1. Introduction
Understanding the statistical relationships between random variables (or features) is a fundamental problem in the fields of statistics and machine learning. Researchers usually visualize the features of a dataset (e.g., using scatter plots) to understand the dependencies between variables. For example, in supervised learning, researchers use visualizations of the features to understand which features are relevant or irrelevant to the values of the target variables, and how the relevant features are related to the target variables. They then select learning algorithms based on the dependence types. However, if the number of features is large, interpretation of visualizations is cumbersome and time-consuming.
Therefore, many dependence measures have been developed to automatically detect dependency relationships. A number of information-theoretic measures have been invented based on the Shannon entropy [
1]. Mutual information (MI) [
2] and its variants have been widely used to analyze categorical or discrete features [
3]. Various types of correlations have been developed to help categorize the relationships between continuous features: Pearson’s correlation coefficient (pCor), Spearman’s rank, Kendall’s tau, distance correlation (dCor) [
4], maximal information coefficient [
5], the randomized dependence coefficient [
6], and etc.
Various applications of dependency measures have been developed. Li et al. [
7] used dCor to screen the features of gene expression data. Zeng et al. [
8] proposed a variant of MI to measure interactions between features to aid feature selection. Chen et al. [
9] applied MI to the representation learning of generative adversarial networks (GANs). Kaul et al. [
10] presented an automated feature generation algorithm based on dCor called AutoLearn.
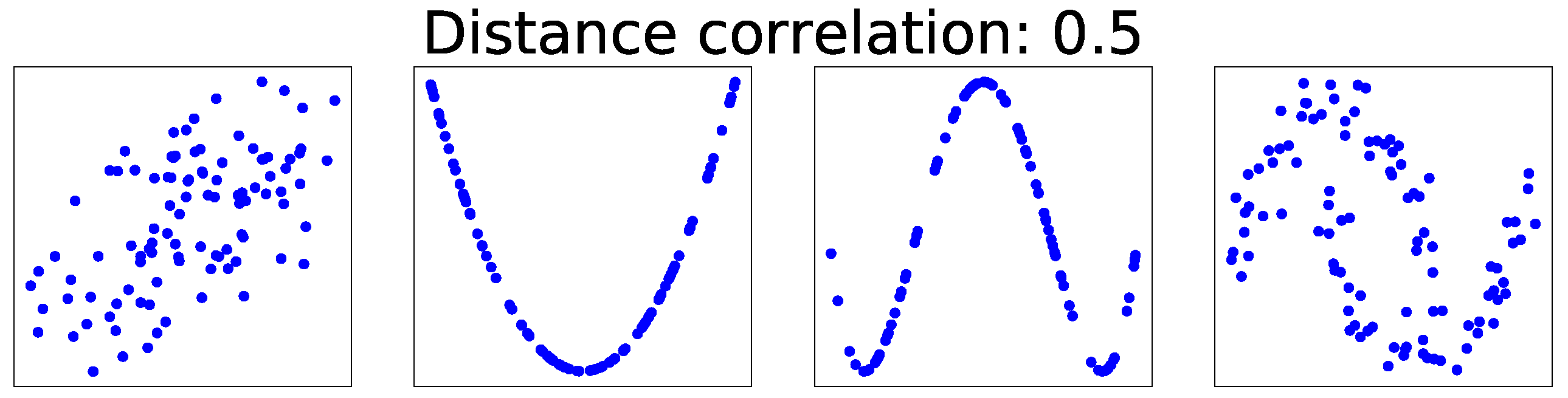
Existing dependency measures are subject to three main limitations. Firstly, they do not consider dependency types. For example, in
Figure 1, the dCor of all feature pairs is 0.5, but the types of dependence are obviously different. Secondly, these measures are human-readable scalars rather than machine-readable vectors. Thirdly, they are not optimized to specific applications.
These three problems can be solved by using visualizations as inputs to convolutional neural networks (CNNs). Firstly, dependency types can easily be classified based on visualizations, as shown in
Figure 1. The visualizations, such as scatter plots, can be regarded as images. Advances in deep learning-based computer vision have proven CNNs to be effective for processing high-dimensional images. CNNs are frequently used for image classification [
11], object detection [
12], and image generation [
13]. Secondly, neural networks are also capable of learning machine-readable representations; for example, word representations [
14] from textual data have achieved great success in natural language processing [
15]. Likewise, dependence representations can be learned by CNNs if an appropriate learning method is applied. Thirdly, when applying an end-to-end (feature learning) approach, CNNs can learn task-specific representations from visualizations, and thus they can be optimized for a specific task (such as text recognition [
16], speech recognition [
17], and self-driving cars [
18]) based on dependency measures. Basically, the usage of CNNs is motivated by the fact that they can extract significant features from visualizations at different hierarchical levels.
Thanks to the above advantages of CNNs, the hand-crafted representations may be replaced with convolutions. A few previous studies used deep neural networks (DNNs) for dependence estimation; for example, Belghazi el al. [
19] used DNNs for estimation of mutual information between high dimensional continuous random variables. Hjelm et al. [
20] proposed an unsupervised learning method, called Deep InfoMax, for representation learning. Similarly to our work, they tried to perform dependence estimation for specific tasks, but used mutual information as estimation target and they focus only on high dimensional variables such as images. To measure statistical dependence, correlations have been widely used in the fields of statistics and machine learning. However, due to a large amount of types of nonlinear dependence, it is very challenging to distinguish the types with a correlation coefficient. When using CNNs for dependence learning, we should consider one of its important disadvantages; CNNs have a huge number of parameters and with small dataset, would run into an overfitting problem, and thus they require much more training data than other existing learning algorithms.
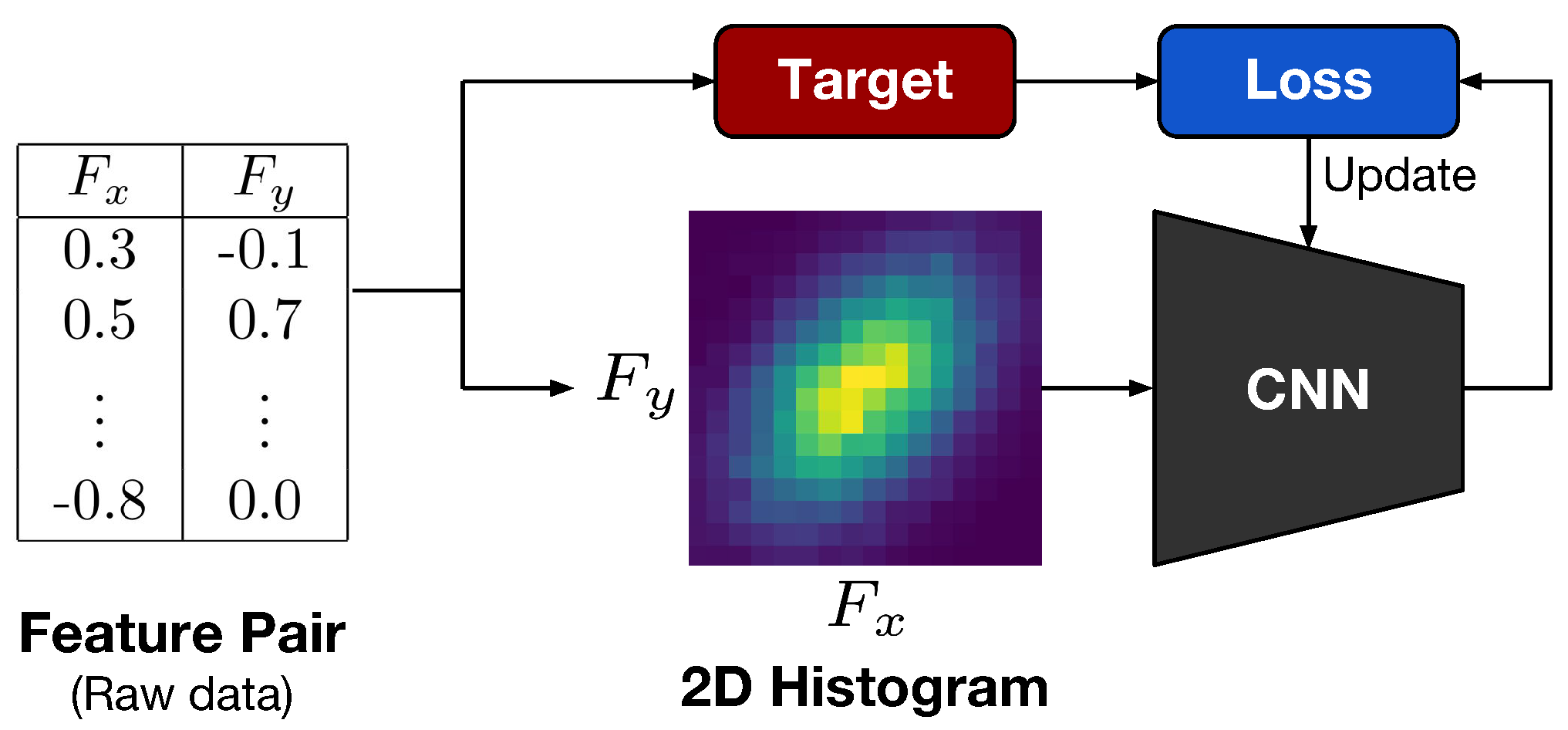
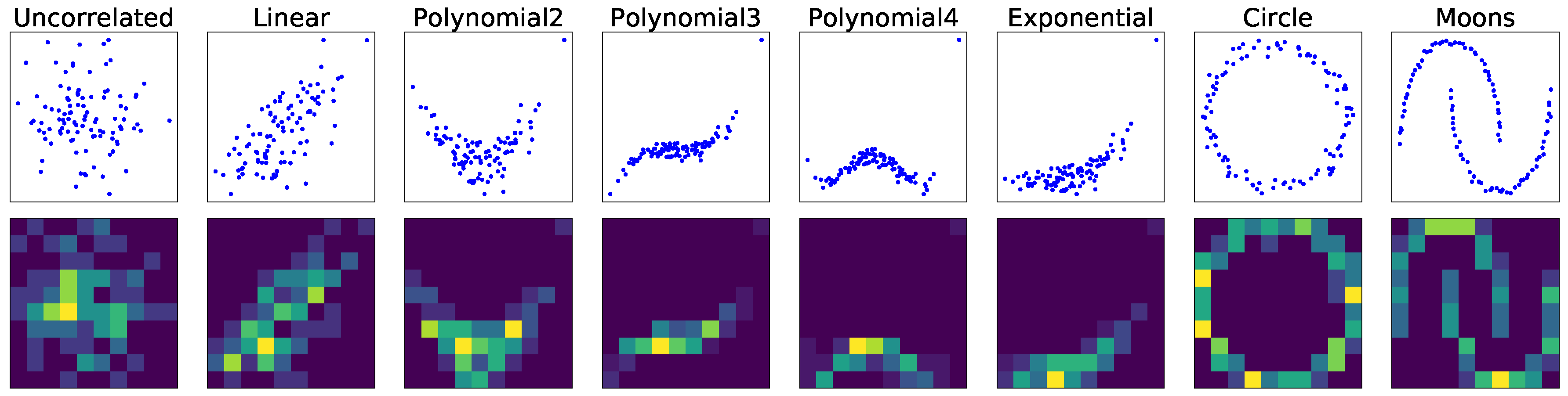
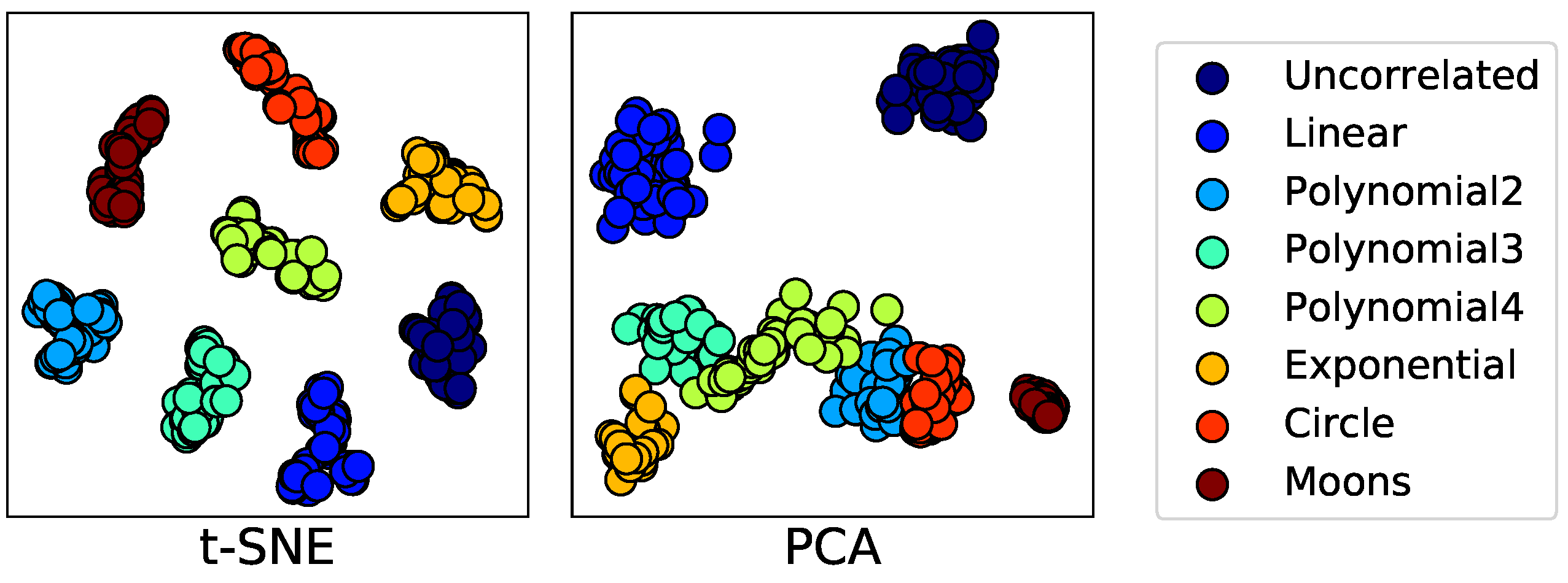
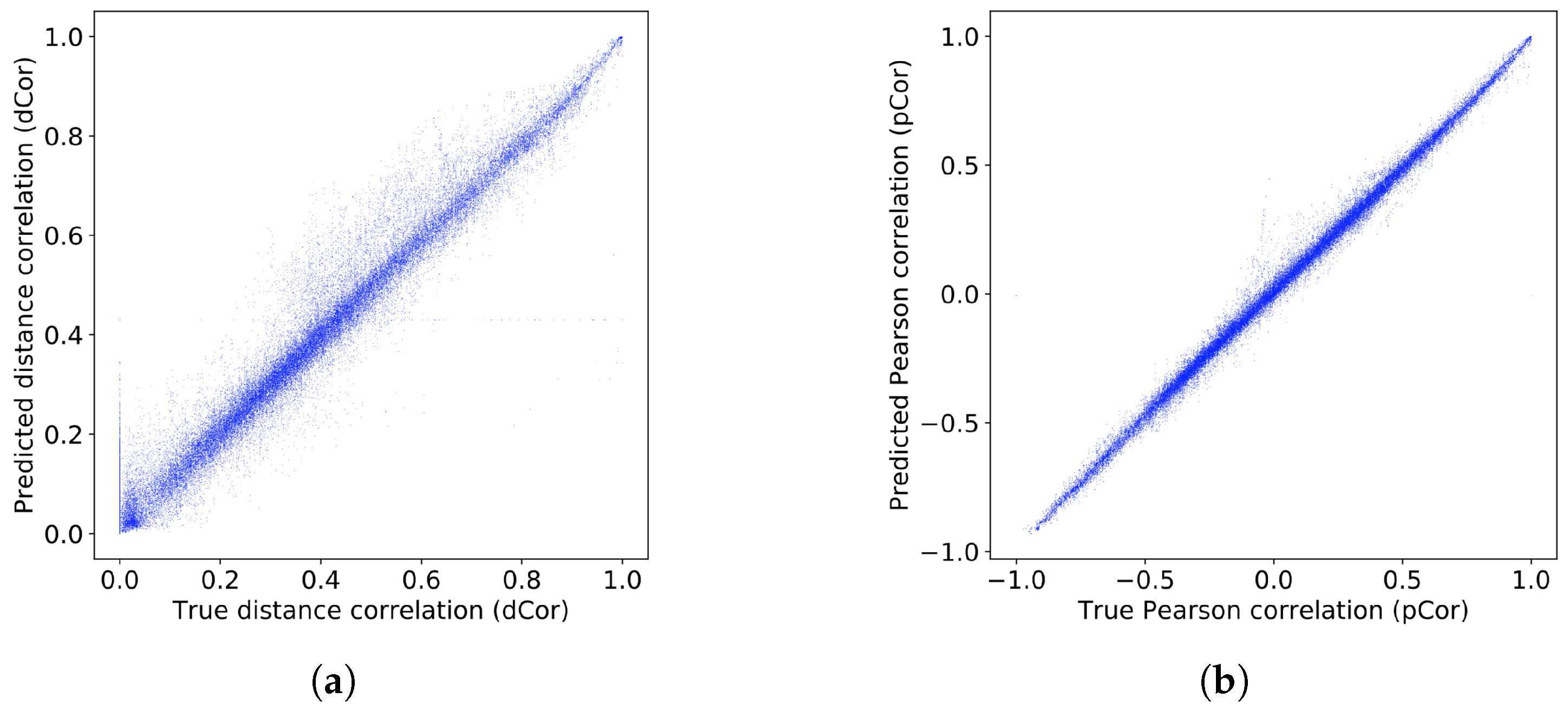
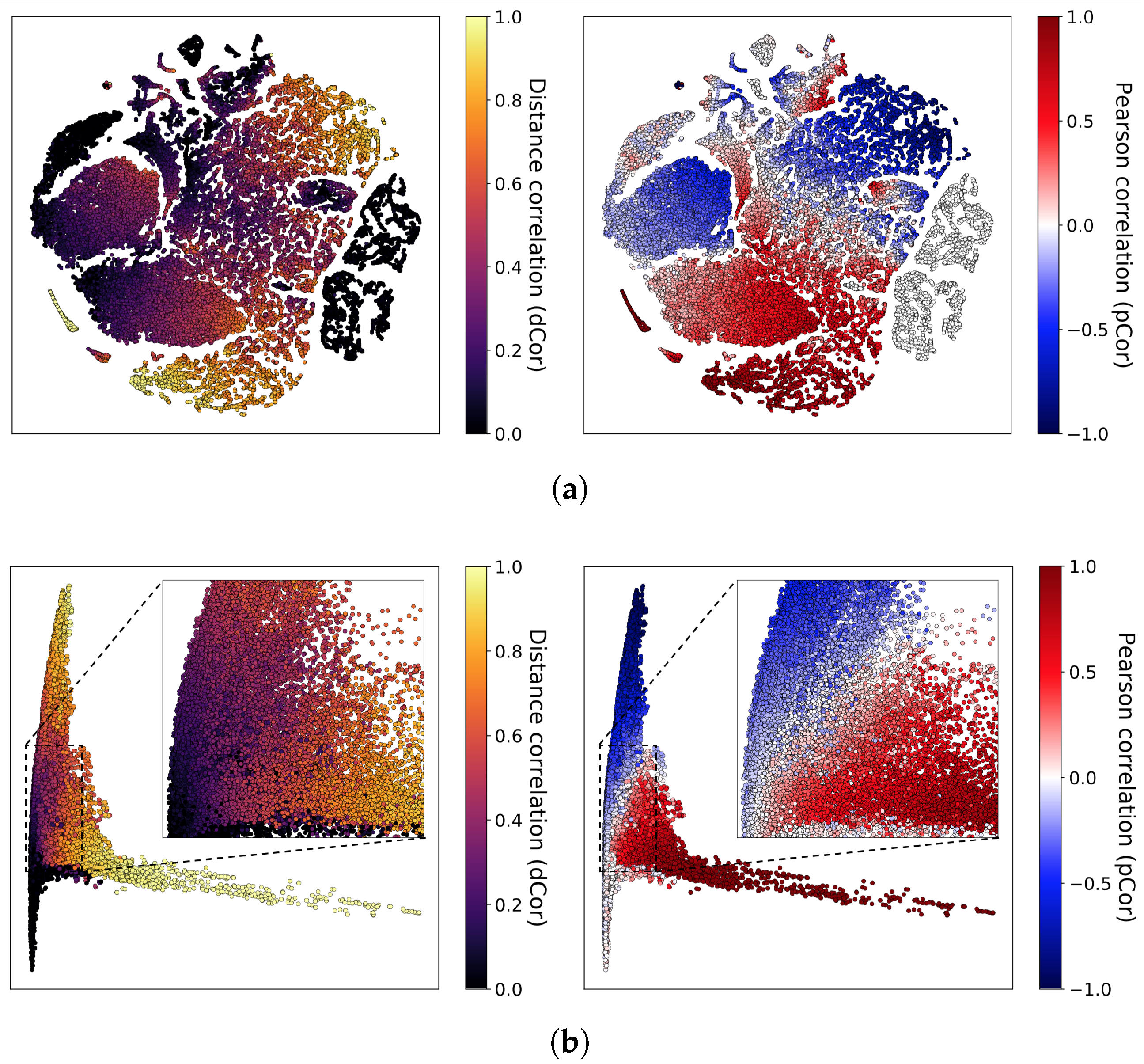
In this paper, we present a dependence representation learning algorithm in which CNNs extract task-specific dependence representations from visualizations of feature pairs. To the best of our knowledge, this is the first method that directly captures dependencies from visualizations of random variables. As the inputs of CNNs, we use 2D histograms, which are compressed scatter plots. In order to show that CNNs can learn dependence representations from 2D histograms, we have conducted three sorts of experiments: In the first experiment, we used a CNN to perfectly classify eight dependence types of a synthetic dataset. Next, we trained the CNN to simultaneously predict Pearson and distance correlations. Our results demonstrated that the CNN can predict the correlations well. Finally, we applied the dependence learning method and found that it outperformed the AutoLearn feature generation algorithm in terms of average classification accuracy and generated half as many features.
4. Feature Generation
As an application of our dependence representation learning method, we have improved the AutoLearn feature generation algorithm [
10], which generates new sets of features that are useful for classification tasks by combining pairs of features from the original ones based on some pre-defined operations; for example, basic arithmetic operations and correlation between two features can be constructed as new features. In this section, we summarize AutoLearn, describe our method, and report the results of our comparison between our method and AutoLearn.
4.1. Background: AutoLearn
4.1.1. Mechanism
The AutoLearn algorithm is composed of three steps:
Preprocessing: shoose candidate features for feature generation by selecting features that are relevant to the classes of interest, as indicated by information gain (IG). If the number of features is less than 500, then use the features with an IG greater than zero; otherwise use the top 2% features.
Feature generation: for all possible feature pairs of candidate features, generate new features by performing regression on input data are with target .
Feature selection: select features that are useful for classification based on stability and IG.
4.1.2. Feature Generation Process
The novelty of AutoLearn is in step 2, where regression is applied to generate new features. The regression algorithm is selected based on the dependence types of the feature pairs. If they are linearly related, then linear ridge regression (LRR) [
31] is used. If the dependency is non-linear, then kernel ridge regression (KRR) is used [
32] with a radial basis function (RBF) kernel. The two types are categorized based on a threshold: if
, then the data are linearly related; otherwise, they are non-linearly related. Note that there is no feature generation if the dCor of the feature pair is zero. Once the appropriate regression algorithm has been identified, the two types of features are generated. The first feature is the predicted value
, which is calculated by inputting
into the regression model. The second feature is the residuals of the regression
.
4.2. Improving Feature Generation
The AutoLearn feature generation process has two limitations: (1) classifying dependence types by setting the dCor threshold to 0.7; and (2) generating new features for almost every feature pair. As shown in
Figure 1, we should not use dCor to specify a criterion for classifying dependence types. AutoLearn generates a huge number of new features because it operates on a strict policy of generating new features for every feature pair unless the features in the pair are independent of each other. If the feature selection in step 3 does not select appropriate features, then useless features are output.
To tackle these two problems, we apply dependence representation learning to feature generation. We train a CNN to predict the usefulness of the features generated before actually generating the features. We predict whether features will be useful directly from the 2D histograms. The CNN is trained to predict whether the generated features with LRR or KRR will be useful for improving the classification accuracy. A target function for feature generation, called
, is described in Algorithm 2. During the training, the target function actually performs the evaluation by applying the classification algorithm to the generated features. If the generated features improve the classification accuracy, then the target function returns 1, otherwise, it returns 0.
| Algorithm 2 Target function for feature generation . |
Input: Feature pair (Fx, Fy), regression algorithm Regr, class label vector C Output: Learning target T - 1:
- 2:
- 3:
- 4:
if then - 5:
return 1 - 6:
else - 7:
return 0 - 8:
end if
|
The objective function for the feature generation
, is composed of two parts,
and
:
is the loss function for predicting the LRR, which is defined by:
and
is the corresponding function for the KRR:
Note that we carry out multi-label classification because both LRR and KRR may be helpful. However, the training is performed using both of
and
:
We add because it helps the CNN learn the dependence representations better.
The prediction probability
, which indicates the usefulness of the generated features based on
and LRR, is defined as:
and the corresponding probability for KRR is defined as:
If one of the probabilities and is greater than some threshold , then new features are generated using the corresponding regression algorithm. If both of the probabilities exceed the threshold, then new features are generated using KRR. No feature is generated for if neither probability is greater than the threshold.
4.3. Experimental Setup
For reliable evaluation of feature generation, we have followed the experimental setup used in the previous study ‘AutoLearn’ as much as possible.
Datasets: we used the same datasets and splits that ware used for the correlation regression (see
Table 1).
Table 4 lists the 13 test datasets used for the evaluation of feature generation and comparison with AutoLearn. The IDs listed in
Table 4 are task IDs of OpenML. Of the 13 datasets, we selected 10 datasets which ware used in the AutoLearn study.
Target function implementation: as for a performance metric of this experiment, it is difficult to directly evaluate the quality of newly generated features. Since AutoLearn used classification accuracy over the original feature space and the augmented feature space to evaluate its empirical results of feature generation, we have also used the same performance metric to compare the results to our results. For more reliable evaluation, we split the samples of the feature vectors into a training set and a test set using the default splits provided by OpenML for each dataset. We also trained the feature generators (LRR and KRR) only using the training samples.
Evaluation method of feature generation: we evaluated feature generation performance using seven classification algorithms, each of which is used in AutoLearn: k-nearest neighbors (KNN), logistic regression (LR), support vector machine (SVM) with linear kernel (SVM-L), SVM with polynomial kernel (SVM-P), random forest (RF), AdaBoost (AB), and decision tree (DT). We also report the average accuracy of seven classifiers. Following AutoLearn, we used the default hyperparameters used by scikit-learn for the classification algorithms. We also report the original accuracy of our algorithm, without feature generation.
Implementation details: The learning rate of the optimizer is . We added dropout layers with a ratio of 0.2 after every convolutional layers and did not decay the weights. To evaluate the newly generated features for the target function, , we simply used the logistic regression functionality provided by scikit-learn with the default hyperparameters. The threshold was set to 0.2 after a small grid search.
4.4. Results
Table 5 lists the average classification accuracy for all of the test datasets. Our method outperforms AutoLearn with respect to the average classification accuracy of the five classification algorithms except for KNN and LR algorithms.
Table 6 lists the total numbers of features generated by each method for the 13 test datasets. AutoLearn generated new features for 88% of the original feature pairs, whereas, our method generated new features for 47% of the original feature pairs. Note that even though our method generated half as many features, it achieved higher classification accuracy than AutoLearn. This suggests that the proposed method produces much better quality features than AutoLearn.
Table 7,
Table 8 and
Table 9 list detailed scores for each dataset. In terms of the average accuracy, our method is superior for all datasets except for the ‘Arcene’ dataset. Moreover, in the tables, the ‘original’ column represents the accuracy of original machine learning algorithm without feature generation. In summary, our method tends to have a relatively high classification accuracy in spite of the fact that the number of generated features is almost half the number of features generated by AutoLearn.