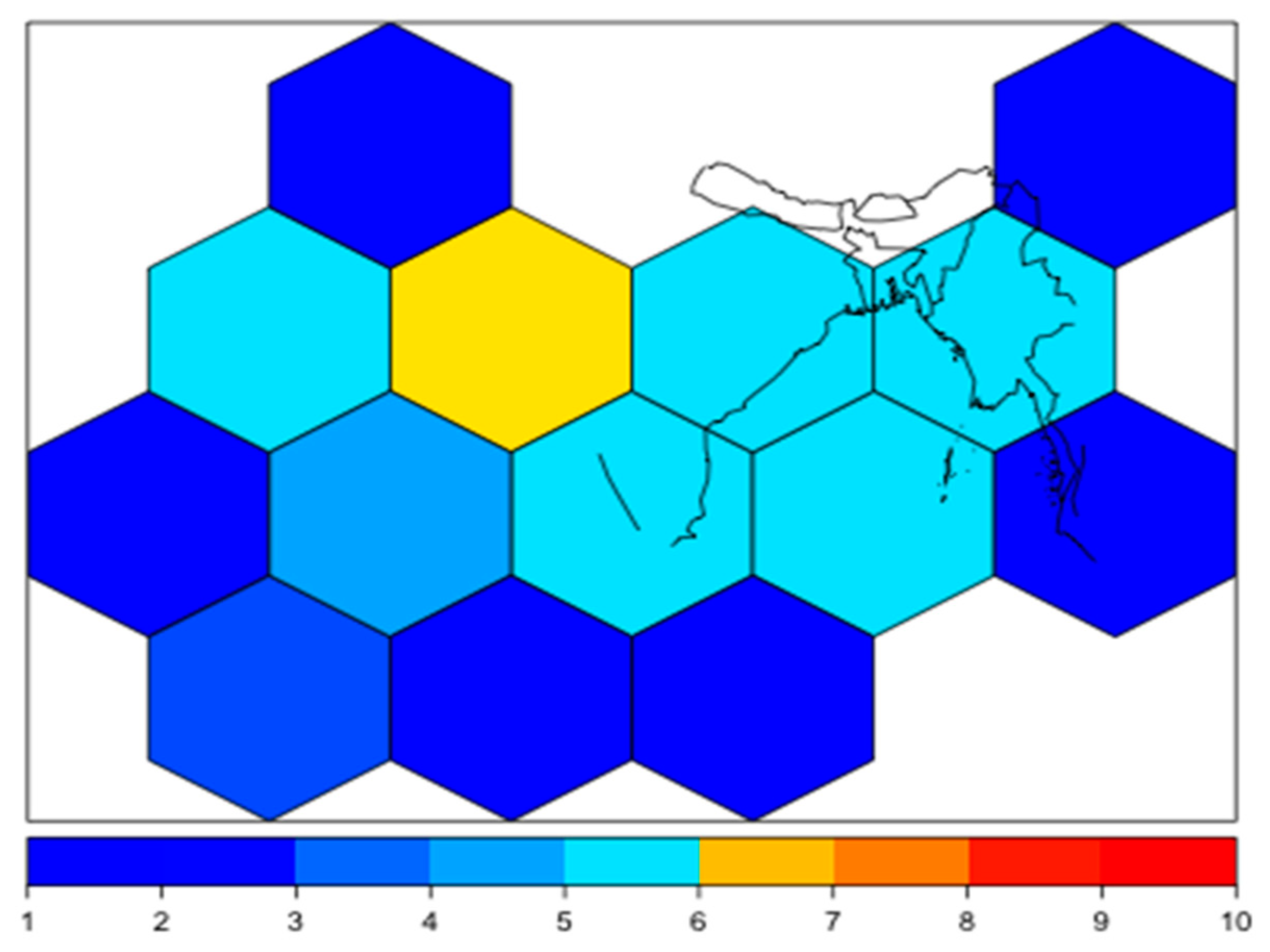
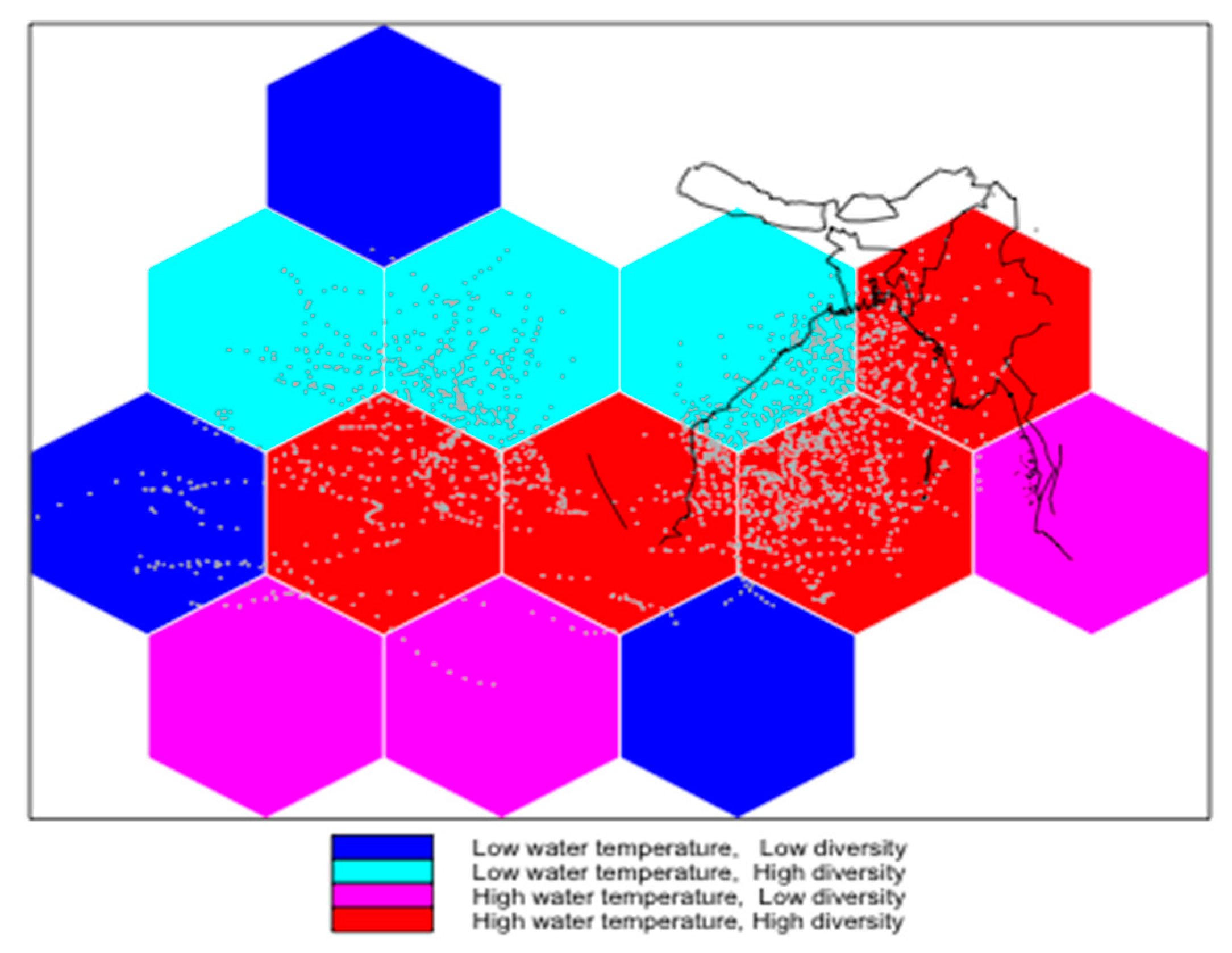
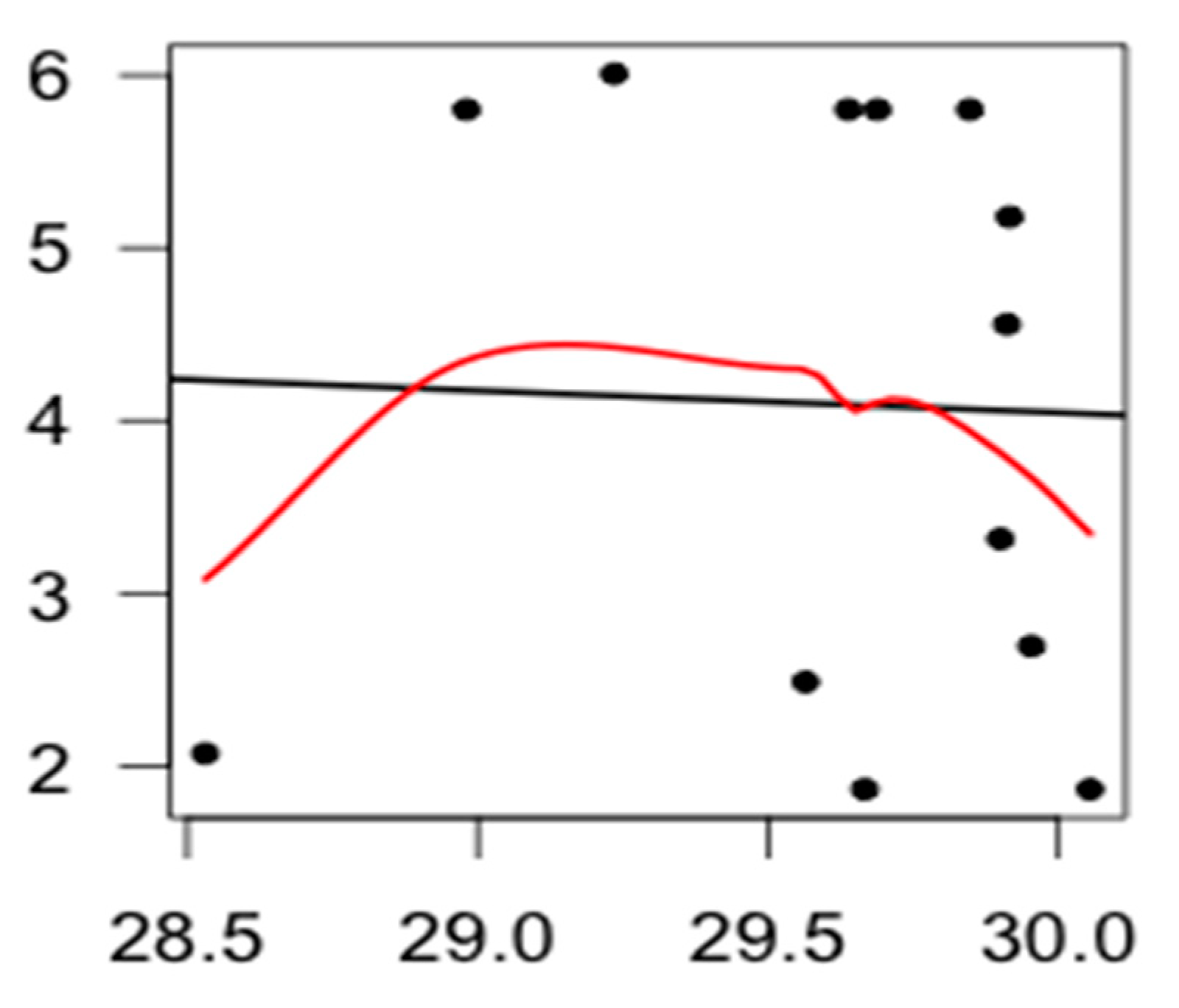
3.2. Spatial Modelling of Bacteria Distribution and Water Temperature
4. Discussion and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Griffiths, R.I.; Whiteley, A.S.; O’Donnell, A.G.; Bailey, M.J. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 2000, 66, 5488–5491. [Google Scholar] [CrossRef] [PubMed]
- Newton, R.J.; Jones, S.E.; Eiler, A.; McMahon, K.D.; Bertilsson, S. A guide to the natural history of freshwater Lake bacteria. Microbiol. Mol. Biol. Rev. 2011, 75, 14–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, J.; Li, H.; Yang, H.; Peng, C.; Peng, Z.; Lu, L. Shift in the microbial community composition of surface water and sediment along an urban river. Sci. Total Environ. 2018, 627, 600–612. [Google Scholar] [CrossRef]
- Guo, X.P.; Yang, Y.; Niu, Z.; Lu, D.P.; Zhu, C.H.; Feng, J.N.; Hou, L. Characteristics of microbial community indicate anthropogenic impact on the sediments along the Yangtze Estuary and its coastal area. China. Sci. Total Environ. 2019, 648, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Pajares, S.; Bohannan, B.J.M.; Souza, V. The role of microbial communities in tropical ecosystems. Front. Microbiol. 2016, 7, 1805. [Google Scholar] [CrossRef] [PubMed]
- Abia, A.L.K.; Ubomba-Jaswa, E.; Momba, M.N.B. Competitive survival of Escherichia coli, Vibrio cholerae, Salmonella typhimurium and Shigella dysenteriae in riverbed sediments. Microb. Ecol. 2016, 72, 881–889. [Google Scholar] [CrossRef] [PubMed]
- Azmuda, N.; Rahman, M.Z.; Madsen, M.S.; Khan, S.I.; Birkeland, N. Prevalence of a Novel Division-Level Bacterial Lineage in Lake Dhanmondi, Dhaka, Bangladesh, as Revealed by Deep Sequencing of 16S rRNA Gene Amplicons. Curr Microbiol. 2012, 65, 356–360. [Google Scholar] [CrossRef]
- Krishnankutty, S.P.; Muraleedharan, M.; Perumal, R.C.; Michael, S.; Benny, J.; Balan, B.; Thomas, G. Next-generation sequencing analysis reveals high bacterial diversity in wild venomous and non-venomous snakes from India. J. Venom. Anim. Toxins Incl. Trop. Dis. 2018, 24, 4–47. [Google Scholar] [CrossRef]
- Wetzel, R.G. Limnology: Lake and River Ecosystems; Academic Press: London, UK, 2001. [Google Scholar]
- Ulrich, N.; Rosenberger, A.; Brislawn, C.; Wright, J.; Kessler, C.; Toole, D.; Lamendella, R. Restructuring of the aquatic bacterial community by hydric dynamics associated with superstorm sandy. Appl. Environ. Microbiol. 2016, 82, 3525–3526. [Google Scholar] [CrossRef]
- Simek, K.; Pernthaler, J.; Weinbauer, M.G.; Hornak, K.; Dolan, J.R.; Nedoma, J.; Amann, R. Changes in bacterial community composition and dynamics and viral mortality rates associated with enhanced flagellate grazing in a mesoeutrophic reservoir. Appl. Environ. Microbiol. 2001, 67, 2723–2733. [Google Scholar] [CrossRef]
- Staley, C.; Unno, T.; Gould, T.J.; Jarvis, B.; Phillips, J.; Cotner, J.B.; Sadowsky, M.J. Application of Illumina next-generation sequencing to characterize the bacterial community of the Upper Mississippi River. J. Appl. Microbiol. 2013, 115, 1147–1158. [Google Scholar] [CrossRef] [PubMed]
- Sagova-Mareckova, M.; Cermak, L.; Novotna, J.; Plhackova, K.; Forstova, J.; Kopecky, J. Innovative methods for soil DNA purification tested in soils with widely differing characteristics. Appl. Environ. Microbiol. 2008, 74, 2902–2907. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, R.G.; Tanaka, Y.; Malla, B.; Bhandari, D.; Tandukar, S.; Inoue, D.; Haramoto, E. Next-generation sequencing identification of pathogenic bacterial genes and their relationship with fecal indicator bacteria in different water sources in the Kathmandu Valley, Nepal. Sci. Total Environ. 2017, 601–602, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Inkinen, J.; Jayaprakash, B.; Santo Domingo, J.W.; Keinänen-Toivola, M.M.; Ryu, H.; Pitkänen, T. Diversity of ribosomal 16S DNA- and RNA-based bacterial community in an office building drinking water system. J. Appl. Microbiol. 2016, 120, 1723–1738. [Google Scholar] [CrossRef]
- Di Bella, J.M.; Bao, Y.; Gloor, G.B.; Burton, J.P.; Reid, G. High throughput sequencing methods and analysis for microbiome research. J. Microbiol. Methods 2013, 95, 401–414. [Google Scholar] [CrossRef]
- Caporaso, G.; Lauber, C.; Walters, W.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Cody, W.L.; Wilson, J.W.; Hendrixson, D.R.; McIver, K.S.; Hagman, K.E.; Ott, C.M.; Schurr, M.J. Skim milk enhances the preservation of thawed −80 degrees C bacterial stocks. J. Microbiol. Methods 2008, 75, 135–138. [Google Scholar] [CrossRef]
- Salipante, S.J.; Kawashima, T.; Rosenthal, C.; Hoogestraat, D.R.; Cummings, L.A.; Sengupta, D.J.; Hoffman, N.G. Performance comparison of Illumina and Ion Torrent next generation sequencing platforms for 16S rRNA-based bacterial community profiling. Appl. Environ. Microbiol. 2014, 80, 7583–7591. [Google Scholar] [CrossRef]
- Begg, A.P.; Todhunter, K.; Donahoe, S.L.; Krockenberger, M.; Slapeta, J. Severe amoebic placentitis in a horse caused by an Acanthamoeba hatchetti isolate identified using next-generation sequencing. J. Clin. Microbiol. 2014, 52, 3101–3104. [Google Scholar] [CrossRef]
- Nowrousian, M. Next-generation sequencing techniques for eukaryotic microorganisms: Sequencing-based solutions to biological problems. Eukaryotic Cell 2010, 9, 1300–1310. [Google Scholar] [CrossRef]
- Vierheilig, J.; Savio, D.; Ley, R.E.; Mach, R.L.; Farnleitner, A.H.; Reischer, G.H. Potential applications of next generation DNA sequencing of 16S rRNA gene amplicons in microbial water quality monitoring. Water Sci. Technol. A J. Int. Assoc. Water Pollut. Res. 2015, 72, 1962–1972. [Google Scholar] [CrossRef] [PubMed]
- Sala-Comorera, L.; Anicet, R.; Blanch, A.; Casanovas-Massana, A.; García-Aljaro, C. Traceability of different brands of bottled mineral water during shelf life, using PCR-DGGE and next generation sequencing techniques. Food Microbiol. 2019, 82, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Mancini, M.V.; Damiani, C.; Accoti, A.; Tallarita, M.; Nunzi, E.; Cappelli, A.; Serrao, A. Estimating bacteria diversity in different organs of nine species of mosquito by next generation sequencing. BMC Microbiol. 2018, 18, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Samarajeewa, A.D.; Hammad, A.; Masson, L.; Khan, I.U.H.; Scroggins, R.; Beaudette, L.A. Comparative assessment of next-generation sequencing, denaturing gradient gel electrophoresis, clonal restriction fragment length polymorphism and cloningsequencing as methods for characterizing commercial microbial consortia. J. Microbiol. Methods 2015, 108, 103–111. [Google Scholar] [CrossRef]
- Ung, P.; Peng, C.; Yuk, S.; Ann, V.; Mith, H.; Tan, R.; Miyanaga, K.; Tanji, Y. Dynamics of bacterial community in Tonle Sap Lake, a large tropical flood-pulse system in Southeast Asia. Sci. Total Environ. 2018, 664, 414–423. [Google Scholar] [CrossRef]
- Zhang, J.; Ding, X.; Guan, R.; Zhu, C.; Xu, C.; Zhu, B.; Lu, Z. Evaluation of different 16S rRNA gene V regions for exploring bacterial diversity in a eutrophic freshwater lake. Sci. Total Environ. 2017, 618, 1254–1267. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhu, C.; Guan, R.; Xiong, Z.; Zhang, W.; Shi, J.; Chen, T. Microbial profiles of a drinking water resource based on different 16S rRNA V regions during a heavy cyanobacterial bloomin Lake Taihu, China. Environ. Sci. Pollut. Res. 2017, 24, 12796–12808. [Google Scholar] [CrossRef]
- Wahiduzzaman, M.; Luo, J.J. A statistical analysis on the contribution of El Niño–Southern Oscillation to the rainfall and temperature over Bangladesh. Meteorol. Appl. Phys. 2020, 1–14. [Google Scholar] [CrossRef]
- Wahiduzzaman, M.; Yeasmin, A. A kernel density estimation approach of North Indian Ocean tropical cyclone formation and the association with convective available potential energy and equivalent potential temperature. Meteorol. Appl. Phys. 2019, 1–10. [Google Scholar] [CrossRef]
- Wahiduzzaman, M.; Oliver, E.C.J.; Wotherspoon, S.J.; Holbrook, N.J. A climatological model of North Indian Ocean tropical cyclone genesis, tracks and landfall. Climate Dyn. 2017, 49, 2585–2603. [Google Scholar] [CrossRef]
- Wahiduzzaman, M.; Yeasmin, A. Statistical forecasting of tropical cyclone landfall activities over the North Indian Ocean rim countries. Atmos. Res. 2019, 227, 89–100. [Google Scholar] [CrossRef]
- Wahiduzzaman, M.; Oliver, E.C.J.; Klotzbach, P.J.; Wotherspoon, S.J.; Holbrook, N.J. A statistical seasonal forecast model of North Indian Ocean tropical cyclones using the Quasi-biennial Oscillation. Int. J. Climatol. 2019, 39, 934–952. [Google Scholar] [CrossRef]
- Wahiduzzaman, M.; Oliver, E.C.J.; Wotherspoon, S.J.; Luo, J.J. Seasonal forecasting of tropical cyclones in the North Indian Ocean region: The role of El Niño Southern Oscillation. Clim. Dyn. 2020, 54, 1571–1589. [Google Scholar] [CrossRef]
- Wheeler, D.C.; Páez, A. Geographically Weighted Regression. In Handbook of Applied Spatial Analysis; Fischer, M., Getis, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 461–486. [Google Scholar]
- Yun, J.; Zhao, F.; Zhang, W.; Yan, H.; Zhao, F.; Ai, D. Monitoring the microbial community succession and diversity of Liangzhou fumigated vinegar during solid-state fermentation with next-generation sequencing. Annals Microbiol. 2019, 69, 279–289. [Google Scholar] [CrossRef]
- Foysal, M.J.; Momtaz, F.; Kawsar, A.Q.M.R.; Rahman, M.M.; Gupta, S.K.; Tay, A.C.Y. Next-generation sequencing reveals significant variations in bacterial compositions across the gastrointestinal tracts of the Indian major carps, rohu (Labeo rohita), catla (Catla catla) and mrigal (Cirrhinus cirrhosis). Lett. Appl. Microbiol. 2020, 70, 173–180. [Google Scholar] [CrossRef]
| Survey Zone | Name of Bacteria |
|---|---|
| Zone-1 (Western side-Khulna) | N1, N3, N2, N4, N6, N7, N10, N9, and N11. |
| Zone-2 (Western side-Satkhira) | N1, N4, N7, N9, N10, and N11. |
| Zone-3 (Middle side-Bhola) | N1, N3, N2, N5, N8, N4, N6, N7, and N10. |
| Zone-4 (Eastern side-Chittagong) | N1, N2, N3, N5, N4, N7, N10, N9, and N11. |
| Zone -5 (Eastern side-Cox’s Bazar) | N1, N3, N2, N4, N5, and N11. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akter, N.; Wahiduzzaman, M.; Yeasmin, A.; Islam, K.S.; Luo, J.-J. Spatial Modelling of Bacterial Diversity over the Selected Regions in Bangladesh by Next-Generation Sequencing: Role of Water Temperature. Appl. Sci. 2020, 10, 2537. https://doi.org/10.3390/app10072537
Akter N, Wahiduzzaman M, Yeasmin A, Islam KS, Luo J-J. Spatial Modelling of Bacterial Diversity over the Selected Regions in Bangladesh by Next-Generation Sequencing: Role of Water Temperature. Applied Sciences. 2020; 10(7):2537. https://doi.org/10.3390/app10072537
Chicago/Turabian StyleAkter, Nabila, Md Wahiduzzaman, Alea Yeasmin, Kazi Saiful Islam, and Jing-Jia Luo. 2020. "Spatial Modelling of Bacterial Diversity over the Selected Regions in Bangladesh by Next-Generation Sequencing: Role of Water Temperature" Applied Sciences 10, no. 7: 2537. https://doi.org/10.3390/app10072537
APA StyleAkter, N., Wahiduzzaman, M., Yeasmin, A., Islam, K. S., & Luo, J.-J. (2020). Spatial Modelling of Bacterial Diversity over the Selected Regions in Bangladesh by Next-Generation Sequencing: Role of Water Temperature. Applied Sciences, 10(7), 2537. https://doi.org/10.3390/app10072537





