Fine-Grained Butterfly Classification in Ecological Images Using Squeeze-And-Excitation and Spatial Attention Modules
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets
2.2. Squeeze-And-Excitation Modules
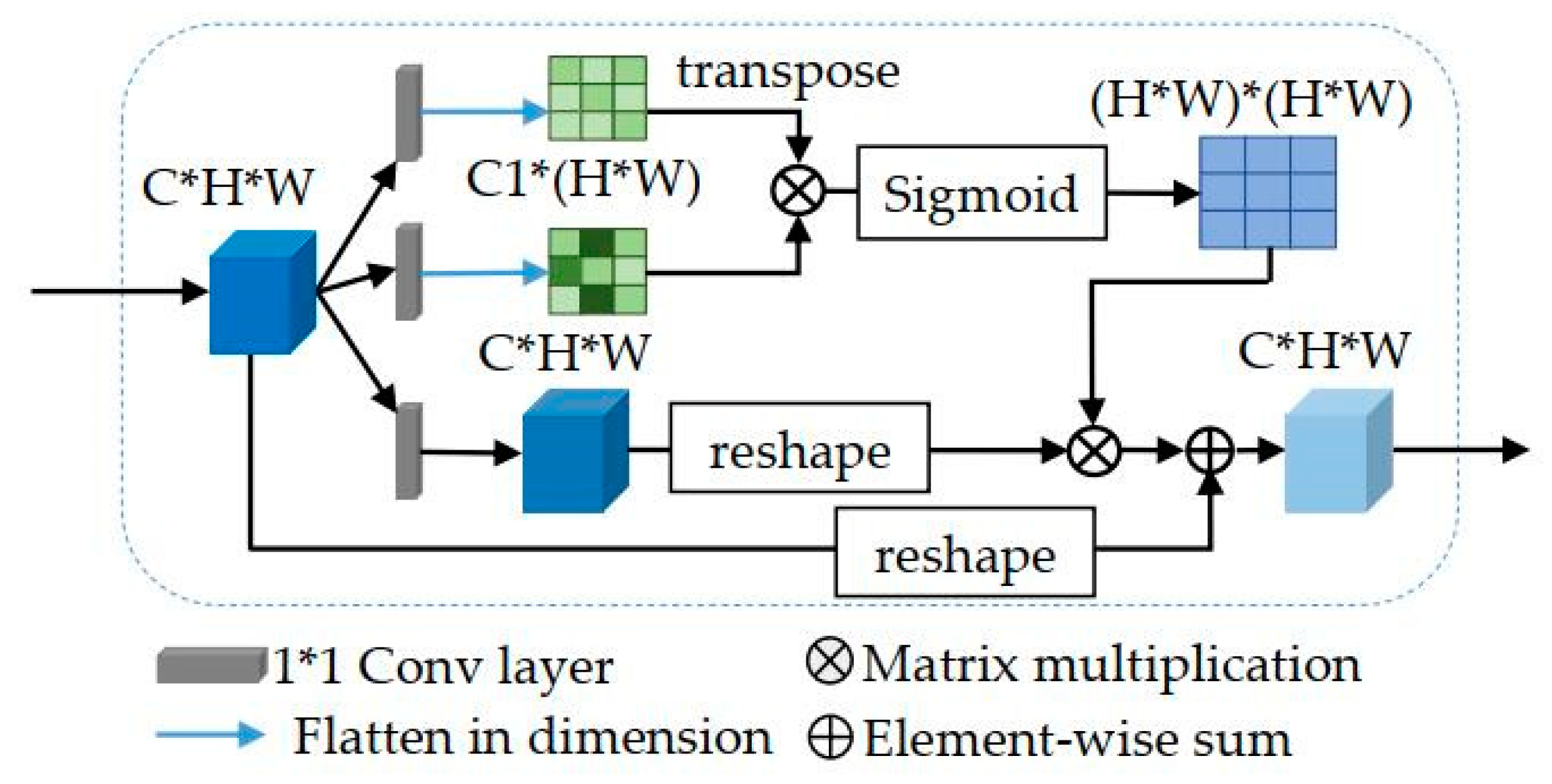
2.3. Spatial Attention Modules
2.4. Overall SESADRN Architecture
3. Experiments and Results
3.1. Performance Comparison with Other Methods
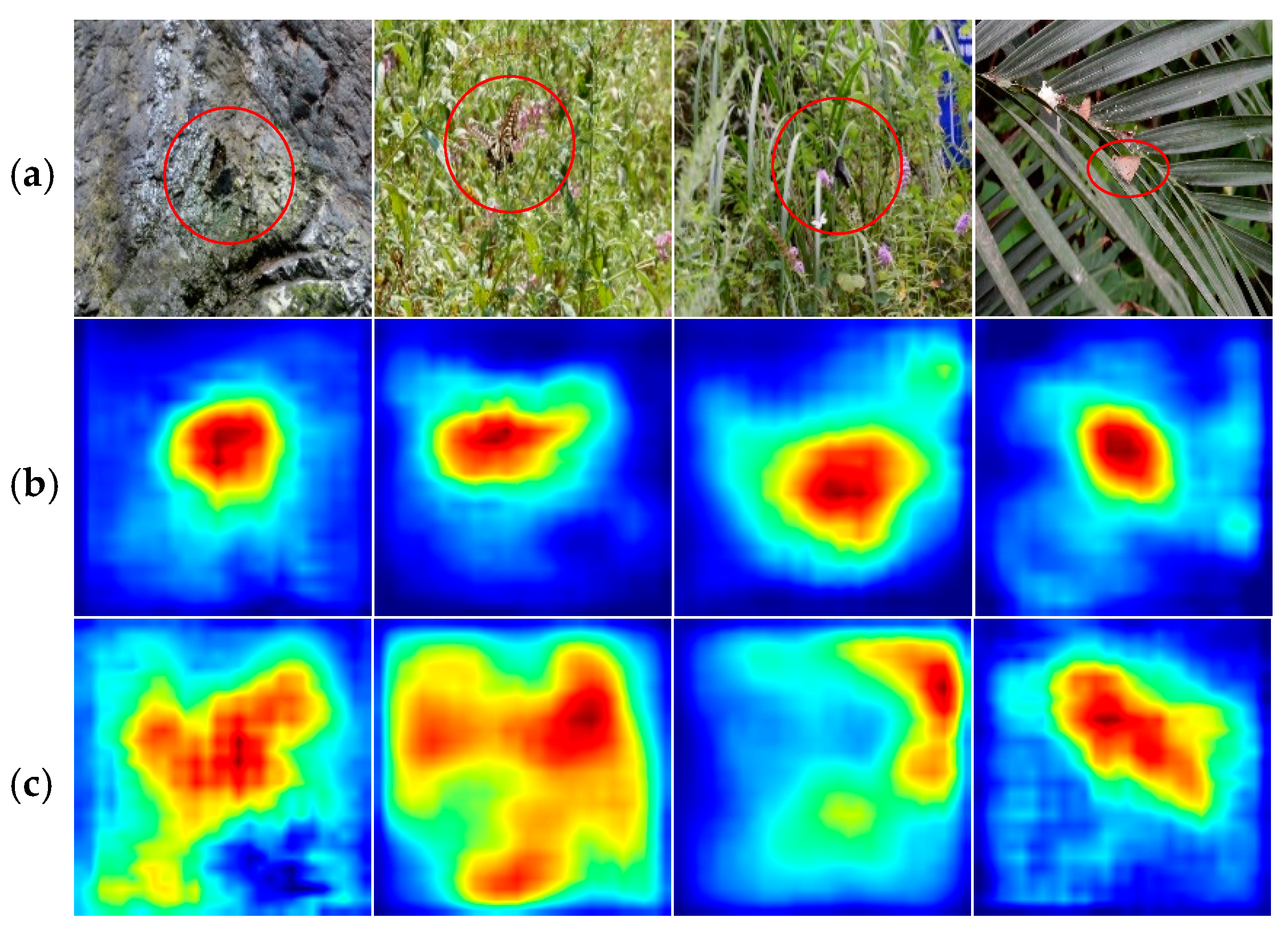
3.2. Classification Analysis Based on Grad-CAM
4. Conclusions and Future Work
- (1)
- Augment the butterfly database by collecting more butterfly photos.
- (2)
- Try to use more appropriate models to classify butterflies, such as introducing a few-shot learning algorithm into the model. We are also going to apply the proposed method to other applications.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shou, J.; Zhou, Y.; Li, Y. Systematic Butterffly Names of the World; Shaanxi Science and Technology Press: Xi’an, China, 2006. [Google Scholar]
- Wang, W.; Zhang, J.; Wang, F. Attention bilinear pooling for fine-grained classification. Symmetry 2019, 11, 1033. [Google Scholar] [CrossRef]
- Andrian, R.; Maharani, D.; Muhammad, M.A.; Junaidi, A. Butterfly identification using gray level co-occurrence matrix (glcm) extraction feature and k-nearest neighbor (knn) classification. Regist. J. Ilm. Teknol. Sist. Inf. 2019, 6, 11–21. [Google Scholar] [CrossRef]
- Li, F.; Xiong, Y. Automatic identification of butterfly species based on HoMSC and GLCMoIB. Vis. Comput. 2018, 34, 1525–1533. [Google Scholar] [CrossRef]
- Xue, A.; Li, F.; Xiong, Y. Automatic identification of butterfly species based on gray-level co-occurrence matrix features of image block. J. Shanghai Jiaotong Univ. 2019, 24, 220–225. [Google Scholar] [CrossRef]
- Kartika, D.S.Y.; Herumurti, D.; Yuniarti, A. Local binary pattern method and feature shape extraction for detecting butterfly image. Int. J. 2018, 15, 127–133. [Google Scholar] [CrossRef]
- Kartika, D.S.Y.; Herumurti, D.; Yuniarti, A. Butterfly image classification using color quantization method on hsv color space and local binary pattern. IPTEK J. Proc. Ser. 2018, 78–82. [Google Scholar] [CrossRef]
- Kaya, Y.; Kayci, L.; Uyar, M. Automatic identification of butterfly species based on local binary patterns and artificial neural network. Appl. Soft Comput. 2015, 28, 132–137. [Google Scholar] [CrossRef]
- Alhady, S.S.N.; Kai, X.Y. Butterfly Species Recognition Using Artificial Neural Network. In Proceedings of the Intelligent Manufacturing & Mechatronics; Hassan, M.H.A., Ed.; Springer: Singapore, 2018; pp. 449–457. [Google Scholar]
- Clément, M.; Kurtz, C.; Wendling, L. Learning spatial relations and shapes for structural object description and scene recognition. Pattern Recognit. 2018, 84, 197–210. [Google Scholar] [CrossRef]
- Li, C.; Zhang, D.; Du, S.; Zhu, Z.; Jia, S.; Qu, Y. A butterfly detection algorithm based on transfer learning and deformable convolution deep learning. Acta Autom. Sin. 2019, 45, 1772–1782. [Google Scholar]
- Zhou, A.; Ma, P.; Xi, T.; Wang, J.; Feng, J.; Shao, Z.; Tao, Y.; Yao, Q. Automatic identification of butterfly specimen images at the family level based on deep learning method. Acta Entomol. Sin. 2017, 60, 1339–1348. [Google Scholar]
- Arzar, N.N.K.; Sabri, N.; Johari, N.F.M.; Shari, A.A.; Noordin, M.R.M.; Ibrahim, S. Butterfly species identification using Convolutional Neural Network (CNN). In Proceedings of the 2019 IEEE International Conference on Automatic Control and Intelligent Systems (I2CACIS), Shah Alam, Malaysia, 29 June 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 221–224. [Google Scholar]
- Lin, Z.; Jia, J.; Gao, W.; Huang, F. Fine-grained visual categorization of butterfly specimens at sub-species level via a convolutional neural network with skip-connections. Neurocomputing 2019, 384, 295–313. [Google Scholar] [CrossRef]
- López-Antequera, M.; Vallina, M.L.; Strisciuglio, N.; Petkov, N. Place and Object Recognition by CNN-based COSFIRE filters. IEEE Access 2019, 7, 66157–66166. [Google Scholar] [CrossRef]
- Almryad, A.S.; Kutucu, H. Automatic identification for field butterflies by convolutional neural networks. Eng. Sci. Technol. Int. J. 2020, 23, 189–195. [Google Scholar] [CrossRef]
- Nie, L.; Wang, K.; Fan, X.; Gao, Y. Fine-grained butterfly recognition with deep residual networks: A new baseline and benchmark. In Proceedings of the 2017 International Conference on Digital Image Computing: Techniques and Applications (DICTA), Sydney, Australia, 29 November–1 December 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 1–7. [Google Scholar]
- Carvajal, J.A.; Romero, D.G.; Sappa, A.D. Fine-tuning based deep convolutional networks for lepidopterous genus recognition. In Proceedings of the Iberoamerican Congress on Pattern Recognition, Lima, Peru, 8–11 November 2016; Springer: Berlin/Heiderbeg, Germany, 2016; pp. 467–475. [Google Scholar]
- Chang, Q.; Qu, H.; Wu, P.; Yi, J. Fine-Grained Butterfly and Moth Classification Using Deep Convolutional Neural Networks. Available online: https://pdfs.semanticscholar.org/4cf2/045b811c9e0807f9c94fc991566a6f5adbf4.pdf (accessed on 28 February 2020).
- Rodner, E.; Simon, M.; Brehm, G.; Pietsch, S.; Wägele, J.W.; Denzler, J. Fine-grained recognition datasets for biodiversity analysis. arXiv 2015, arXiv:1507.00913. [Google Scholar]
- Zhu, L.Q.; Ma, M.Y.; Zhang, Z.; Zhang, P.Y.; Wu, W.; Wang, D.D.; Zhang, D.X.; Wang, X.; Wang, H.Y. Hybrid deep learning for automated lepidopteran insect image classification. Orient. Insects 2017, 51, 79–91. [Google Scholar] [CrossRef]
- Xie, J.; Hou, Q.; Shi, Y.; Lv, P.; Jing, L.; Zhuang, F.; Zhang, J.; Tan, X.; Xu, S. The automatic identification of butterfly species. J. Comput. Res. Dev. 2018, 55, 1609–1618. [Google Scholar]
- Zhao, R.; Li, C.; Ye, S.; Fang, X. Butterfly recognition based on faster R-CNN. In Journal of Physics: Conference Series; IOP Publishing: Bristol, UK, 2019; Volume 1176, p. 32048. [Google Scholar]
- Wang, J.; Markert, K.; Everingham, M. Learning models for object recognition from natural language descriptions. In Proceedings of the British Machine Vision Conference, London, UK, 7–10 September 2009. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Park, Y.J.; Tuxworth, G.; Zhou, J. Insect classification using squeeze-and-excitation and attention modules-a benchmark study. In Proceedings of the 2019 IEEE International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–29 September 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 3437–3441. [Google Scholar]
- Kitada, S.; Iyatomi, H. Skin lesion classification with ensemble of squeeze-and-excitation networks and semi-supervised learning. arXiv 2018, arXiv:1809.02568. [Google Scholar]
- Cao, Y.; Xu, J.; Lin, S.; Wei, F.; Hu, H. GCNet: Non-local networks meet squeeze-excitation networks and beyond. arXiv 2019, arXiv:1904.11492. [Google Scholar]
- Gong, L.; Jiang, S.; Yang, Z.; Zhang, G.; Wang, L. Automated pulmonary nodule detection in CT images using 3D deep squeeze-and-excitation networks. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 1969–1979. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Shen, L.; Albanie, S.; Sun, G.; Wu, E. Squeeze-and-Excitation Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019. [Google Scholar]
- Peng, C.; Zhang, X.; Yu, G.; Luo, G.; Sun, J. Large kernel matters—Improve semantic segmentation by global convolutional network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4353–4361. [Google Scholar]
- Chen, X.; Lin, L.; Liang, D.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.-H.; Chen, Y.-W.; Tong, R.; Wu, J. A dual-attention dilated residual network for liver lesion classification and localization on CT images. In Proceedings of the 2019 IEEE International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–25 September 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 235–239. [Google Scholar]
- Yu, F.; Koltun, V.; Funkhouser, T. Dilated residual networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 472–480. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Cardiff, UK, 9–12 September 2019. [Google Scholar]
- Wang, F.; Jiang, M.; Qian, C.; Yang, S.; Li, C.; Zhang, H.; Wang, X.; Tang, X. Residual attention network for image classification. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 3156–3164. [Google Scholar]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 2921–2929. [Google Scholar]








| Datasets | Species | Quantity | Train Dataset | Val Dataset | Test Dataset |
|---|---|---|---|---|---|
| China Butterfly Dataset | Speyeria aglaja | 61 | 36 | 13 | 12 |
| Achillidesbianor Cramer | 43 | 25 | 10 | 8 | |
| Limenitis helmanni | 35 | 21 | 7 | 7 | |
| Aphantopus hyperanthus | 35 | 21 | 7 | 7 | |
| Elymnias hypermnestra | 28 | 16 | 7 | 5 | |
| Suastus gremius | 26 | 15 | 6 | 5 | |
| Celastrina oreas | 25 | 15 | 5 | 5 | |
| Papilio paris | 23 | 13 | 6 | 4 | |
| Papilio xuthus | 23 | 13 | 6 | 4 | |
| Eurema hecabe | 22 | 13 | 5 | 4 | |
| Papilio alcmenor Felder | 21 | 12 | 5 | 4 | |
| Iambrix salsala | 20 | 12 | 4 | 4 | |
| Polygonia c-aureum | 20 | 12 | 4 | 4 | |
| Pyrgusmaculatus Bremeret Grey | 18 | 10 | 5 | 3 | |
| Abisara echerius | 13 | 7 | 4 | 2 | |
| Neptis themis Leech | 13 | 7 | 4 | 2 | |
| Thecla betulae | 11 | 6 | 3 | 2 | |
| Albulina orbitula | 10 | 6 | 2 | 2 | |
| Menelaides protenor | 10 | 6 | 2 | 2 | |
| Euploea midamus | 10 | 6 | 2 | 2 | |
| Leeds Butterfly Dataset | Danaus plexippus | 82 | 49 | 17 | 16 |
| Heliconius charitonius | 93 | 55 | 20 | 18 | |
| Heliconius erato | 61 | 36 | 13 | 12 | |
| Junonia coenia | 90 | 54 | 18 | 18 | |
| Lycaena phlaeas | 88 | 52 | 19 | 17 | |
| Nymphalis antiopa | 100 | 60 | 20 | 20 | |
| Papilio cresphontes | 89 | 53 | 19 | 17 | |
| Pieris rapae | 55 | 33 | 11 | 11 | |
| Vanessa atalanta | 90 | 54 | 18 | 18 | |
| Vanessa cardui | 84 | 50 | 18 | 16 | |
| Total | 1299 | 768 | 280 | 251 |
| Model | Accuracy | Weighted Precision | Weighted Recall | Weighted F1-Score |
|---|---|---|---|---|
| SE-Resnet50 [30] | 0.873 | 0.874 | 0.862 | 0.886 |
| RAN56 [35] | 0.861 | 0.859 | 0.847 | 0.870 |
| RAN92 [35] | 0.892 | 0.889 | 0.873 | 0.905 |
| DADRN [32] | 0.932 | 0.932 | 0.924 | 0.940 |
| SESADRN | 0.956 | 0.954 | 0.948 | 0.960 |
| Species | Number of Test Samples | SE-Resnet50 | RAN56 | RAN92 | DADRN | SESADRN |
|---|---|---|---|---|---|---|
| Danaus plexippus | 16 | 1 | 1 | 1 | 1 | 1 |
| Heliconius charitonius | 18 | 0.944 | 0.944 | 1 | 0.944 | 1 |
| Heliconius erato | 12 | 1 | 1 | 1 | 1 | 1 |
| Junonia coenia | 18 | 0.889 | 1 | 1 | 1 | 1 |
| Lycaena phlaeas | 17 | 0.941 | 1 | 1 | 1 | 1 |
| Nymphalis antiopa | 20 | 0.85 | 0.9 | 0.95 | 0.95 | 0.95 |
| Papilio cresphontes | 17 | 1 | 1 | 1 | 1 | 1 |
| Pieris rapae | 11 | 0.818 | 0.909 | 0.818 | 1 | 1 |
| Vanessa atalanta | 18 | 0.944 | 1 | 1 | 1 | 1 |
| Vanessa cardui | 16 | 0.938 | 1 | 1 | 1 | 1 |
| Speyeria aglaja | 12 | 0.917 | 0.583 | 0.917 | 0.917 | 1 |
| Achillidesbianor Cramer | 8 | 0.5 | 0.625 | 0.5 | 0.625 | 0.75 |
| Limenitis helmanni | 7 | 0.571 | 0.571 | 0.429 | 0.714 | 0.714 |
| Aphantopus hyperanthus | 7 | 1 | 0.714 | 0.857 | 0.714 | 1 |
| Elymnias hypermnestra | 5 | 0.8 | 0.8 | 0.8 | 1 | 1 |
| Suastus gremius | 5 | 1 | 1 | 1 | 1 | 1 |
| Celastrina oreas | 5 | 0.8 | 0.8 | 1 | 1 | 1 |
| Papilio paris | 4 | 0.5 | 0.25 | 0.5 | 0.75 | 0.5 |
| Papilio xuthus | 4 | 1 | 0.75 | 1 | 1 | 1 |
| Eurema hecabe | 4 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Papilio alcmenor Felder | 4 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 |
| Iambrix salsala | 4 | 1 | 0.5 | 1 | 1 | 1 |
| Polygonia c-aureum | 4 | 1 | 1 | 1 | 1 | 1 |
| Pyrgusmaculatus Bremeret Grey | 3 | 0.667 | 0.667 | 0.333 | 1 | 1 |
| Abisara echerius | 2 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Neptis themis Leech | 2 | 0 | 0 | 0 | 1 | 1 |
| Thecla betulae | 2 | 0.5 | 0.5 | 0.5 | 1 | 1 |
| Albulina orbitula | 2 | 1 | 1 | 1 | 1 | 1 |
| Menelaides protenor | 2 | 1 | 1 | 1 | 1 | 1 |
| Euploea midamus | 2 | 0.5 | 0 | 0 | 0 | 1 |
| Average | 0.873 | 0.861 | 0.892 | 0.932 | 0.956 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xin, D.; Chen, Y.-W.; Li, J. Fine-Grained Butterfly Classification in Ecological Images Using Squeeze-And-Excitation and Spatial Attention Modules. Appl. Sci. 2020, 10, 1681. https://doi.org/10.3390/app10051681
Xin D, Chen Y-W, Li J. Fine-Grained Butterfly Classification in Ecological Images Using Squeeze-And-Excitation and Spatial Attention Modules. Applied Sciences. 2020; 10(5):1681. https://doi.org/10.3390/app10051681
Chicago/Turabian StyleXin, Dongjun, Yen-Wei Chen, and Jianjun Li. 2020. "Fine-Grained Butterfly Classification in Ecological Images Using Squeeze-And-Excitation and Spatial Attention Modules" Applied Sciences 10, no. 5: 1681. https://doi.org/10.3390/app10051681
APA StyleXin, D., Chen, Y.-W., & Li, J. (2020). Fine-Grained Butterfly Classification in Ecological Images Using Squeeze-And-Excitation and Spatial Attention Modules. Applied Sciences, 10(5), 1681. https://doi.org/10.3390/app10051681




