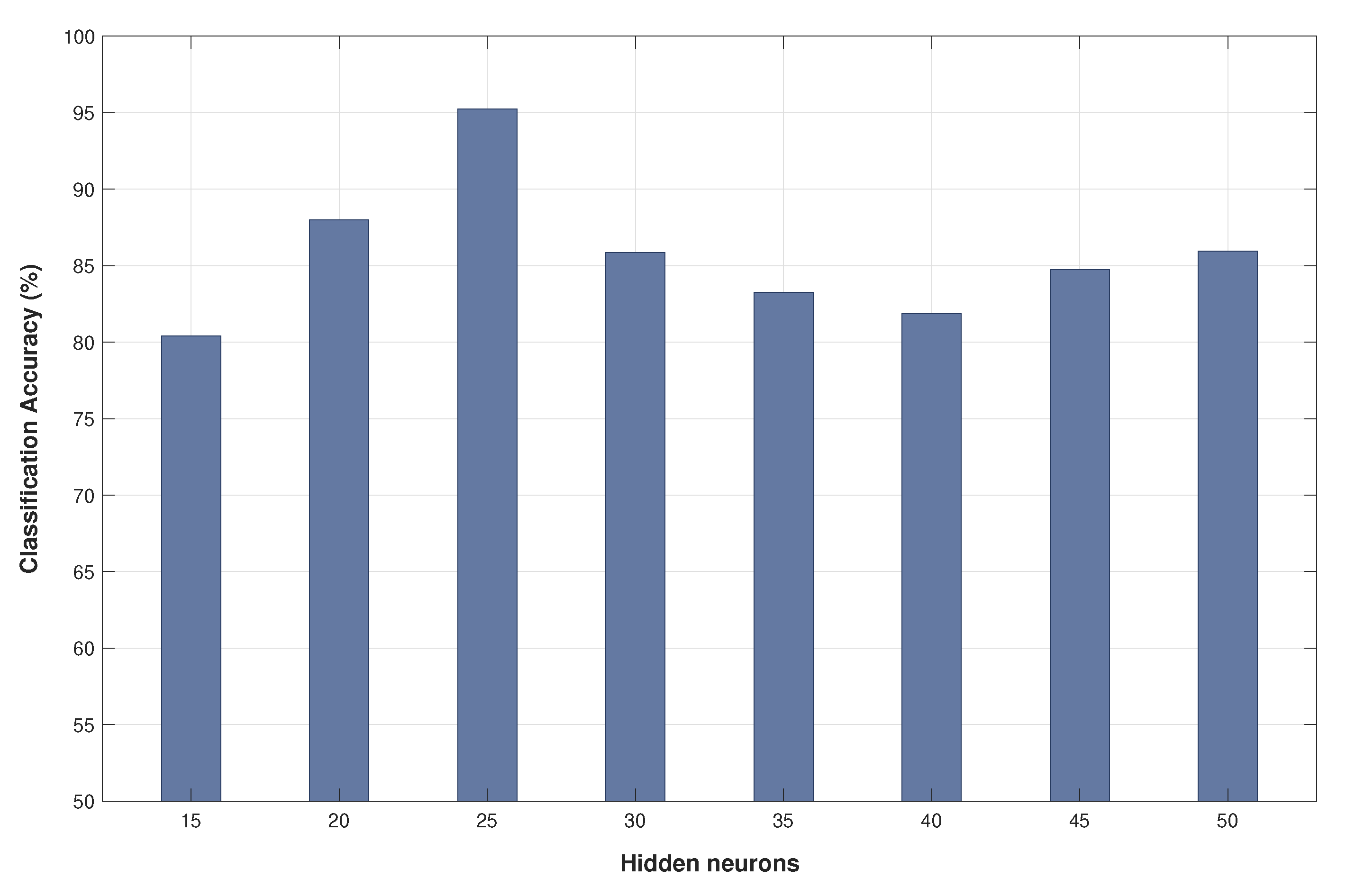1. Introduction
Epilepsy is a common neurological disorder usually described by seizures which are recurrent in nature. This disorder can be produced by different brain disorders, such as brain tumors, intracranial hemorrhages and brain malformations [
1], and depending on the affected area, a disorder may generate, apart from epileptic seizures, malfunctions in motion and patient perception [
2].
An epileptic seizure is a period of time where the patient experiences a set of symptoms with different levels of severity: uncontrolled shaking movements of the body with loss of consciousness (generalized tonic-clonic seizure), shaking movements of a specific part of the body with different levels of consciousness (focal seizure), or short moments of focal seizures with impairment of awareness (absence seizure). Epileptic seizures can be originated by abnormal, synchronous, or even excessive brain neural activity, causing a temporary disruption to the way that the brain normally works. Anyway, what happens to someone during a seizure depends on the affected part of the brain and how far the seizure activity disseminates in the brain.
Epilepsy can be classified primarily into two types: generalized and partial (or focal) epilepsy [
3]. Generalized onset seizures affect, at the same time, both sides of the brain or groups of cells on both sides of the brain. On the other hand, focal onset seizures (the term focal is used instead of partial to be more accurate when talking about where seizures begin) usually start in one area or group of cells on one side of the brain.
The activity of the brain is usually registered using either electroencephalography (EEG) or functional magnetic resonance imaging (fMRI). Although fMRI has better spatial resolution, the use of the multidimensional time series generated from electroencephalogram EEG is more popular, as it allows high precision time measurements, is functionally fast and is relatively cheap.
The EEG epileptogenic source’s localization has been studied for decades [
4,
5]; however, the methods were not implemented in clinical practices until recently. Nowadays, the EEG is considered a noninvasive and useful test to assess whether a pharmacoresistant patient can benefit from the resective epilepsy surgery [
6]. As the resective surgery aims to remove surgically the brain sections involved in the focal onset epilepsy, it is important to distinguish precisely between “focal signals,” those recorded in brain areas where first ictal signals are detected, and “non-focal signals,” those registered from brain areas not related to the seizure onset [
7]. Many patients with epilepsy may require EEG signals to be recorded from deep structures of the brain using intracranial electrodes.
Usually, the focus localization is essentially made using registers acquired monitoring the patient 24 h a day, during a stay of several days in an epilepsy monitoring unit (EMU). In these kind of units, apart from scalp EEG (and intracranial electrodes to record signals from deep structures of the brain, in many patients), the epilepsy patients are recorded on video, along with their speech and movements. Thus, all data are collected targeting the evaluation of its seizure disorder, seeking to gather data before a seizure starts, during one and during recovery. The evaluation of this information can be used to locate candidate areas for the epileptogenic focus, although in some cases it is not enough to locate the epileptogenic focus precisely prior to the surgery.
The visual analysis of the EEG recordings of seizures with intracranial electrodes can help in locating the seizure. However, visual inspection is a hard and time consuming process that can be affected by the clinician subjectivity. In addition, it is not easy to determine the seizure source by a direct visual inspection of the EEG signal recordings.
To select candidate areas from EEG signals could be helpful a computerized analysis of the EEG [
8,
9]. As with other pathologies [
10,
11,
12], machine learning has been applied in epilepsy at many works [
13,
14,
15] to classify EEG signals as normal versus epileptic or seizure versus inter-ictal. However, the most challenging classification problem is focal (F) versus non-focal (NF). The classification of seizure from normal and seizure-free signals has achieved a 100% classification accuracy. However, this goal has not been achieved to date for the classification of focal and non-focal EEG signals. Neurosurgeons have difficulties determining the brain region responsible of the initial seizure discharge, so this kind of classification may serve as a tool to help epileptologists to resect the epileptogenic area. Compared with signals of the epileptogenic areas, the signals from non-epileptogenic areas are more nonlinear, less random and more nonstationary.
Many machine learning systems have been developed to classify and detect the epileptogenic source signals. Sharma et al. [
13] used entropies derived from the coefficients of the wavelet transform of the EEG signals to feed a least squares-support vector machine (LS-SVM) model to distinguish focal and non-focal EEG signals. In [
16], Sharma et al. also used the LS-SVM classifier to feed the entropies derived from some subbands decomposed using tunable-Q wavelet transform (TQWT). In [
17], Sharma et al. utilized empirical mode decomposition (EMD) and entropy for the classification of focal and non-focal EEG signals. In this work, intrinsic mode functions (IMFs) from focal and non-focal EEG signals were extracted using EMD, and then the entropies were fed the input of a LS-SVM classifier. Das et al. [
18] also used entropy-based features from the EMD, DWT (discrete wavelet transform) and EMD-DWT domains, along with a k-nearest neighbor (k-NN) classifier model. In turn, Zeng et al. [
19] used features derived from euclidean measures obtained from the phase space reconstruction (PSR) of several IMFs, obtained using EMD as well. Bhattacharyya et al. [
20] proposed the decomposition of the EEG signal into rhythms using the empirical wavelet transform (EWT), and then used some area measures from them as input for a LS-SVM classifier model, to recognize focal and non-focal EEG signals. Another work of Bhattacharyya et al. [
15] also used multivariate subband entropy measures from TQWT along with multivariate fuzzy entropy in combination with a LS-SVM classifier model. Chatterjee et al. [
21] also used SVM and k-NN classifiers fed by multifractal, detrended fluctuation analysis (MFDFA) based feature sets. Singh et al. [
22] used features derived from DFT-based rhythms of the EEG to fed the LS-SVM classifier. Taran et al. [
23] proposed the use of spectral moment based features extracted from the modes of the clustering VMD (CVMD) and extreme learning machine (ELM) classifiers. Deivasigamani et al. [
24] utilized features extracted from the dual tree complex wavelet transform (DT-CWT) to fed an adaptive neurons fuzzy interference system (ANFIS).
However, these works tend to require a considerable computational load, especially the most recent ones. As an example, San-Segundo et al. [
25] proposed a deep neural network (DNN) made up of two convolutional layers for feature extraction and three fully connected layers for classification. In this work, authors increased the classification accuracy a little at the expense of increasing, considerably, its computational needs. In turn, Daoud et al. [
26] used both a deep convolutional autoencoder and an unsupervised learning scheme merging a deep convolutional variational autoencoder and a K-means algorithm.
This progressive increase of the computational demand could impede the jump of using the epileptogenic source localization during surgery. This application needs a real-time implementation, and could be used as a help-decision tool by neurosurgeons to refine the localization of an epileptogenic area during resective epilepsy surgery. Note that the recent technology is mature enough to implement machine learning processes in real-time [
27,
28].
Thus, the goals of this work were to assess the possibility of locating the epileptogenic focus in real-time, and study the simplification of the classification process to reduce its computational needs as much as possible while maintaining similar classification accuracy to previous works. We also studied the resource usage and performance of the real-time application on a recent Xilinx FPGA reconfigurable device.
The main contributions of this work are:
The proposal of an automatic classification procedure optimized to classify in real-time the location of the epileptogenic focus from EEG inter-ictal signals. It is conceived to be used in a portable device as a decision-assisting tool by neurosurgeons during surgery.
The proposed feature set and the classifier model have been selected to minimize both the number of features and the computational cost, while preserving the classification accuracy at a level similar to that in previous works.
The classification procedure has been implemented using a reconfigurable logic FPGA device. This hardware implementation computes the whole procedure, accepts the EEG raw input signal and delivers the classification result. Two designs have been implemented, using single and double floating point precision following the IEEE 754 standard for floating-point arithmetic.
The analysis of the resource usage of this kind of implementation, its accuracy with respect a Matlab implementation and how fast the device can deliver results (maximum frequency of operation).
The rest of the paper is organized as follows.
Section 2 introduces the dataset, and the analyzed features and classifier models. The details of the hardware implementation and the proposed computational method are described in
Section 3. Results of the analysis and discussion are presented in
Section 4 and
Section 5, respectively. Finally,
Section 6 concludes the paper.
3. Hardware Implementation and Computational Method
This section discusses the implementation details. We will leave for
Section 4 the reasons for and how this computational procedure was chosen. At this point, all that matters to know that the classifier model used was a perceptron with 25 neurons in the single hidden layer, 5 neurons in the input layer and 2 neurons in the output layer (Algorithm 2). The 5 extracted features (
Table 2) were computed following Algorithm 1. Details about feature selection and classifier model selection are left for
Section 4.
The proposed hardware implementation was conceived as an intellectual property (IP) core using Xilinx Vivado HLS and the Xilinx Vivado Design Suite 2016.2 [
42]. It provides a signal interface definition that enables it as a standalone module, being able to also be used as a peripheral of a more complex system on chip (SoC), embedded microprocessor, etc. Further, this approach offers the capability of customization for specific needs in many different hardware applications.
3.1. Working Modes
The IP core perform two different working modes:
- -
Initialization mode. The initialization of the IP core consists of the load of the matrices and , using the external signal interface. These matrices are essentially weights and bias of the neural network, along with normalization and denormalization values and vectors. All together, these matrices allow the IP core to perform a proper classification. Note that the calculation of these matrices is achieved out of this IP core, and the results are transferred to it during this initialization process. Once the initialization is complete, the core can change to another working mode, never before.
- -
Run mode or on-line mode. In this mode, the input data is fed into the IP core. Then, several features are extracted, and in turn, fed into the neural network system. The output is computed according to the initialized network topology. The IP core, when running in this mode, computes and serves the corresponding output before accepting a new input.
3.2. IP Core Signal Interface
In Xilinx FPGAs, external core signal interfaces are used to follow proprietary protocol specifications, such as AXI4 [
43,
44], AXI4-Lite or AXI4-Stream [
44,
45]. In this work the AXI4 protocol was selected to permit specifying arrays as arguments. However, note that the protocol interface has almost no influence on performance when the core is running in the on-line mode. The reason is that the reported performance refers to the complete epileptic focus classification task, while the load of one input vector by iteration implies a negligible overhead. Therefore, for replication purposes, it can be expected to achieve similar results, for the running mode, regardless of the protocol specification implemented for the core signal interface (e.g., AXI4-Lite, AXI4 or AXI4-Stream).
Figure 1 outlines the external signal interface of the IP core, where signal lines are represented by thin black arrows and buses, and bunches of signals are represented by white thick arrows.
Signal START indicates when the core can start processing data, the READY signal indicates when the core is ready to accept new inputs, the IDLE signal indicates when the core is idle and the DONE signal indicates when the core operation has been completed. Altogether, these signals constitute the block-level interface, controlling the core independently of the port-level I/O protocol.
On the other hand, the input and output data ports implement a handshake data flow protocol. Lines A_TVALID, A_TREADY and the bus A_TDATA integrate the input data port, while B_TVALID, B_TREADY and the bus B_TDATA integrate the output data port. The TDATA bus is the payload, while TVALID and TREADY lines signal when the information pass across the interface. These signals integrate a two-way flow control mechanism that enable master and slave to control the data transmission rate rate across the interface.
Finally, the MODE signal is used as application-signaling. It requests the entering on initialization mode or on-line mode.
3.3. System Parameterization
The definition of the SLFN neural network topology was conceived parametrically to achieve a flexible design with minimal code modifications. The main parameters are:
- –
L: Is the length, in samples, of the input signal window.
- –
IN: Is the number of neurons in the input layer.
- –
: Is the number of neurons in the hidden layer.
- –
ON: Is the number of neurons in the output layer.
- –
FT: Boolean determining the type of IEEE 754 floating-point arithmetic precision: single or double.
3.4. Computation of the RT-EPI Algorithm
The proposed IP hardware core implements a real-time epileptogenic focus classification of a different EEG input window signal each iteration.
3.4.1. Algorithm Description
Algorithm 1 shows the steps in which perform the feature computation. This computational procedure follows the expressions described in
Table 2 to extract features from the input signal, and then, to compose the input data pattern to be used as input to the neural network.
| Algorithm 1: Feature extraction pseudocode. |
| Input:→ Input signal. |
| Output:→ Output features vector. |
| 1: | . |
| 2: | . |
| 3: | . |
| 4: | . |
| 5: | . |
| 6: | . |
| 7: | . |
| 8: | . |
| 9: | . |
| 10: | . |
| 11: | . |
| 12: | . |
| 13: | . |
| 14: | . |
| 15: | . |
| 16: | . |
| (*) Filter coefficients: B[0] = 0.0001; B[1] = 0.0006; B[2] = 0.0014; B[3] = 0.0019; B[4] = 0.0014; B[5] = 0.0006; B[6] = 0.0001; A[0] = 1.0000; A[1] = −4.1069; A[2] = 7.2450; A[3] = −6.9795; A[4] = 3.8569; A[5] = −1.1558; A[6] = 0.1464. |
Algorithm 2 shows the computational procedure implementing the classification. Note that it is mandatory to the previous IP core so that the implementation may be fully functional and begin accepting any input data. The initialized structures are the normalization and denormalization parameters (
), plus the weights and biases of the hidden and output layers (
). Further, the first step of the algorithm is the normalization of the data input pattern. Then, the outputs of the hidden and output layers are calculated. Although the hyperbolic tangent sigmoid function has been used as an activation function, the proposed algorithm allows the use of a wide range of activation functions (including piece-wise linear activation functions). The results of the output layer are then denormalized to obtain the final output.
| Algorithm 2: Classification of an input pattern. pseudocode |
- Input:
→ Input data pattern. → Input normalization vectors. → Range normalizing input. → Hidden layer weights matrix. → Hidden layer bias. → Output weights matrix. → Output layer bias. → Output denormalization vectors. - Output:
|
| 1: | . |
| 2: | . |
| 3: | . |
| 4: | . |
| (: number of hidden neurons, : number of input neurons, : number of output neurons.) |
The pseudocode in Algorithms 1 and 2 show all the matrices and vectors involved in each step of the computation, with their respective dimensions.
The computational procedure was implemented using a sequential architecture. This architecture minimizes both the usage of memory and arithmetic slices. And, although the throughput results can be improved using a parallel architecture, the use of a sequential architecture enabled us to establish a standard machine to be used as a reference in subsequent works.
3.4.2. Design Considerations
The proposed design allows the definition of IEEE 754 floating point units using single or double data type precision. This selection together with the parametric definition of the SLFN neural network permits one to test the design in different conditions with few code modifications.
The activation function implemented in this design was the hyperbolic tangent sigmoid.
On the other hand, the proposed design uses pipelining. The main reasons for that is that pipelining alleviates the great latency involving the use of floating point operations, and the suitability of the algorithm for its use, since most of the steps of Algorithms 1 and 2 can be implemented with for loops or nested for loops.
The pipelining technique helps to optimize the initiation interval, defined as the number of clock cycles that must occur before a new input can be applied. Thus, the initiation interval becomes the parameter to optimize, and the effort must focus on approximating it to one as much closely as possible. To carry this out, we used the PIPELINE optimization pragma directive in each step of the computational procedure implemented using for loops. This generates a pipeline design with an initiation interval as low as possible, which dramatically reduces the total latency of the loop implementation.
In addition, the clock period target was set to 4 ns. This forces the compiler to obtain the fastest hardware implementation.
Take into account that both the above design considerations and the computational procedure described in Algorithms 1 and 2 must be followed to replicate the implementation results in this work.
4. Results
The analysis described in this work was carried out using the Bern-Barcelona dataset (
Section 2.1). The first 50 focal and non-focal register pairs of the dataset were used. As all considered features are univariate, and 40 s of EEG signals are available in each register pair (20 s × 2), a total of 4000 s of EEG signals were considered in this study.
In order to better compare our results with the results provided by previous works, we used five-fold cross validation in our experimental procedure. In this approach, the EEG signals are divided randomly into five equal portions. Four out five portions were considered for training and the rest, half for validation and half for testing.
All the feature extraction algorithms and classification models were implemented using Matlab R2018a and the “Statistics and Machine Learning Toolbox”.
The first goal was to find the classification procedure with the lowest computing cost, because of the real-time implementation aim. Thus, we had to be find (1) a set of features with the minimum number of features and minimum computing cost, using (2) the simplest classifier model, and (3) the optimum segment length. A minimum classification accuracy of 95% was required, a value above the average of related works in the bibliography.
To do so, all considered features,
Table 1, were ranked by discrimination capacity and by computational cost. Then, different classification procedures were checked in a loop. At each iteration of the loop, a combination of features were selected (taking into account the ranks); then, fed to 24 classification models,
Table 3; and finally the classification accuracies for different segment lengths were calculated. The loop finished when the smallest set of features reached 95% of classification accuracy on at least one of the classification models.
Once the classification procedure was defined, it was coded in C using Xilinx Vivado HLS and the Xilinx Vivado Design Suite 2016.2 [
42]. These tools were also used to carry out synthesis, simulations and cosimulations. The Xilinx Virtex-7 XC7VX1140T FPGA device was selected for synthesis and implementation, because it is a biggest Virtex-7 device that permits implement the application without resource restrictions.
The coded design is parameterizable, and follows a pipelined and sequential architecture that computes (Algorithms 1 and 2) all the classification procedure from accepting the raw signal to the delivery of the classifier output.
The reported analyses were conducted using two different arithmetic precisions: a 32-bit floating-point algorithm (“single” design), and 64-bit floating-point algorithm (“double” design). Both implementation designs used the IEEE 754 standard.
4.1. Set of Features and Classification Model
The RelieF algorithm was used as feature extraction algorithm. RelieF returns a rank of features and its weights to represent the discrimination capacity of these features. These ranks and weights were used to select the relevant set of features, along with the computational cost criteria, optimizing the real-time implementation of the application. The number of selected features was defined as the minimum set of features that allowed us to obtain a minimum threshold of 95% classification accuracy on at least one of the classification models (
Table 3).
To determine the best classification model, 24 classification models were tested.
Table 3 lists all these models along with their prediction speed and memory usage characteristics. Note that only those classification models with low or medium speed and memory usage were chosen, due to the importance of minimizing these parameters in the real-time implementation.
All classification models were trained with several sets of features, seeking to determine the feature set and classification model at the same time. However, the evaluation of the classification models depends on a third parameter: the window length.
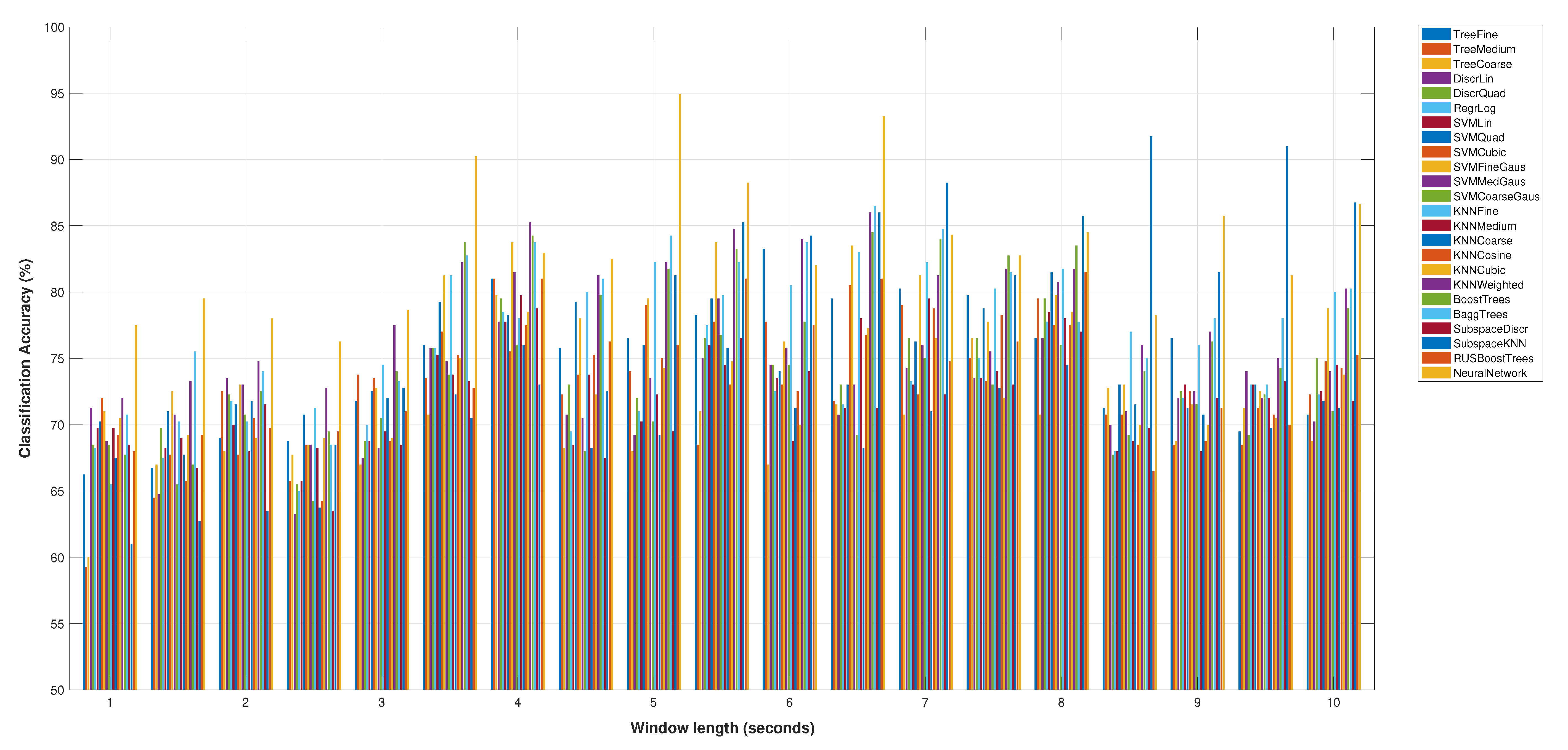
Figure 2 shows the accuracy results for the selected sets of features (
Table 2), for all the classification models and the variations of the window length from 1 to 10 s. Note that this figure only shows the best score for the neural network classification model.
Figure 3 details the classification accuracy obtained using different numbers of neurons for the hidden layer.
Thus, five features were finally selected, those in
Table 2, to be used with a neural network as the classification model. As it can be seen, the computational cost of the five selected features is low (case of features derived from the temporal domain) or medium (in the case of frequency domain features, where it is necessary to compute the Fourier transform).
This minimum set of features achieves a classification accuracy of 95% using a Neural Network and five seconds of segment length. In addition, the optimum number of neurons in the hidden layer of the neural network is 25, as it can be shown in the analysis of
Figure 3.
Thus, from that point we will assume that the implementation is done by a neural network of type perceptron with just one hidden layer of 25 hidden neurons and two output neurons in the output layer (provided the two classes of this classification problem). In addition, the neural network will have five input neurons, because five is the dimensionality of the selected number of features, which, in turn, would be computed from window lengths of 5fiveseconds of the input signal.
4.2. Hardware Resources Analysis
Table 4 gathers all the resource analysis results. It also shows resource usage as a percentage of occupation of the Xilinx XC7VX1140T FPGA, intending to provide an idea of the design occupancy in current FPGAs.
It can be seen that the design demands near the same DSP48E slices for both designs, 133 for single precision and 137 for double precision. Obviously, this slight variation has been achieved thanks to the pipelined design.
The required number of flip-flops (FF) and look up tables (LUT) were not high, presenting only small variations between designs.
On the other hand, as is natural, the amount of block RAM doubled for double arithmetic precision with respect to single arithmetic precision. That is due to the use of 64-bit representation of double precision and 32-bit of single precision. Then, block RAM requirement halves according the amount of memory needed for its representation.
4.3. Hardware Performance and Accuracy
Mean absolute error (MAE) was used to measure the accuracy of the results of each design with respect to the Matlab implementation. MAE measures the average magnitude of the errors without considering the direction of its deviation, taking into account the absolute differences between them.
The accuracy shown in
Table 4 is the maximum of the accuracy obtained for the results of each output neuron. As it can be seen, the accuracy for the single design is very low, but the accuracy for the double design is so low that, in practice, it indicates that results in this case are similar to the results obtained in its Matlab implementation. It is natural, because both implementations use double precision data types.
In turn, the number of clock cycles shows a strong dependency of the data type (
Table 4), and the numbers of minimum allowable cycles reported for both designs were similar.
The maximum frequency of operation is obtained from the minimum clock period and the required number of clock cycles. It is represented in
Table 4 for both data type designs. As it can be seen, the computation of all the classification procedure, from accepting raw signals to the delivery of the classification output can be done at a rhythm of 2.53 Hz for the “double” design and 3.55 Hz for the “single” designs (3.55 classification outputs by second).
5. Discussion
The analysis described in this paper was carried out using the Bern-Barcelona dataset. Thus, all related works in the bibliography, selected for the sake of comparison, use the same database. That provides a more comparable framework, given that the use of works using other datasets may expose significant differences when performing the same classification method. As an example, San-Segundo et al. [
25] shows that the focal-nonfocal (F-NF) classification accuracy may differ more than 20% when the same methods applies to the Bern-Barcelona dataset [
7] and the Epileptic Seizure Recognition dataset [
46]. In this example, the nature of the signals in the dataset, mainly the difference between signal lengths (only 1s for the latter), makes the difference. Thus, note that the Bern-Barcelona dataset is the logical selection when facing just the F-NF problem, provided its longer signal length (20 s) and its specialization in inter-ictal signals (recordings of seizure activity and three hours after the last seizure activity are excluded).
Table 5 compares our results with previous works for the task of classifying the focal and non-focal signals using the Bern-Barcelona EEG dataset. It details the obtained classification accuracy, summarizes the extracted features, and indicates the type of classifier used in each work.
Note that the purpose of this work was not to beat the accuracy results of previous works in the bibliography; our goal was to obtain, at the same time, the best classification procedure having the lowest possible computational load (for feature extraction and classification), aiming at its real-time implementation. Despite this, we obtained pretty good accuracy results. The 95.5% classification accuracy obtained in this work surpasses many other related works [
13,
15,
16,
17,
18,
20,
21,
22,
47], while some other works [
19,
23,
24,
48] surpasses this result by a maximum of 1.5% classification accuracy ( [
19] achieved 97% classification accuracy). Thus, when not considering [
25], the comparison with the other related works can be considered pretty good in light of the great simplification achieved for the feature extraction process. In turn, San-Segundo et al. [
25] obtained to 98.6% classification accuracy (3.1% more than this work), but at the expense of using a computationally intensive tool, a deep neural network, which is far away from the simplicity sought in this work; that is the reason to exclude this work from comparison from this point forward.
As it can be seen in
Table 5, extracted features were used to proceed from computationally intensive processes, such as the decomposition of EEG signals using empirical mode decomposition (EMD) to extract intrinsic mode functions (IMFs). Thus, Sharma et al. [
17] obtained 87.0% classification accuracy using five entropy features extracted from IMFs; Das et al. [
18] also used entropy-based features from the EMD, DWT (discrete wavelet transform) and EMD-DWT domains, achieving 89.4% classification accuracy; and Zeng et al. [
19] arrived to a 97% classification accuracy using features derived from Euclidean measures obtained from the phase space reconstructions (PSRs) of several IMFs obtained using EMD.
The wavelet transform has been also a computationally intensive process used in related works as the basis of the feature extraction process. Thus, Sharma et al. [
16] obtained several entropy features from the tunable-Q wavelet transform (TQWT), reporting a 95.0% classification accuracy; Bhattacharyya et al. [
20] obtained 90.0% classification accuracy using as features, projections of the reconstructed phase space (RPS) from the rhythm separation achieved using the empirical wavelet transform (EWT); Sharma et al. [
13] obtained 94.25% classification accuracy from various wavelet based entropies; Bhattacharyya et al. [
15] obtained 84.67% classification accuracy using TQWT based multivariate sub-band fuzzy entropy with LS-SVM classifiers; and Deivasigamani et al. [
24] obtained 96.0% classification accuracy based on a set of features extracted from the dual tree complex wavelet transform (DT-CWT) and using an adaptive neuron fuzzy interference system (ANFIS).
Other works use variational mode decomposition as the basis of feature extraction, such as Rahman et al. [
47], who obtained a 95.2% classification accuracy using features such as refined composite multi scale dispersion entropy (RCMSDE), refined composite multiscale fuzzy entropy (RCMSFE) and autoregressive model (AR) coefficients extracted from variational mode decomposition (VMD), DWT and VMD-DWT domains; or Taran et al. [
23], who obtained 96.0% classification accuracy using spectral moment based features extracted from the modes of the clustering VMD (CVMD) and extreme learning machine classifiers.
Note that the computational cost of the feature extraction process in previous works is greater than the computational cost of the feature set proposed in our work. The only work in the bibliography having a computational cost comparable to that of our work is Singh et al. [
22], that obtained a 89.7% of classification accuracy deriving features from DFT-based rhythms of the EEG. In the same way, we have to compute the DFT too. Nevertheless, we obtain a better classification accuracy (5.8% more).
Thus, despite its simplicity, a classification procedure that performs better than most of related works, or, in the worst case, got surpassed by a maximum of 1.5% of classification accuracy (not considering [
25]) was achieved.
However, this work does not propose just an optimum feature set, but an optimum classification procedure, as a whole. Thus, the proposed feature set,
Table 2, can be combined optimally with a neural network classifier model when five seconds of segment length are used (
Section 4.1). Our analysis indicates that this, altogether, guarantees the best accuracy performance with a minimum computational cost. The proposed neural network is of perceptron type, with just one hidden layer of 25 hidden neurons, five input neurons (the dimensionality of the proposed feature set) and two output neurons in the output layer.
On the other hand, the FPGA real-time implementation of the classification procedure, following Algorithms 1 and 2, has been done using a sequential architecture. The benefits of using this architecture are the minimization of the memory usage and the number of arithmetic hardware blocks. Anyway, to improve the throughput results this computation can be easily parallelized.
The proposed hardware design allows the definition of floating point arithmetic units of single or double data type precision (following the IEEE 754 standard for floating-point arithmetic). As it was expected, both designs offer a great MAE accuracy. “Double” design achieves an accuracy similar to the Matlab environment implementation: 3.94 , while the “float” design offers a great accuracy: 1.00 . MAE accuracy was measured using the Matlab implementation as a reference.
The analysis demonstrates that the proposed hardware implementation does not uses many resources. Both designs need no more than 137 DSP slices, while BRAM usage is 95 MB for the “single” design (doubling to 185 MB the requirements for the “double” design, provided that, obviously, the 64-bit double precision data types doubles the memory needs of the single 32-bit data type).
Note that the computational needs of the implementation in a Virtex 7 Xilinx FPGA device requires a reduced portion of its total resources; see
Table 4. In fact, this application can be executed even on a small and cost-effective Xilinx XC6SLX100 Spartan-6 FPGA, assuring a low-cost of implementation.
Regarding the performance, it has been shown that the proposed implementation can perform all computation tasks at a maximum of 3.55 Hz when using the single data types, or 2.53 Hz when using double data types. That means that the single design can deliver outputs at a rhythm of 3.55 times by second.
However, note that this 3.55 Hz of classification frequency (2.53 Hz for double design) is only effective after the first 5 s of acquisition time due to the 5 s segmentation. This implies a minimum latency time to achieve the first result from the beginning of an acquisition without artifacts of the EEG signal. From this 5 s latency, the proposed implementation is capable of handling overlapped time windows, delivering results at the maximum classification frequency. Thus, for the single design, a result will be provided each seconds after the first 5 s window length.
Thus, we have shown that an adequate selection of the set of features, classifier model and length of the window segment, allows one to obtain good classification accuracy results (above the average of previous related works) while maintaining a low computational load for the whole classification procedure. It enables us to move the classification procedure to the real-time field, embedded in a logic-reconfigurable FPGA.
The proposed implementation can be carried out on a small portable device embedding a fast classification engine of epileptogenic focus. This device can serve as a help decision tool to assist neurosurgeons to refine the localization of the epileptogenic area during the resective epilepsy surgery in those cases where greater precision or confirmation were needed.